中国科学院微生物研究所,中国微生物学会,中国菌物学会
文章信息
- 邢庆花, 张英杰, 廖子亚, 赵百锁. 2021
- Qinghua Xing, Yingjie Zhang, Ziya Liao, Baisuo Zhao. 2021
- 嗜盐细菌中四氢嘧啶和羟基四氢嘧啶的生物合成及其生物学功能
- Ectoine and hydroxyectoine: biosynthesis and its biological function in halophilic bacteria
- 微生物学报, 61(6): 1428-1440
- Acta Microbiologica Sinica, 61(6): 1428-1440
-
文章历史
- 收稿日期:2020-09-18
- 修回日期:2020-12-15
- 网络出版日期:2021-04-01
2. 重庆市辅仁中学校, 重庆 400066
2. Chongqing Furen High School, Chongqing 400066, China
栖息于高盐环境中的嗜盐菌,通过从外界环境中吸收或自身生物合成来积累一些易溶的、低分子量的有机溶质以抵御外界的渗透胁迫。这些在生理pH范围内不带电荷,且能与细胞内体系相容,而不影响其它生物大分子功能的溶质被称为“相容性溶质”[1-3]。这种在高盐环境下嗜盐菌为求得生存繁殖而依赖的策略则被称为“相容性溶质机制” (compatible solutes strategy)[1, 4-5],该机制对嗜盐菌适应外界长期的高盐或短时间内的较大盐波动具有很强的灵活性。相容性溶质主要包括甜菜碱类(甘氨酸甜菜碱、脯氨酸甜菜碱等)、四氢嘧啶类(四氢嘧啶和羟基四氢嘧啶)、糖类和糖苷类(海藻糖、葡萄糖苷等)、氨基酸类(谷氨酸、脯氨酸和谷氨酰胺等)和乙酰二氨基酸类(乙酰鸟氨酸、乙酰赖氨酸等)[4-6],其中,甜菜碱类和四氢嘧啶类是最重要的两类相容性溶质。最近,本研究组翔实地报道了嗜盐古菌和嗜盐细菌中甜菜碱类的生物合成及其生物学功能[7],与甜菜碱类在嗜盐菌中的广泛存在性不同,四氢嘧啶类主要存在于嗜盐细菌中,只有极少数古菌中发现含有四氢嘧啶类合成基因。因而作为其姊妹篇,本综述仅针对嗜盐细菌(特指适宜生长的盐度在3.0% NaCl (W/V)以上的纯培养标准菌株)中四氢嘧啶类相容性溶质的生物合成及其生物学功能进行了全面的归纳整理、提炼分析及综合述评等。期望本文能抛砖引玉,为从事嗜盐细菌的学者提供一些新的科研思路,也可激发更多工作者对嗜盐微生物的兴趣。
四氢嘧啶(Ectoine,1,4,5,6-四氢-2-甲基-4-嘧啶羧酸,C6H10N2O2,MW=142.16)于1985年首次在极端嗜盐光合细菌盐绿需盐红螺菌(Halorhodospira halochloris) (原命名为Ectothiorho dospira halochloris)中发现[8],而羟基四氢嘧啶(Hydroxyectoine,1,4,5,6-四氢-2-甲基-5-羟基-4-嘧啶羧酸,C6H10N2O3, MW=158.16)在微小链霉菌(Streptomyces parvulus)中发现[9]。随后,这两种物质在许多嗜盐细菌中被陆续报道,且与其盐适应策略有关[4, 10]。四氢嘧啶和羟基四氢嘧啶均为中性的、非离子的、低分子量的、易结合水的环状氨基酸衍生物(图 1)。四氢嘧啶类与甜菜碱类的功能相似,它们在盐、碱、热、冷冻、干燥等不良条件下可以稳定细胞结构,保持生物大分子(生物膜、蛋白质、酶和核酸)的活性[11-12]。此外,丰富的C、N含量使得四氢嘧啶类可以作为贮存能源,为细胞生长提供碳源或氮源[3, 9]。

|
| 图 1 四氢嘧啶(A)和羟基四氢嘧啶(B)的结构示意图 Figure 1 Structures of ectoine (A) and hydroxyectoine (B). |
1 嗜盐细菌中四氢嘧啶类的生物合成
高盐或盐波动条件下,大多数嗜盐细菌常常从外部吸收四氢嘧啶类相容性溶质来维持细胞内的高渗透压,当环境中缺乏甜菜碱和四氢嘧啶等相容性溶质时,一些嗜盐细菌可以从头合成四氢嘧啶类进行渗透调节[13-14]。随着在模式菌株伸长盐单胞菌(Halomonas elongata) DSM 2581T中四氢嘧啶合成酶(ectoine synthase,EctC)的发现[13],四氢嘧啶合成的3个关键转化酶及生物合成路径也在其他嗜盐细菌中得到验证[5, 14-17]。四氢嘧啶的生物合成分为三步(图 2):首先,以L-天冬氨酸-β-半醛(L-aspartate-β-semialdehyde,ASA)为底物,由2,4-二氨基丁酸转氨酶(diaminobutyric acid aminoransferase,EctB)催化,生成L-2,4-二氨基丁酸(L-2,4-diaminobutyrate,DABA);其次,在2,4-二氨基丁酸乙酰基转移酶(diaminobutyric acid acetyltransferase, EctA)的催化下,将DABA乙酰化生成N-乙酰-L-2,4-二氨基丁酸(N-γ-acetyl-L-2,4-diaminobutyrate,ADABA);最后,由四氢嘧啶合成酶(ectoine synthase,EctC)将ADABA环化形成四氢嘧啶。羟基四氢嘧啶则是由四氢嘧啶羟化酶(ectoine hydroxylase,EctD)将四氢嘧啶羟基化而生成[13-17]。
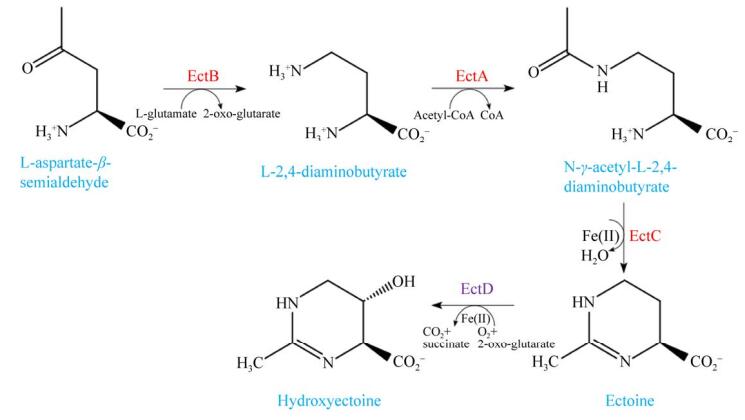
|
| 图 2 嗜盐细菌中四氢嘧啶和羟基四氢嘧啶的合成途径 Figure 2 Synthetic pathways of ectoine and hydroxyectoine in halophilic bacteria. |
在嗜盐细菌中,编码四氢嘧啶生物合成相关酶的3个基因通常位于ectABC或ectABC-ask基因簇中,而羟基四氢嘧啶合成基因ectD则在基因组中单独存在[16]。四氢嘧啶类合成相关基因对于高盐条件下嗜盐细菌维持正常的生理活动至关重要,ectA缺失突变体H. elongata KB1无法合成四氢嘧啶前体物质ADABA,与野生型菌株相比,该突变体在最低必需培养基上对NaCl的最大耐受浓度由5%降为3%,进一步的实验表明将四氢嘧啶合成基因ectC敲除后,H. elongata WUB01 (ΔectC)和H. elongata WUB02 (ΔectA,ΔectC)分别在10%和4% NaCl的条件下无法生存[18]。四氢嘧啶生物合成基因簇ectABC或ectABC-ask作为一个操纵子转录,不同来源的嗜盐细菌中ectABC启动子序列的位置及数量存在明显差异,其转录调控机制十分复杂,但均表现出了对高盐的依赖性以及对盐度波动的敏感性。H. elongata DSM 2581T中ectABC上游的启动子区promA受盐度的诱导[19];需盐色盐杆菌(Chromohalobacter salexigens) DSM 3043T中ectA上游存在4个启动子PectA1-4,其中PectA1、PectA3、PectA4为渗透调节启动子[20];嗜碱甲基微菌(Methylomicrobium alcaliphilum) 20ZT中ectABC-ask的转录起始于两个启动子ectAp1p2[21]。