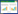扩展功能
文章信息
- 许莹, 董胜男, 张青阳, 何浩洋, 张部昌, 刘静
- XU Ying, DONG Shengnan, ZHANG Qingyang, HE Haoyang, ZHANG Buchang, LIU Jing
- 植物毒素thaxtomins生物合成及其分子调控研究进展
- Biosynthesis and molecular regulation of phytotoxin thaxtomins: a review
- 微生物学通报, 2023, 50(3): 1281-1290
- Microbiology China, 2023, 50(3): 1281-1290
- DOI: 10.13344/j.microbiol.china.220528
-
文章历史
- 收稿日期: 2022-05-28
- 接受日期: 2022-07-27
- 网络首发日期: 2022-09-06
2. 安徽大学物质科学与信息技术研究院, 安徽 合肥 230601
2. Institution of Physical Science and Information Technology, Anhui University, Hefei 230601, Anhui, China
疮痂病链霉菌是引起马铃薯疮痂病的主要致病菌之一,自疮痂病发生以来,其对马铃薯的危害及整个马铃薯产业的影响受到了广泛的关注。随着国内外研究者对疮痂病病原、致病毒素和防治等方面的研究逐步深入,目前已取得了较好的研究进展[1-3]。已报道的马铃薯疮痂病菌有20余种,最常见的是疮痂病链霉菌(Streptomyces scabies)、酸性疮痂病链霉菌(Streptomyces acidiscabies)和肿痂链霉菌(Streptomyces turgidiscabies),其中疮痂病链霉菌是马铃薯疮痂病的主要致病菌,能通过皮孔、伤口或幼嫩的块茎进入组织,产生次级代谢物thaxtomins,thaxtomins通过阻断植物细胞扩张和分裂过程中纤维素的合成而引发疮痂病。疮痂病的症状包括块茎表面结痂状、隆起或凹陷性病变,严重影响马铃薯的产量和质量,导致巨大的经济损失[4-5]。除马铃薯外,疮痂病链霉菌还能侵染萝卜、甜菜、胡萝卜等主要作物,甚至包括中药植物防风草[6]。
鉴于疮痂病链霉菌在农业上的重要影响,其中thaxtomins生物合成过程的分子调控得到越来越多的关注,并取得了较好的进展[3-4, 7-9]。本文综述了thaxtomins的结构特征、生物合成基因簇和生物合成途径,并重点介绍了疮痂病链霉菌中thaxtomins生物合成的分子调控机制等方面的研究进展,有利于深入认知疮痂病链霉菌次级代谢调控网络和马铃薯疮痂病的致病机理,以期为将来开发新型马铃薯疮痂病的防治策略提供理论指导。
1 Thaxtomins的结构性质及应用Thaxtomins是一类由疮痂病链霉菌及少数其他病原链霉菌产生、具有独特2, 5-二酮哌嗪结构的化合物,依据二酮哌嗪主链上特定位置是否存在N-甲基和羟基,可将thaxtomins家族分为thaxtomin A、B、C和D等共计11个结构类似物[4, 10]。Thaxtomin A是thaxtomins家族中代谢量最多、活性最显著且结构最具代表性的产物,于1989年首次分离于感染疮痂病的马铃薯切片中[11],因此,thaxtomin A在农业上可作为筛选剂用于筛选具有抗疮痂病的马铃薯品种。Thaxtomin A的特征结构单元是l-4-硝基吲哚和C-羟基-哌嗪二酮环[12] (图 1),其中l-4-硝基吲哚为thaxtomins家族在所有微生物次级代谢产物中所独有的化学结构单元,C-羟基-哌嗪二酮环则存在很多具有生物活性的天然产物[4]。由于thaxtomins家族化合物具有非常显著的抑制植物纤维合成酶的活性[13-15],因此可作为植物毒素在纳摩尔水平抑制常见双子叶和单子叶杂草的生长,是一类具有良好活性和巨大潜力的新型天然除草剂[16-18]。不同结构的thaxtomin能够抑制植物纤维合成酶的活性则不同,因而呈现出不同的植物毒性,其中thaxtomin A的活性最强[13-15]。然而thaxtomin A具体的分子模型和特异靶点目前并不清楚[13-15]。
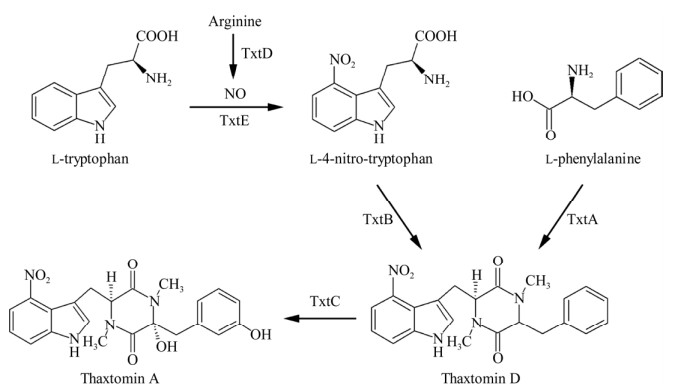
引起马铃薯疮痂病的疮痂病链霉菌、酸性疮痂病链霉菌和肿痂链霉菌等都能够通过次级代谢产生thaxtomins家族化合物,原因在于负责thaxtomin A及其结构类似物的生物合成基因簇(thaxtomin biosynthetic gene cluster, txt cluster)具有高度保守性[19]。整个txt cluster是由txtC、txtH、txtB、txtA、txtR、txtE和txtD共计7个基因依次排列组成的,长度大约为18.3 kb,共计包括3个转录单元:txtA-txtB-txtH-txtC、txtE-txtD和txtR[19] (图 2)。其中,txtR编码AraC/XylS家族转录调控蛋白[20],剩余6个基因分别编码thaxtomin生物合成各步骤所需的催化酶:txtA和txtB分别编码非核糖体肽合成酶(non-ribosomal peptide synthetase, NRPS)[21];txtC编码细胞色素P450单加氧酶(cytochrome P450, CYP450)[22];txtD编码一氧化氮合酶[23];txtE编码一种独特的CYP450[24-25];txtH编码MbtH样蛋白家族的小分子蛋白[26]。