盐度对于四氢嘧啶类合成的诱导在转录水平上得以体现[11, 19],与0.6% NaCl条件下相比,当生长盐浓度提高至12%时,H. elongata中ectA、ectB和ectC基因的转录水平分别上调了4.4倍、4.5倍和2.0倍[22]。在盐反硝化枝芽孢杆菌(Virgibacillus halodenitrificans) PDB-F2中,四氢嘧啶类生物合成基因的转录水平随着盐度的增加呈上升趋势,与无盐条件下相比,在12% NaCl条件下,ectA、ectB和ectC分别上调了334倍、160倍和188倍;而盐度高于10% NaCl后ectD的转录水平也开始显著提高,在12% NaCl和15% NaCl下分别上调了9.4倍和20.4倍[23]。当生长盐浓度由3.5% NaCl升至14.6% NaCl时,C. salexigens DSM 3043T中羟基四氢嘧啶合成酶基因ectD上调了2.79倍[24]。15%与0% NaCl条件下相比,Nocardiopsis gilva的ectB、ectA、ectC、ectD基因分别上调了7.4倍、5.3倍、7.6倍和22.9倍[25]。同时,盐度也影响着四氢嘧啶相关合成酶的活性,与无NaCl时相比,5.8% NaCl条件下的M. alcaliphilum 20Z中EctA、EctB、EctC 3种酶的活性分别提高了15.6倍、4.4倍、12倍[21]。这些结果表明,盐度对于四氢嘧啶生物合成基因的表达不可或缺,两种相容性溶质的合成受到渗透胁迫的调节。值得注意的是,羟基四氢嘧啶合成基因的表达不仅受到盐度的诱导,高温条件下ectD的转录水平及产物羟基四氢嘧啶的合成明显提升,温度胁迫下羟基四氢嘧啶对嗜盐细菌的保护能力也相应增强[20, 24]。
2 四氢嘧啶来源于多种类型的嗜盐细菌中度嗜盐菌(moderately halophiles)是指最适生长盐浓度为3%–15% NaCl的嗜盐菌,而当嗜盐菌的最适生长盐浓度大于15%时为极端嗜盐菌(extreme halophiles)。中度嗜盐菌的盐适应范围极宽,四氢嘧啶类在中度嗜盐细菌中的研究和应用十分广泛,是其最重要的相容性溶质之一[26]。截止笔者撰写时,我们从JGI-IMG (https://img.jgi.doe.gov/cgi-bin/mer/main.cgi)和EZBioCloud (https://www.ezbiocloud.net)数据库中共获得67株含有四氢嘧啶合成基因的标准菌株,这些嗜盐细菌属于6个主要的系统发育分支:变形菌门(Proteobacteria,占总数约61.2%)、放线菌门(Actinobacteria,约11.2%)、厚壁菌门(Firmicutes,约23.9%)、脱铁杆菌门(Deferribacteres,约1.5%)、螺旋体门(Spirochaetes,约1.5%)。其中包括本课题组测序分析了全基因组序列的变形菌门Halomonas urumqiensis BZ-SZ-XJ27T,厚壁菌门Alkalicoccus saliphilus DSM 15402T、Alkalicoccus halolimnae BZ-SZ-XJ29T及Bacillus urumqiensis BZ-SZ-XJ18T[27-30]。系统发育分析表明,这67株嗜盐细菌在亲缘关系上相距甚远(图 3),其最适生长和耐受盐浓度范围广、差别较大。其中,Salinicoccus albus DSM 19776T耐受盐浓度区间最广(表 1),为1%–30% NaCl;Halorhodospira halophila SL1T耐受盐浓度最高,达35% NaCl。
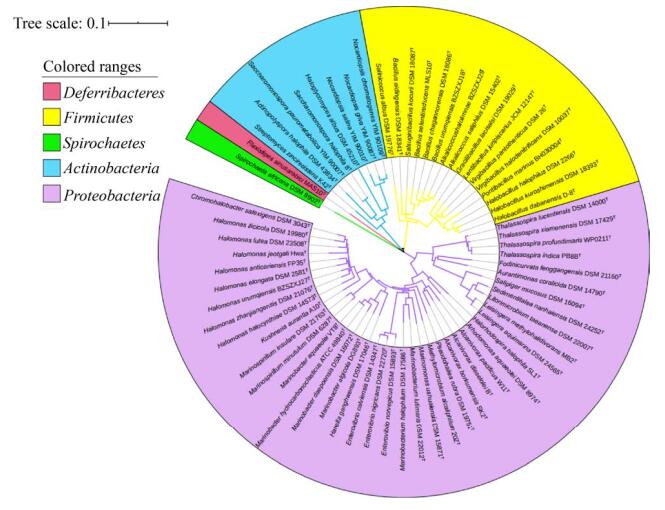
|
| 图 3 基于16S rRNA基因序列的四氢嘧啶合成嗜盐细菌的亲缘图 Figure 3 Phylogenetic relationship based on 16s rRNA gene sequences of halophilic bacteria that synthesize ectoine. The scale bar indicates the phylogenetic distance. |
| Species | Accession number | Na+ opt./% (W/V) |
Na+/%(W/V) | pH opt. | pH |
| Actinopolyspora halophila DSM 43834T | AQUI01000002 | 15.0-20.0 | 12.0-30.0 | 7.5 | 6.0-8.0 |
| Alcanivorax borkumensis SK2T | AM286690 | 3.0-10.0 | 1.0-12.5 | ND | ND |
| Alcanivorax dieselolei B-5T | CP003466 | 3.0-7.5 | 1.0-15.0 | ND | ND |
| Alcanivorax pacificus W11-5T | AJGP01000039 | 3.0-5.0 | 0.5-12.0 | 7.5 | ND |
| Alkalicoccus saliphilus DSM 15402T | AJ493660 | 15.0 | 1.0-20.0 | 9.0 | 7.0-10.0 |
| Alkalicoccus halolimnae BZ-SZ-XJ29T | KX618877 | 8.3-12.0 | 4.3-24.0 | 7.5 | 6.0-10.5 |
| Arhodomonas aquaeolei DSM 8974T | M26631 | 15.0 | 6.0-20.0 | 6.5-7.5 | 6.0-8.0 |
| Aurantimonas coralicida DSM 14790T | ATXK01000033 | 3.2 | ND | ND | ND |
| Bacillus aidingensis DSM 18341T | DQ504377 | 12.0 | 8.0-33.0 | 7.2 | 6.0-9.5 |
| Bacillus chagannorensis DSM 18086T | AM492159 | 7.0 | 3.0-20.0 | 8.5 | 5.8-11.0 |
| Bacillus selenitireducens MLS10T | ABHZ01000019 | 2.4-6.0 | 2.0-22.0 | ND | 8.5-10 |
| Bacillus urumqiensis BZ-SZ-XJ18T | KM066107 | 6.3 | 1.3-25.3 | 8.5-9.5 | 6.5-10.0 |
| Chromohalobacter salexigens DSM 3043T | CP000285 | 7.5-10.0 | 0.9-25.0 | 7.5 | 5.0-10.0 |
| Enterovibrio calviensis DSM 14347T | AF118021 | 2.5-3.5 | 1.5-12.0 | ND | ND |
| Enterovibrio nigricans DSM 22720T | AM942722 | ND | 0.9-6.0 | ND | ND |
| Enterovibrio norvegicus DSM 15893T | AJ316208 | ND | 2.0-8.0 | ND | ND |
| Flexistipes sinusarabici MAS10T | CP002858 | 3.0 | 3.0-18.0 | 6.0-8.0 | ND |
| Fodinicurvata fenggangensis DSM 21160T | JMLV01000013 | 5.0 | 1.5-20.0 | 7.5 | 6.5-8.5 |
| Gracilibacillus lacisalsi DSM 19029T | DQ664540 | 5.0-7 .0 | 1.0-18.0 | 7.5-8.0 | 5.5-10.0 |
| Hahella ganghwensis DSM 17046T | AQXX01000025 | 4.0-6 .0 | 1.0-10.0 | 7.0-8.0 | 5.0-10.0 |
| Halobacillus halophilus DSM 2266T | HE717023 | 5.9-8.8 | 3.0-12.0 | 7.5 | 5.5-10.0 |
| Halobacillus kuroshimensis DSM 18393T | AB195680 | 6 .0 | 0.5-25.0 | 7.5-8.0 | 5.5-10.0 |
| Haloglycomyces albus DSM 45210T | AZUQ01000001 | 8.0-12.0 | 3.0-18 .0 | 7.0-7.5 | 5.0-9.0 |
| Halomonas anticariensis FP35T | ASTJ01000004 | 7.5 | 0.5-15.0 | ND | 6.0-9.0 |
| Halomonas elongata DSM 2581T | FN869568 | 3.5-8.0 | 3.5-20.0 | 7.2 | 5.0-9.0 |
| Halomonas halocynthiae DSM 14573T | AJ417388 | 3.0-6.0 | 0.5-15.0 | 7.5-8.5 | 5.0-11.0 |
| Halomonas ilicicola DSM 19980T | EU218533 | 10.0 | 2.0-17.5 | 6.5 | 6.0-9.0 |
| Halomonas jeotgali HwaT | AMQY01000015 | 10.0 | 5.0-25.0 | 7.0-8.0 | 5.0-10.0 |
| Halomonas lutea DSM 23508T | ARKK01000003 | 5.0-10.0 | 1.0-20.0 | 7.5 | 5.0-9.0 |
| Halomonas urumqiensis BZ-SZ-XJ27T | KM066108 | 8.3 | 1.3-25.0 | 6.0-10.0 | 8.0-8.5 |
| Halomonas zhanjiangensis DSM 21076T | FJ429198 | 3.0-5.0 | 1.0-20.0 | 7.5 | 6.0-10.5 |
| Halorhodospira halophila SL1T | CP000544 | 15.0-35.0 | 3.0-35.0 | 7.4-7.9 | ND |
| Kushneria aurantia A10T | AM941746 | 10.0 | 5.0-17.5 | 7.0-8.0 | 5.5-8.5 |
| Leisingera aquimarina DSM 24565T | AXBE01000001 | ND | 1.0-7 .0 | 6.5-8.0 | 5.5-9.0 |
| Leisingera methylohalidivorans MB2T | CP006773 | 3.5 | 1.5-6.5 | 7.7 | 6.0-8.8 |
| Lentibacillus juripiscarius JCM 12147T | AB127980 | 10.0 | 3.0-30 .0 | 7.0 | 5.0-9.0 |
| Litorimicrobium taeanense DSM 22007T | jgi.1055387 | 2.0-4.0 | 1.0-6.0 | 6.5-7.5 | 6.0-8.0 |
| Marinobacter aquaeolei VT8T | 640721707 | 5.0 | 0-20.0 | 7.3 | 5.0-10.0 |
| Marinobacter daepoensis DSM 16072T | AY517633 | 2.0-6.0 | 0-18.0 | 7.0-8.0 | 5.5-ND |
| Marinobacter hydrocarbonoclasticus ATCC 49840T | FO203363 | 3.5 | 0.5-20.0 | 7.0-7.5 | 6.0-9.5 |
| Marinobacterium halophilum DSM 17586T | AY563030 | ND | 3.0-12 .0 | ND | 5.3-8.8 |
| Marinobacterium lutimaris DSM 22012T | jgi.1107934 | 2.0-5.0 | 1.0-10.0 | 6.5-7.5 | 6.0-8.0 |
| Marinomonas ushuaiensis DSM 15871T | AJ627909 | 6.0 | 1.0-6.0 | 8.0 | 7.0-12.5 |
| Marinospirillum insulare DSM 21763T | 2576860715 | 2.0-3.0 | 0.5-10 .0 | 8.0 | 6.5-10.0 |
| Marinospirillum minutulum DSM 6287T | AB006769 | 3.0 | 0.5-9.0 | 8.0 | 7.0-10.5 |
| Nocardiopsis chromatogenes YIM 90109T | AY619715 | 5.0-8 .0 | 0-18.0 | 7.2 | 6.0-9.0 |
| Nocardiopsis gilva YIM 90087T | AY619712 | 5.0-8.0 | 0-18.0 | 7.2 | 6.0-9.0 |
| Nocardiopsis salina YIM 90010T | AY373031 | 10 .0 | 3.0-20.0 | 7.2 | 6.0-9.0 |
| Pontibacillus marinus BH030004T | AVPF01000156 | 2.0-5.0 | 1.0-9.0 | 7.0-7.5 | 6.0-9.0 |
| Pseudohaliea rubra DSM 19751T | AUVB01000024 | 3.5 | 0.7-4.2 | 8.0 | 5.0-9.0 |