|
| 图 2 疮痂病链霉菌thaxtomins生物合成基因簇及共转录单元示意图 Figure 2 Genetic organization and co-transcriptional unit of gene cluster for the biosynthesis of thaxtomins in Streptomyces scabies. |
|
|
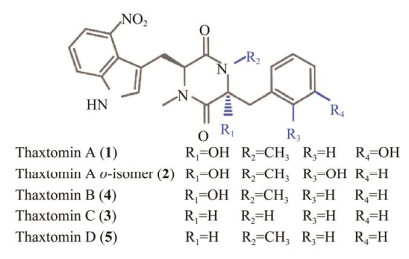
Thaxtomins生物合成途径并不烦琐复杂,首先从两个简单的氨基酸:l-色氨酸(l-tryptophan, Trp)和l-精氨酸(L-arginine, Arg)开始,Arg经一氧化氮合酶TxtD氧化生成瓜氨酸和一氧化氮(nitric oxide, NO)[23];然后,Trp经独特的CYP450 TxtE和NO催化生成l-4-硝基色氨酸,TxtE是目前已知的唯一能够直接硝化碳氢键的酶[24-25];接着,l-4-硝基色氨酸和另一氨基酸l-苯丙氨酸(l-phenylalanine, Phe)分别经非核糖体肽合成酶TxtB和TxtA催化,并连接形成具有2, 5-二酮哌嗪结构的thaxtomin D[21, 27];最后,thaxtomin D经CYP450 TxtC催化后引入两个羟基至分子骨架上而生成最终的thaxtomin A[22, 26, 28-29] (图 3)。目前关于MbtH样家族蛋白TxtH对thaxtomins生物合成的作用机制并不十分清晰,已有研究表明,疮痂病链霉菌中TxtH与thaxtomin A生物合成产量之间存在显著的正相关性,作用机制可能是TxtH发挥分子伴侣作用而促进TxtA和TxtB两个蛋白的正确折叠[26, 30]。
Thaxtomin A作为天然除草剂在农业上具有巨大的开发潜力,然而thaxtomin A在本源链霉菌宿主中的产量却非常低,限制了其广泛推广应用[10]。对目标化合物进行异源表达,改变遗传背景能够有效地提高目标化合物的产量。例如,Jiang等[31]将疮痂病链霉菌87.22中txt cluster克隆至白色链霉菌(Streptomyces albus) J1074中进行异源表达,成功将thaxtomin A在摇瓶水平上的产量从9 mg/L提高至91 mg/L;近期,Li等再次通过异源表达策略将txt cluster克隆至5个常用的链霉菌异源表达宿主,其中微白黄链霉菌(Streptomyces albidoflavus) J1074中thaxtomin A的产量最高,继而选择微白黄链霉菌作为异源宿主对thaxtomin A进行代谢工程系列改造,最终将thaxtomin A在摇瓶水平上的产量提高至728 mg/L[12]。S. albus和S. albidoflavus在常用链霉菌中具有较小规模的基因组,因此,利用其进行thaxtomin A的合成时能够减少其他竞争途径而使thaxtomin A的合成量提高。
3 Thaxtomins生物合成的调控Thaxtomins生物合成过程中受关键转录调控因子的调控,包括途径特异性转录调控和全局多效调控,而这些转录调控因子在疮痂病链霉菌的初级代谢和次级代谢中起着至关重要的作用。
3.1 途径特异性转录调控TxtR是thaxtomins生物合成基因簇内唯一的簇内专一性调控因子(cluster-situated regulator, CSR),能够激活thaxtomins生物合成基因簇的表达[20]。疮痂病链霉菌87.22缺失txtR基因后,菌株不再产生thaxtomins化合物,对烟草幼苗的毒力作用几乎消失,而且txtA、txtB和txtC等生物合成基因的表达明显下降,说明TxtR对thaxtomins生物合成具有显著的转录激活作用[20] (图 4)。TxtR能够结合其配体分子纤维二糖(cellobiose),纤维二糖是纤维素的基本结构单元,由两个葡萄糖通过β-1, 4-糖苷键连接而形成[20]。当纤维二糖存在时,txtR和thaxtomins生物合成基因的转录水平均得到大幅提升,因而纤维二糖可作为thaxtomins生物合成的诱导剂[20]。TxtR是AraC/XylS家族蛋白中第一个受到纤维二糖调节的成员[20, 32]。

|
| 图 4 疮痂病链霉菌中TxtR和CebR调控thaxtomins生物合成的模式 Figure 4 Regulatory mode of TxtR and CebR on thaxtomins biosynthesis in Streptomyces scabies. |
|
|