| Saccharomonospora halophila 8T | AICX01000084 | 10.0 | 10.0-30.0 | ND | 6.0-9.0 |
| Saccharomonospora paurometabolica YIM 90007T | 2510465783 | 10.0 | 5.0-20.0 | ND | ND |
| Salinicoccus albus DSM 19776T | EF177692 | 10.0 | 1.0-30.0 | 8.5 | 6.0-10.0 |
| Salipiger mucosus DSM 16094T | ARRM01000007 | 3.0-6 .0 | 0.5-20 .0 | ND | 6.0-10.0 |
| Salsuginibacillus kocurii DSM 18087T | AM492160 | 10.0 | 3.0-20 .0 | 8.5 | 5.8-10.0 |
| Sedimentitalea nanhaiensis DSM 24252T | AXBG01000021 | 1.0-4.0 | 0.6-6.0 | 7.0-8.5 | 6.0-9.3 |
| Streptomyces zinciresistens K42T | AGBF01000432 | 4.0 | 4.0-7.0 | 7.5 | 5.0-11.0 |
| Spirochaeta africana Z-7692T | AGRZ01000060 | 4.7-8.2 | 3.0-10.0 | 8.8-9.8 | 8.0-10.8 |
| Thalassospira indica PB8BT | 2774822901 | 2.0-4.0 | 0-15.0 | 6.0-7.0 | 5.0-11.0 |
| Thalassospira lucentensis DSM 14000T | AM294944 | ND | 2.0-10.0 | 7.2 | ND |
| Thalassospira profundimaris WP0211T | AY186195 | 3.0-4.0 | 2.0-8.0 | ND | ND |
| Thalassospira xiamenensis DSM 17429T | CP004388 | 2.0-4.0 | 0.5-10.0 | ND | ND |
| The blue background shown in Table 1 indicates the halophilic bacterium can synthesize both ectoine and glycine betaine. The white background shown in Table 1 the halophilic bacterium can synthesize ectoine, but not glycine betaine. ND: data were not shown in the published paper. | |||||
嗜盐细菌的盐适应机制极为复杂,在应对外界盐度变化时,几种相容性溶质相辅相成,作为最为重要的2种相容性溶质,四氢嘧啶类和甜菜碱类密切相关。我们得到的67株四氢嘧啶合成菌株中就有51株同时具备甘氨酸甜菜碱合成能力(表 1蓝色背景),包括两株极端嗜盐细菌嗜盐放线多孢菌(Actinopolyspora halophila) DSM 43834T (15.0–20.0[Na+]opt./% (W/V))和嗜盐红螺菌(Halorhodospira halophila) SL1T (15.0–35.0[Na+]opt./% (W/V)),相对于其他菌株,二者对高盐度具有更强的适应力,其胞内四氢嘧啶类和甘氨酸甜菜碱的合成与渗透调控极有可能存在着特殊的联系。同时,有12株四氢嘧啶类合成嗜盐细菌中不含有甘氨酸甜菜碱合成基因(表 1白色背景),它们的盐度耐受范围均不超过10% NaCl,进一步佐证了2种相容性溶质在嗜盐机制中联系密切。但是,在众多来源的嗜盐细菌中,2种相容性溶质的合成如何达到动态平衡,渗透调节过程中怎样相互作用,不同来源的嗜盐细菌中二者又存在怎样的差异呢?关于二者在嗜盐菌中的异同是很重要的科学问题,仍需更多的理论和实验数据来验证。
3 四氢嘧啶类在盐适应及盐胁迫中起重要的作用四氢嘧啶类的生物学功能在多种类型的嗜盐细菌中均有报道,但由于“嗜盐细菌”并不是分类学意义上的专有名词,且不同来源的嗜盐细菌在生理特性上具有一定的差异,四氢嘧啶类与其他相容性溶质在盐适应和盐胁迫过程中的功能也有所区别。因而我们仅选取了部分代表性的四氢嘧啶类合成嗜盐细菌进行了分析归纳,对四氢嘧啶及羟基四氢嘧啶在盐适应过程中的作用进行系统的阐述。
作为革兰氏阴性嗜盐细菌的模式菌株,伸长盐单胞菌在高盐条件下以四氢嘧啶类作为主要的相容性溶质,H. elongata通过积累四氢嘧啶获得了广泛的耐盐性,甚至可以在盐饱和的盐水(> 30% NaCl)中存活,其四氢嘧啶类的生物合成得到广泛而深入的研究[22, 31-35]。G?ller等构建的NaCl敏感突变体H. elongata SAA4与野生型相比,虽然在2% NaCl下具有相同的生长速率(0.23 h–1),但当盐度达到4% NaCl时,突变体的生长严重受阻,ect基因的转录量不再增加,且生长速率也降为野生型的一半。而将此突变体在以四氢嘧啶和羟基四氢嘧啶为唯一碳源的培养基上进行培养,8% NaCl条件下,H. elongata SAA4能够生长繁殖[31-32]。此外,四氢嘧啶合成受阻菌株H. elongata CHR62和H. elongata CHR63分别在5.9%、11.7% NaCl条件下不能生长,而相同条件下能够合成四氢嘧啶的野生型菌株却长势良好[33-34]。高盐条件下,盐反硝化枝芽孢杆菌同样以四氢嘧啶和羟基四氢嘧啶作为主要的相容性溶质,无盐条件下生长的V. halodenitrificans菌株中没有检测到四氢嘧啶类物质,随着盐度的增加,细胞内四氢嘧啶类浓度逐渐增加;在12% NaCl下,四氢嘧啶浓度最高(468.1 nmol/mg干重),羟基四氢嘧啶的浓度则在15% NaCl条件下达到最大值(89.1 nmol/mg干重)。同时,V. halodenitrificans的细胞生长浓度也随盐度的增加而增加,在12% NaCl条件下其OD578值最高达1.0;即使在细胞生长严重受阻的15% NaCl盐度下,其OD578值仍高于未添加NaCl的实验组[23]。四氢嘧啶类也是需盐色盐杆菌应对渗透胁迫的主要相容性溶质[24]:14.6% NaCl条件下生长的C. salexigens DSM 3043胞内四氢嘧啶和羟基四氢嘧啶总含量约是3.5% NaCl条件下的2.8倍,而菌体OD600值则由1.8增至3.6,提高了2倍。通过基因组测序分析,12% NaCl条件下,泛酸枝芽孢杆菌(Virgibacillus pantothenticus) PDB-F2也以四氢嘧啶类作为主要的相容性溶质,外源添加四氢嘧啶类对菌体的渗透保护作用大于其他相容性溶质,且菌体对羟基四氢嘧啶的偏好性要高于四氢嘧啶[36]。四氢嘧啶类还在多种类别的嗜盐细菌中发挥着主要作用:Brevibacterium sp. JCM 6894中四氢嘧啶和羟基四氢嘧啶的总量随胞外盐度线性增加,当盐度增至14.6% NaCl时,二者占总相容性溶质的55%以上[6]。在Nocardiopsis gilva YIM 90087T中,四氢嘧啶类的积累随着盐度的升高而增加,当盐度由0% NaCl升至15% NaCl后,N. gilva YIM 90087T胞内四氢嘧啶含量提高了6.6倍;而0% NaCl或5% NaCl条件下其无法合成羟基四氢嘧啶,15% NaCl其胞内羟基四氢嘧啶的含量为1.17 mg/g CDW。值得注意的是,在10%和15%的NaCl浓度下,四氢嘧啶的浓度分别是羟基四氢嘧啶的42.6倍和28.3倍[25]。以上结果表明,二者相比,四氢嘧啶在嗜盐细菌的盐适应机制中起主要作用,随着盐度的升高,羟基四氢嘧啶的功能不断增强,它们分工协作,相辅相成,这或许正是众多嗜盐细菌在高盐条件下更青睐以四氢嘧啶类作为主要相容性溶质的重要原因。
四氢嘧啶类相容性溶质的重要作用不只局限于平衡嗜盐细菌细胞内外渗透压上,二者也可作为细胞稳定剂提高菌体对于高温、干燥、冻融处理,甚至是紫外线辐射、细胞毒素等压力的抵抗作用[11, 37],而且由于独特的羟基化属性,羟基四氢嘧啶对热和干燥下的细胞具有额外的应力保护[24]。此外,四氢嘧啶类在细胞营养物质缺乏时还可以作为碳/氮源供嗜盐细菌利用[3, 11]。
4 四氢嘧啶类合成基因在生物学技术中的应用目前,随着四氢嘧啶类独特的生物学功能被不断的深入挖掘,四氢嘧啶类合成基因在生物学技术应用领域也越来越受到广泛的关注[26, 38]。以下就其在基因工程菌的构建、转基因农作物的应用等方面的研究进行概述。
(1) 四氢嘧啶类合成基因应用于构建基因工程菌。Louis等(1997)首次将嗜盐海球菌Marinococcus halophilus DSM 20408T中的四氢嘧啶合成基因簇ectABC在大肠杆菌Escherichia coli中成功表达,重组工程菌E. coli (pOSM1)在0%–5% NaCl条件下正常生长,且伴随着盐度的升高,E. coli (pOSM1)胞内的四氢嘧啶浓度不断增加,并在5% NaCl条件下达到最大干重浓度1 mmol/g[16]。同样的情况在其他来源的嗜盐细菌中得到验证,含有来自M. alcaliphilum 20ZT的ectABC基因簇的重组大肠杆菌在1%–4% NaCl条件下能够生长,积累的四氢嘧啶从20 μmol/g (细胞干重)提升至280 μmol/g (细胞干重)[21]。将克劳氏芽孢杆菌Bacillus clausii中的ectABC基因簇导入大肠杆菌E. coli M15中,在M15的培养物中检测到了四氢嘧啶[39]。此外,来自达班湖喜盐芽孢杆菌Halobacillus dabanensis、喜盐涅斯特连科氏菌Nesterenkonia halobia和N. gilva的ectABC基因同样在大肠杆菌中成功表达[25, 40-41]。将H. elongata BK-AG25的ectABC基因导入E. coli BL21中,重组大肠杆菌胞内和胞外四氢嘧啶浓度分别在1.78%和0.28% NaCl时达到最大值,并且随着盐度的升高胞内四氢嘧啶浓度不断增加,胞外浓度不断减少,说明高盐条件下,重组子通过积累四氢嘧啶以抵御渗透胁迫[42]。Zhang等将嗜碱芽孢杆菌Bacillus alcalophilus DTY1中的ectABC基因簇片段转入蜡质芽孢杆菌Bacillus cereus Z中,与野生型相比,宿主的耐盐度提高了20%,在5.0% NaCl条件下生成118.6 mg/g四氢嘧啶,当盐浓度从1.0%升至5.0%时,四氢嘧啶的产量提高了1.7倍[43]。同时非嗜盐转基因工程菌异源表达生产四氢嘧啶类也成为了研究热点:Zhang等将ectABC基因导入大肠杆菌中,重组工程菌E. coli ET08四氢嘧啶产量为36.5 g/L,超过了先前的报道[44]。除大肠杆菌外,Canter将ectABC基因转入谷氨酸棒杆菌(Corynebacterium glutamicum)中,转基因工程菌C. glutamicumectABC成功合成四氢嘧啶,产量高达65.3 g/L (56 h)[45]。而在代表性嗜盐细菌海球菌属(Marinococcus)中四氢嘧啶产量达70.4–135 mg/g,短杆菌属(Brevibacterium)中达150–160 mg/g,盐单胞菌属(Halomonas)中达155–358 mg/g[46-47]。四氢嘧啶的异源表达产量远高于天然生产菌株的产量,并且打破了“细菌泌乳”工艺生产中由于嗜盐细菌的自身特性所造成的发酵周期长、设备腐蚀严重等局限性,为四氢嘧啶的工业化生产提供了新的策略。
(2) 四氢嘧啶类合成基因应用于转基因农作物。土壤盐分是造成农业用地流失和农作物减产的主要环境因素之一,因而四氢嘧啶合成基因在转基因经济作物中具有极高的应用价值。Nakayama等将H. elongata OUT30018中的ectABC基因转化到烟草BY2中,在转基因烟草细胞中四氢嘧啶积累量达79 nmol/g,同时其耐渗透胁迫的能力是空白对照组的1.6倍;FDA染色法检测烟草细胞的活性时发现,在3% NaCl条件下,野生型烟草细胞未被染色,而转基因烟草细胞FDA染色量超过70%,这表明四氢嘧啶类的合成有效提高了转基因烟草细胞的活性[48]。Moghaieb等同样将H. elongata的ectABC基因导入到了烟草细胞(Nicotiana tabaccum)中,并在0%、0.6%、1.2% NaCl的不同条件下将其培养7 d,随着盐度的增加,转基因烟草中四氢嘧啶的积累量逐渐提高至50 µmol/g (细胞鲜重)。结果表明,NaCl对烟草植株的干重、茎秆粗度、光合作用效率、叶片膨压、氮吸收等均有损害,但与野生型相比,转基因烟草植株所受的伤害较小,四氢嘧啶的合成有效提高了宿主的耐盐性能[49]。Rai等的转基因烟草实验验证了Moghaieb等的结论,四氢嘧啶合成基因的表达赋予了烟草耐盐性和温度胁迫应力[50-51]。为了进一步研究四氢嘧啶合成基因在转基因作物中的应用广泛性,Moghaieb等又将ectABC基因转入番茄(Lycopersicon esculentum)中,一方面,随着盐度的增加,转基因番茄的根和叶中的四氢嘧啶不断积累,且在0.6%、1.2%、1.8%的盐度下,转基因番茄的生物量是野生型的两倍多;另一方面,四氢嘧啶基因的导入有效提高了番茄的光合速率和根活力,并有效降低了盐胁迫对转基因番茄造成的伤害[52]。
不耐盐菌体中四氢嘧啶类生物合成基因的导入,显著提高了宿主对高盐高渗环境的应对能力,使其在盐胁迫下可以稳定生长并发挥其生理功能,作为分子生物学的有力工具更加广泛地应用于生产实践。长期以来,产量低、生产成本高一直制约着四氢嘧啶类在各领域的实际应用,因而四氢嘧啶类的高效异源表达为其合成与工业化生产提供了更多可能性。同时,四氢嘧啶生物合成基因在植物中的成功表达增强了转基因作物的耐盐碱性,对干旱盐碱土地的治理具有深远的意义。