bld基因家族在链霉菌次级代谢和形态分化的过程中具有非常重要的调控意义,包括bldA (tRNA)、bldK (ABC转运系统)、bldN (σ factor)、bldG (anti-anti-σ factor)及bldB/bldC/bldD/bldH/ bldM (转录调控因子)[33-34]。Bignell等在疮痂病链霉菌中通过对9个bld家族同源基因进行缺失研究(表 1),发现突变株ΔbldA和ΔbldC中thaxtomin A的生物合成均被完全阻断,检测不到thaxtomin A的产量;突变株ΔbldD、ΔbldG和ΔbldH中thaxtomin A的产量也显著低于原始菌株;而突变株ΔbldB、ΔbldKA和ΔbldN中thaxtomin A的产量却有不同程度的提高[35]。通过毒力实验分析这些bld基因缺失突变株对萝卜幼苗和马铃薯块茎的致病情况发现,ΔbldB、ΔbldKA和ΔbldN的致病表型相似于出发菌株;而ΔbldA、ΔbldC、ΔbldD、ΔbldG和ΔbldH的致病表型如组织坏死或根茎发育迟缓等情况,相较于出发菌株均有明显减少,这与各突变株中thaxtomin A的含量相一致(表 1)[35]。再通过实时荧光定量聚合酶链式反应(real-time fluorescence quantitative polymerase chain reaction, RT-qPCR)分析ΔbldA、ΔbldC、ΔbldD、ΔbldG和ΔbldH这5个突变株中thaxtomin生物合成相关基因的转录情况发现,各突变株中txtA和txtD的表达量均显著下降;而txtR的表达量在ΔbldH中无明显变化,在其他突变株中均显著下降[35]。
| Gene | Protein | Predicted function | Phenotype of gene mutant in S. scabies |
| scab54521 | BldA | Leucyl tRNA | Thaxtomin A production abolished |
| scab25271 | BldB | Small DNA binding protein | Thaxtomin A production increased |
| scab47901 | BldC | MerR-type DNA binding protein | Thaxtomin A production abolished |
| scab75171 | BldD | DNA binding transcriptional regulator | Thaxtomin A production decreased |
| scab40861 | BldG | Anti-anti-sigma factor | Thaxtomin A production decreased |
| scab57831 | BldH | AraC/XylS family transcriptional regulator | Thaxtomin A production decreased |
| scab31541 | BldKA | ABC transport system integral membrane protein | Thaxtomin A production slightly increased |
| scab36231 | BldM | Orphan response regulator | This mutant was not studied |
| scab39121 | BldN | Extracytoplasmic function sigma factor | Thaxtomin A production increased |
BldB[36]、BldC[37]、BldD[38-40]和BldH[41]作为典型的转录调控因子,已知能够调控thaxtomin生物合成基因和CSR基因的表达,然而更加深入的调控机制并不清楚。例如,BldB、BldC、BldD和BldH对thaxtomin生物合成基因和CSR基因的调控作用是直接还是间接,具体的结合位点是否与其他链霉菌中已鉴定的保守结合位点一致,BldD在疮痂病链霉菌中行使功能是否同样依赖于第二信使c-di-GMP的介导作用等。
3.2.2 LacI/GalR家族转录调控因子CebRCebR是链霉菌中调控纤维素转化利用的转录抑制因子,属于LacI/GalR家族蛋白[42-43]。2015年,Francis等在疮痂病链霉菌中利用PREDetector软件预测得到txtA-txtR基因间隔区中存在一段与CebR保守结合位点高度相似的回文序列(命名为cbstxtR-A:5ʹ-CGGGAGCGCTCCCA-3ʹ),EMSA分析发现CebR蛋白的确能够与cbstxtR-A结合,并特异性应答其配体分子纤维二糖和纤维三糖(cellotriose)[44-45];疮痂病链霉菌中cebR (scab57761)基因缺失后,txt cluster中生物合成基因txtA和txtB及CSR基因txtR的转录水平显著提高,thaxtomin A的产量大幅度提升10倍以上,而且ΔcebR突变株对拟南芥和马铃薯的致病活性也明显增强,说明疮痂病链霉菌CebR对thaxtomin A的生物合成具有直接的转录抑制作用(图 4)[44]。