5 展望嗜盐细菌的盐适应机制是极为复杂的一个过程,在盐波动下往往需要1种以上的相容性溶质共同发挥作用以抵御外界的高渗透环境。四氢嘧啶和羟基四氢嘧啶作为一类极为重要的相容性溶质,在此过程中起着重要的作用。二者与其他相容性溶质如何相互影响,盐适应过程中四氢嘧啶类又具有怎样的独特性?为何一些四氢嘧啶合成菌株可以同时合成甘氨酸甜菜碱,而另一些菌株却不含有甜菜碱合成基因,究竟嗜盐细菌更偏向于利用哪类相容性溶质作为渗透保护剂,这种倾向与外界盐度又有何联系?此外,虽然羟基四氢嘧啶是四氢嘧啶的衍生物,化学结构上密切相关,但二者的渗透保护功能却不尽相同,这是否与其微小的结构差异有关,它们在渗透调节中的作用又有何规律?目前,虽然我们对嗜盐细菌中四氢嘧啶类的生物合成过程及其生理功能有了一定的了解,但是当外界盐度升高时,细胞如何感知外界盐度的变化、信息如何传递到细胞内、生物合成及转运的调控机制如何触发、以及四氢嘧啶类物质的转运与代谢等诸多环节尚不明确。这些问题对于我们认识和比较不同类别嗜盐细菌之间的亲缘关系至关重要,更是四氢嘧啶类合成基因资源开发与应用的关键,有待相关研究者的深入探究。
四氢嘧啶和羟基四氢嘧啶表现出了巨大的应用价值和商业前景,其生物合成基因在构建基因工程菌、现代食品工业、化妆品领域、医学应用、污水处理和石油污染生物修复、微生物电池等方面得到广泛应用,但在转基因植物开发领域仍处于初步探索状态。我国盐碱土地面积辽阔,四氢嘧啶类基因的研究和应用可为增强干旱盐碱地区农作物的抗逆性提供优质资源。因此,有望本综述可以促进四氢嘧啶类生物合成基因在代谢组学、蛋白组学、分子进化等生物技术领域更为深入的研究,促进四氢嘧啶类相容性溶质在干旱盐碱环境的改良和治理等领域更为广泛的应用。
| [1] | Brown AD. Microbial water stress. Bacteriological Reviews, 1976, 40(4): 803-846. DOI:10.1128/br.40.4.803-846.1976 |
| [2] | Piubeli F, Salvador M, Argandoña M, Nieto JJ, Bernal V, Pastor JM, Cánovas M, Vargas C. Insights into metabolic osmoadaptation of the ectoines-producer bacterium Chromohalobacter salexigens through a high-quality genome scale metabolic model. Microbial Cell Factories, 2018, 17(1): 2. DOI:10.1186/s12934-017-0852-0 |
| [3] | Becker J, Wittmann C. Microbial production of extremolytes-high-value active ingredients for nutrition, health care, and well-being. Current Opinion in Biotechnology, 2020, 65: 118-128. DOI:10.1016/j.copbio.2020.02.010 |
| [4] | Galinski EA. Compatible solutes of halophilic eubacteria: molecular principles, water-solute interaction, stress protection. Experientia, 1993, 49(6/7): 487-496. |
| [5] |
Zhao BS, Yang LF, Wang L, Lu WD, Yang SS. Study progress on compatible solutes in moderately halophilic bacteria. Acta Microbiologica Sinica, 2007, 47(5): 937-941.
(in Chinese) 赵百锁, 杨礼富, 王磊, 卢伟东, 杨苏声. 中度嗜盐菌相容性溶质机制的研究进展. 微生物学报, 2007, 47(5): 937-941. DOI:10.3321/j.issn:0001-6209.2007.05.037 |
| [6] | Nagata S, Adachi K, Sano H. NMR analyses of compatible solutes in a halotolerant Brevibacterium sp.. Microbiology, 1996, 142(12): 3355-3362. DOI:10.1099/13500872-142-12-3355 |
| [7] |
Zhang YJ, Liao ZY, Zhao BS. Glycine betaine: biosynthesis and biological function in halophilic microorganisms. Acta Microbiologica Sinica, 2020, 60(6): 1074-1089.
(in Chinese) 张英杰, 廖子亚, 赵百锁. 嗜盐菌中甘氨酸甜菜碱的合成途径及其生物学功能. 微生物学报, 2020, 60(6): 1074-1089. |
| [8] | Galinski EA, Pfeiffer HP, Trüper HG. 1,4,5,6-Tetrahydro-2-methyl-4-pyrimidinecarboxylic acid: a novel cyclic amino acid from halophilic phototrophic bacteria of the genus Ectothiorhodospira. European Journal of Biochemistry, 1985, 149(1): 135-139. DOI:10.1111/j.1432-1033.1985.tb08903.x |
| [9] | Inbar L, Lapidot A. The structure and biosynthesis of new tetrahydropyrimidine derivatives in actinomycin D producer Streptomyces parvulus. Use of 13C-and 15N-labeled L-glutamate and 13C and 15N NMR spectroscopy. Journal of Biological Chemistry, 1988, 263(31): 16014-16022. DOI:10.1016/S0021-9258(18)37550-1 |
| [10] |
Zhao BS, Li J. Biodiversity of culture-dependent haloalkaliphilic microorganisms. Acta Microbiologica Sinica, 2017, 57(9): 1409-1420.