2016年,Jourdan等发现疮痂病链霉菌CebR调控thaxtomins生物合成的另一路径:cebEFG是与cebR位置相邻且转录方向相反的操纵子,编码负责纤维二糖和纤维三糖吸收利用的相关转运蛋白(CebEFG-MsiK ABC转运蛋白系统),其中CebE已被证实能够直接结合纤维二糖和纤维三糖[46-48];疮痂病链霉菌中cebE (scab57751)基因缺失后,thaxtomin A的产量大幅度下降,ΔcebE突变株对萝卜苗的致病活性也明显减弱;RT-qPCR分析发现ΔcebR突变株中cebE和cebF的表达水平显著提高,进一步EMSA分析发现CebR蛋白能够直接结合cebEFG启动子DNA并特异性应答纤维二糖分子,而且cebEFG启动子序列具有14 bp的CebR保守结合位点,说明CebR能够通过直接调控cebEFG转运蛋白基因的表达介导纤维二糖和纤维三糖的吸收过程,而最终影响thaxtomin A的生物合成(图 4)[46]。2018年,Jourdan等[49]再进一步发现CebR能够直接调控其上游非邻近的bglC (scab57721)基因的功能,bglC基因编码一种β-葡萄糖苷酶(又称纤维二糖酶),属糖基水解酶GH1家族蛋白,能够水解结合于末端非还原性的β-葡萄糖苷键,同时释放出β-葡萄糖和相应的配基;CebR通过直接作用于bglC启动子区保守结合位点cbsbglC而调控bglC的转录,而且CebR对bglC的转录抑制作用同样应答其配体分子纤维二糖;遗传学分析证实bglC基因的缺失能够引起疮痂病链霉菌在不同碳源情况下thaxtomin A产量和致病活性的相应变化,说明CebR还能够通过直接调控BglC介导的葡萄糖水解途径而影响thaxtomin A的生物合成(图 4)。
4 总结与展望马铃薯疮痂病是制约全世界马铃薯产业积极向好发展的重要因素,其致病菌次级代谢产生的毒素thaxtomins危害马铃薯块茎表面,对马铃薯和其他重要作物的生产种植都具有严重影响[4-6]。本文聚焦于thaxtomins生物合成过程和分子调控机制的研究,从途径特异性调控和全局多效调控的角度阐述thaxtomins生物合成的关键调控因子。一方面有利于在理论上全面深刻地认知毒素thaxtomins生物合成的分子调控网络,为未来通过代谢调控的策略防治疮痂病的发生提供新的方向;另一方面,有利于挖掘开发相关转录调控元件结合高效的代谢工程手段进行thaxtomin A高产设计,为thaxtomin A作为新型天然除草剂在农业上的广泛应用提供理论支持。例如,Li等通过代谢工程手段将thaxtomin A生物合成基因簇txt cluster克隆至微白黄链霉菌J1074中异源表达,结合转录抑制因子CebR失活和启动子优化等联合策略的改造,最终将thaxtomin A的产量在小型发酵罐水平上提高至1 973 mg/L,这是截至目前已报道的thaxtomin A合成产量的最高水平[12]。
目前,疮痂病链霉菌及其代谢产物thaxtomins生物合成的分子调控研究虽然取得了一定的进展,但是相对于其他研究较为成熟的模式链霉菌如天蓝色链霉菌(Streptomyces coelicolor)[33, 50]和委内瑞拉链霉菌(Streptomyces venelazue)[51-52]等,其次级代谢的分子调控机制研究还不够深入,很多问题尚不清晰。例如,TxtR作为thaxtomins生物合成基因簇唯一的途径特异性调控因子[20],其精确的作用靶点和转录激活的机制有待深入解析,TxtR能够应答配体分子纤维二糖,具体应答机制如何,以及是否与CebR应答纤维二糖的模式相同等;又如已知BldB、BldC、BldD和BldH能够调控thaxtomin生物合成基因和CSR基因的表达[35],然而BldB、BldC、BldD和BldH对thaxtomin生物合成基因和CSR基因的调控作用是直接还是间接,具体的结合位点是否与其他链霉菌中已鉴定的保守结合位点一致,BldD在疮痂病链霉菌中行使功能是否同样依赖于第二信使c-di-GMP的介导作用等。这些问题都有待研究者们深入地探讨解析。
| [1] |
LERAT S, SIMAO-BEAUNOIR AM, BEAULIEU C. Genetic and physiological determinants of Streptomyces scabies pathogenicity[J]. Molecular Plant Pathology, 2009, 10(5): 579-585. DOI:10.1111/j.1364-3703.2009.00561.x |
| [2] |
WELLER DM, RAAIJMAKERS JM, GARDENER BB, THOMASHOW LS. Microbial populations responsible for specific soil suppressiveness to plant pathogens[J]. Annual Review of Phytopathology, 2002, 40: 309-348. DOI:10.1146/annurev.phyto.40.030402.110010 |
| [3] |
LORIA R, KERS J, JOSHI M. Evolution of plant pathogenicity in Streptomyces[J]. Annual Review of Phytopathology, 2006, 44: 469-487. DOI:10.1146/annurev.phyto.44.032905.091147 |
| [4] |
LORIA R, BIGNELL DRD, MOLL S, HUGUET-TAPIA JC, JOSHI MV, JOHNSON EG, SEIPKE RF, GIBSON DM. Thaxtomin biosynthesis: the path to plant pathogenicity in the genus Streptomyces[J]. Antonie Van Leeuwenhoek, 2008, 94(1): 3-10. DOI:10.1007/s10482-008-9240-4 |
| [5] |
HILTUNEN LH, KELLONIEMI J, VALKONEN JPT. Repeated applications of a nonpathogenic Streptomyces strain enhance development of suppressiveness to potato common scab[J]. Plant Disease, 2017, 101(1): 224-232. DOI:10.1094/PDIS-07-16-1020-RE |
| [6] |
SANTOS-CERVANTES ME, FELIX-GASTELUM R, HERRERA-RODRÍGUEZ G, ESPINOZA-MANCILLAS MG, MORA-ROMERO AG, LEYVA-LÓPEZ NE. Characterization, pathogenicity and chemical control of Streptomyces acidiscabies associated to potato common scab[J]. American Journal of Potato Research, 2017, 94(1): 14-25. DOI:10.1007/s12230-016-9541-5 |
| [7] |
KING RR, CALHOUN LA. The thaxtomin phytotoxins: sources, synthesis, biosynthesis, biotransformation and biological activity[J]. Phytochemistry, 2009, 70(7): 833-841. DOI:10.1016/j.phytochem.2009.04.013 |
| [8] |
BIGNELL DRD, FYANS JK, CHENG Z. Phytotoxins produced by plant pathogenic Streptomyces species[J]. Journal of Applied Microbiology, 2014, 116(2): 223-235. DOI:10.1111/jam.12369 |
| [9] |
LI YT, LIU JY, DÍAZ-CRUZ G, CHENG ZL, BIGNELL DRD. Virulence mechanisms of plant-pathogenic Streptomyces species: an updated review[J]. Microbiology: Reading, England, 2019, 165(10): 1025-1040. DOI:10.1099/mic.0.000818 |
| [10] |
WANG LQ, WANG MY, FU YD, HUANG PJ, KONG DK, NIU GQ. Engineered biosynthesis of thaxtomin phytotoxins[J]. Critical Reviews in Biotechnology, 2020, 40(8): 1163-1171. DOI:10.1080/07388551.2020.1807461 |
| [11] |
KING RR, LAWRENCE CH, CLARK MC, CALHOUN LA. Isolation and characterization of phytotoxins associated with Streptomyces scabies[J]. Journal of the Chemical Society, Chemical Communications, 1989(13): 849. DOI:10.1039/c39890000849 |
| [12] |
LI ZL, HUANG PJ, WANG MY, WANG X, WANG LQ, KONG DK, NIU GQ. Stepwise increase of thaxtomins production in Streptomyces albidoflavus J1074 through combinatorial metabolic engineering[J]. Metabolic Engineering, 2021, 68: 187-198. DOI:10.1016/j.ymben.2021.10.008 |
| [13] |