(in Chinese) 赵百锁, 李俊. 可培养盐碱菌多样性的研究进展. 微生物学报, 2017, 57(9): 1409-1420. |
| [11] | Czech L, Hermann L, Stöveken N, Richter AA, Höppner A, Smits SHJ, Heider J, Bremer E. Role of the extremolytes ectoine and hydroxyectoine as stress protectants and nutrients: genetics, phylogenomics, biochemistry, and structural analysis. Genes, 2018, 9(4): 177. DOI:10.3390/genes9040177 |
| [12] | Hahn MB, Smales GJ, Seitz H, Solomun T, Sturm H. Ectoine interaction with DNA: influence on ultraviolet radiation damage. Physical Chemistry Chemical Physics, 2020, 22(13): 6984-6992. DOI:10.1039/D0CP00092B |
| [13] | Peters P, Galinski EA, Trüper HG. The biosynthesis of ectoine. FEMS Microbiology Letters, 1990, 71(1/2): 157-162. |
| [14] | Richter AA, Kobus S, Czech L, Hoeppner A, Zarzycki J, Erb TJ, Lauterbach L, Dickschat DS, Bremer E, Smits SHJ. The architecture of the diaminobutyrate acetyltransferase active site provides mechanistic insight into the biosynthesis of the chemical chaperone ectoine. Journal of Biological Chemistry, 2020, 295(9): 2822-2838. DOI:10.1074/jbc.RA119.011277 |
| [15] | Hillier HT, Altermark B, Leiros I. The crystal structure of the tetrameric DABA-aminotransferase EctB, a rate-limiting enzyme in the ectoine biosynthesis pathway. The FEBS Journal, 2020, 287(21): 4641-4658. DOI:10.1111/febs.15265 |
| [16] | Louis P, Galinski EA. Characterization of genes for the biosynthesis of the compatible solute ectoine from Marinococcus halophilus and osmoregulated expression in Escherichia coli. Microbiology, 1997, 143(4): 1141-1149. DOI:10.1099/00221287-143-4-1141 |
| [17] | Gunde-Cimerman N, Plemenitaš A, Oren A. Strategies of adaptation of microorganisms of the three domains of life to high salt concentrations. FEMS Microbiology Reviews, 2018, 42(3): 353-375. DOI:10.1093/femsre/fuy009 |
| [18] | Witt EMHJ, Davies NW, Galinski EA. Unexpected property of ectoine synthase and its application for synthesis of the engineered compatible solute ADPC. Applied Microbiology and Biotechnology, 2011, 91(1): 113-122. DOI:10.1007/s00253-011-3211-9 |
| [19] | Stiller LM, Galinski EA, Witt EMHJ. Engineering the salt-inducible ectoine promoter region of Halomonas elongata for protein expression in a unique stabilizing environment. Genes, 2018, 9(4): 184. DOI:10.3390/genes9040184 |
| [20] | Calderón MI, Vargas C, Rojo F, Iglesias-Guerra F, Csonka LN, Ventosa A, Nieto JJ. Complex regulation of the synthesis of the compatible solute ectoine in the halophilic bacterium Chromohalobacter salexigens DSM 3043T. Microbiology, 2004, 150(9): 3051-3063. DOI:10.1099/mic.0.27122-0 |
| [21] | Mustakhimov II, Reshetnikov AS, Glukhov AS, Khmelenina VN, Kalyuzhnaya MG, Trotsenko YA. Identification and characterization of EctR1, a new transcriptional regulator of the ectoine biosynthesis genes in the halotolerant methanotroph Methylomicrobium alcaliphilum 20Z. Journal of Bacteriology, 2010, 192(2): 410-417. DOI:10.1128/JB.00553-09 |
| [22] | Kindzierski V, Raschke S, Knabe N, Siedler F, Scheffer B, Pflüger-Grau K, Pfeiffer F, Oesterhelt D, Marin-Sanguino A, Kunte HJ. Osmoregulation in the halophilic bacterium Halomonas elongata: a case study for integrative systems biology. PLoS One, 2017, 12(1): e0168818. DOI:10.1371/journal.pone.0168818 |
| [23] | Tao P, Li H, Yu YJ, Gu JD, Liu YD. Ectoine and 5-hydroxyectoine accumulation in the halophile Virgibacillus halodenitrificans PDB-F2 in response to salt stress. Applied Microbiology and Biotechnology, 2016, 100(15): 6779-6789. DOI:10.1007/s00253-016-7549-x |
| [24] | Salvador M, Argandoña M, Naranjo E, Piubeli F, Nieto JJ, Csonka LN, Vargas C. Quantitative RNA-seq analysis unveils osmotic and thermal adaptation mechanisms relevant for ectoine production in Chromohalobacter salexigens. Frontiers in Microbiology, 2018, 9: 1845. DOI:10.3389/fmicb.2018.01845 |
| [25] | Han J, Gao QX, Zhang YG, Li L, Mohamad OAA, Rao MPN, Xiao M, Hozzein WN, ALkhalifah DHM, Tao Y, Li WJ. Transcriptomic and ectoine analysis of halotolerant Nocardiopsis gilva YIM 90087T under salt stress. Frontiers in Microbiology, 2018, 9: 618. DOI:10.3389/fmicb.2018.00618 |
| [26] |
Zhao BS, Yang LF, Song L, Wang H. Biotechnology applications in moderately halophilic eubacteria. Microbiology, 2007, 34(2): 359-362.
(in Chinese) 赵百锁, 杨礼富, 宋蕾, , 等. 中度嗜盐菌在生物技术中的应用. 微生物学通报, 2007, 34(2): 359-362. DOI:10.3969/j.issn.0253-2654.2007.02.038 |
| [27] | Zhang SS, Pan J, Lu WD, Yan YC, Wang HS, Wiegel J, Zhao BS. Halomonas urumqiensis sp. nov., a moderately halophilic bacterium isolated from a saline-alkaline lake. International Journal of Systematic and Evolutionary Microbiology, 2016, 66(5): 1962-1969. DOI:10.1099/ijsem.0.000975 |
| [28] | Zhang SS, Li ZJ, Yan YC, Zhang CL, Li J, Zhao BS. Bacillus urumqiensis sp. nov., a moderately haloalkaliphilic bacterium isolated from a salt lake. International Journal of Systematic and Evolutionary Microbiology, 2016, 66(6): 2305-2312. DOI:10.1099/ijsem.0.001028 |
| [29] | Qiu DC, Liao ZY, Lu WD, Wang HS, Li J, Zhao BS. Draft genome sequence of Alkalicoccus saliphilus DSM 15402T, a haloalkaliphilic bacterium isolated from a mineral pool. Microbiology Resource Announcements, 2019, 8(24): e00266-19. |
| [30] | Zhang YJ, Qiu DC, Liao ZY, Zhao BS. Draft genome sequence of Alkalicoccus halolimnae BZ-SZ-XJ29T, a moderately halophilic bacterium isolated from a salt lake. Microbiology Resource Announcements, 2020, 9(27): e00500-20. DOI:10.1128/MRA.00500-20 |
| [31] | Göller K, Ofer A, Galinski EA. Construction and characterization of an NaCl-sensitive mutant of Halomonas elongata impaired in ectoine biosynthesis. FEMS Microbiology Letters, 1998, 161(2): 293-300. DOI:10.1111/j.1574-6968.1998.tb12960.x |
| [32] | Göller K, Stein M, Galinski E, Kunte HJ. Characterization of ectoine synthesis and its regulation in the halophilic bacterium Halomonas elongata. Comparative Biochemistry and Physiology Part A: Molecular & Integrative Physiology, 2000, 126(S1): 61. |
| [33] | Cánovas D, Vargas C, Iglesias-Guerra F, Csonka LN, Rhodes D, Ventosa A, Nieto JJ. Isolation and characterization of salt-sensitive mutants of the moderate halophile Halomonas elongata and cloning of the ectoine synthesis genes. Journal of Biological Chemistry, 1997, 272(41): 25794-25801. DOI:10.1074/jbc.272.41.25794 |
| [34] | Cánovas D, Vargas C, Calderón MI, Ventosa A, Nieto JJ. Characterization of the genes for the biosynthesis of the compatible solute ectoine in the moderately halophilic bacterium Halomonas elongata DSM 3043. Systematic and Applied Microbiology, 1998, 21(4): 487-497. DOI:10.1016/S0723-2020(98)80060-X |
| [35] | Nakayama H, Kawamoto R, Miyoshi K. Ectoine production from putrefactive non-volatile amines in the moderate halophile Halomonas elongata. IOP Conference Series: Earth and Environmental Science, 2020, 439: 012001. DOI:10.1088/1755-1315/439/1/012001 |
| [36] | Zhou Y, Sun ZY, Li H, Qian CJ, Wu X, Tang HZ, Ali F, Liu YD. Investigation of compatible solutes synthesis and transport of Virgibacillus halodenitrificans PDB-F2 with complete genome analysis. International Biodeterioration & Biodegradation, 2017, 122: 165-172. |
| [37] | Bownik A, Stępniewska Z. Ectoine as a promising protective agent in humans and animals. Archives of Industrial Hygiene and Toxicology, 2016, 67(4): 260-265. DOI:10.1515/aiht-2016-67-2837 |
| [38] | Zhao BS, Yan YC, Chen SL. How could haloalkaliphilic microorganisms contribute to biotechnology?. Canadian Journal of Microbiology, 2014, 60(11): 717-727. DOI:10.1139/cjm-2014-0233 |
| [39] | Anburajan L, Meena B, Sreelatha T, Vinithkumar NV, Kirubagaran R, Dharani G. Ectoine biosynthesis genes from the deep sea halophilic eubacteria, Bacillus clausii NIOT-DSB04:its molecular and biochemical characterization. Microbial Pathogenesis, 2019, 136: 103693. DOI:10.1016/j.micpath.2019.103693 |
| [40] | Zhao B, Lu W, Yang L, Zhang B, Wang L, Yang SS. Cloning and characterization of the genes for biosynthesis of the compatible solute ectoine in the moderately halophilic bacterium Halobacillus dabanensis D-8T. Current Microbiology, 2006, 53(3): 183-188. DOI:10.1007/s00284-005-0396-0 |
| [41] | Zhang B, Bao X, Wang L, Yang SS. Cloning and characterization of the gene cluster for biosynthesis of ectoine from Nesterenkonia halobia DSM 20541. The Journal of Microbiology, 2008, 46(3): 309-318. DOI:10.1007/s12275-008-0001-x |
| [42] | Parwata IP, Wahyuningrum D, Suhandono S, Hertadi R. Heterologous ectoine production in Escherichia coli: optimization using response surface methodology. International Journal of Microbiology, 2019, 2019: 5475361. |
| [43] | Zhang W, Wei HL, Gao HW, Huang GH. Cloning and characterization of ectABC cluster from Bacillus alcalophilus DTY1. Chinese Journal of Biotechnology, 2008, 24(3): 395-400. DOI:10.1016/S1872-2075(08)60021-7 |
| [44] | Zhang H, Zhao M, Dong YS, Cao YY, Ma YQ, Luo ZS, Li S, Xu H. Metabolic engineering of Escherichia Coli for ectoine production with the fermentation strategy of supplementing amino donor. Applied & Industrial Microbiology, 2020, 1: 1-17. |
| [45] | Gießelmann G, Dietrich D, Jungmann L, Kohlstedt M, Jeon EJ, Yim SS, Sommer F, Zimmer D, Mühlhaus T, Schroda M, Jeong KJ, Becker J, Wittmann C. Metabolic engineering of Corynebacterium glutamicum for high-level ectoine production: design, combinatorial assembly, and implementation of a transcriptionally balanced heterologous ectoine pathway. Biotechnology Journal, 2019, 14(9): e1800417. DOI:10.1002/biot.201800417 |
| [46] | Pastor JM, Salvador M, Argandoña M, Bernal V, Reina-Bueno M, Csonka LN, Iborra JL, Vargas C, Nieto JJ, Cánovas M. Ectoines in cell stress protection: uses and biotechnological production. Biotechnology Advances, 2010, 28(6): 782-801. DOI:10.1016/j.biotechadv.2010.06.005 |
| [47] | Cantera S, Lebrero R, Rodríguez S, García-Encina PA, Muñoz R. Ectoine bio-milking in methanotrophs: a step further towards methane-based bio-refineries into high added-value products. Chemical Engineering Journal, 2017, 328: 44-48. DOI:10.1016/j.cej.2017.07.027 |
| [48] | Nakayama H, Yoshida K, Ono H, Murooka Y, Shinmyo A. Ectoine, the compatible solute of Halomonas elongata, confers hyperosmotic tolerance in cultured tobacco cells. Plant Physiology, 2000, 122(4): 1239-1247. DOI:10.1104/pp.122.4.1239 |
| [49] | Moghaieb REA, Tanaka N, Saneoka H, Murooka Y, Ono H, Morikawa H, Nakamura A, Nguyen NT, Suwa R, Fujita K. Characterization of salt tolerance in ectoine-transformed tobacco plants (Nicotiana tabaccum): photosynthesis, osmotic adjustment, and nitrogen partitioning. Plant, Cell & Environment, 2006, 29(2): 173-182. |
| [50] | Rai M. Biosynthesis of 1,4,5,6-tetrahydro-2-methyl-4-pyrimidinecarboxylic acid (ectoine) from Marinococcus halophilus into tobacco. Journal of New Seeds, 2001, 2(1): 15-42. DOI:10.1300/J153v02n01_02 |
| [51] | Rai M, Pal MD, Sumesh KV, Jain V, Sankaranarayanan A. Engineering for biosynthesis of ectoine (2-methyl 4-carboxy tetrahydro pyrimidine) in tobacco chloroplasts leads to accumulation of ectoine and enhanced salinity tolerance. Plant Science, 2006, 170(2): 291-306. DOI:10.1016/j.plantsci.2005.08.019 |
| [52] | Moghaieb REA, Nakamura A, Saneoka H, Fujita K. Evaluation of salt tolerance in ectoine-transgenic tomato plants (Lycopersicon esculentum) in terms of photosynthesis, osmotic adjustment, and carbon partitioning. GM Crops, 2011, 2(1): 58-65. DOI:10.4161/gmcr.2.1.15831 |
 2021, Vol. 61
2021, Vol. 61