BISCHOFF V, COOKSON SJ, WU S, SCHEIBLE WR. Thaxtomin A affects CESA-complex density, expression of cell wall genes, cell wall composition, and causes ectopic lignification in Arabidopsis thaliana seedlings[J]. Journal of Experimental Botany, 2009, 60(3): 955-965. DOI:10.1093/jxb/ern344 |
| [14] |
KING RR, LAWRENCE CH, GRAY JA. Herbicidal properties of the thaxtomin group of phytotoxins[J]. Journal of Agricultural and Food Chemistry, 2001, 49(5): 2298-2301. DOI:10.1021/jf0012998 |
| [15] |
SCHEIBLE WR, FRY B, KOCHEVENKO A, SCHINDELASCH D, ZIMMERLI L, SOMERVILLE S, LORIA R, SOMERVILLE CR. An Arabidopsis mutant resistant to thaxtomin A, a cellulose synthesis inhibitor from Streptomyces species[J]. The Plant Cell, 2003, 15(8): 1781-1794. DOI:10.1105/tpc.013342 |
| [16] |
STRANGE RN. Phytotoxins produced by microbial plant pathogens[J]. Natural Product Reports, 2007, 24(1): 127-144. DOI:10.1039/B513232K |
| [17] |
ZHANG HB, WANG QP, NING X, HANG H, MA J, YANG XD, LU XL, ZHANG JB, LI YH, NIU CW, SONG HR, WANG X, WANG PG. Synthesis and biological evaluations of a series of thaxtomin analogues[J]. Journal of Agricultural and Food Chemistry, 2015, 63(14): 3734-3741. DOI:10.1021/jf506153t |
| [18] |
WOLFE JC, NEAL JC, HARLOW CD. Selective broadleaf weed control in turfgrass with the bioherbicides Phoma macrostoma and thaxtomin A[J]. Weed Technology, 2016, 30(3): 688-700. DOI:10.1614/WT-D-15-00159.1 |
| [19] |
HUGUET-TAPIA JC, LEFEBURE T, BADGER JH, GUAN DL, PETTIS GS, STANHOPE MJ, LORIA R. Genome content and phylogenomics reveal both ancestral and lateral evolutionary pathways in plant-pathogenic Streptomyces species[J]. Applied and Environmental Microbiology, 2016, 82(7): 2146-2155. DOI:10.1128/AEM.03504-15 |
| [20] |
JOSHI MV, BIGNELL DRD, JOHNSON EG, SPARKS JP, GIBSON DM, LORIA R. The AraC/XylS regulator TxtR modulates thaxtomin biosynthesis and virulence in Streptomyces scabies[J]. Molecular Microbiology, 2007, 66(3): 633-642. DOI:10.1111/j.1365-2958.2007.05942.x |
| [21] |
HEALY FG, WACH M, KRASNOFF SB, GIBSON DM, LORIA R. The txtAB genes of the plant pathogen Streptomyces acidiscabies encode a peptide synthetase required for phytotoxin thaxtomin A production and pathogenicity[J]. Molecular Microbiology, 2000, 38(4): 794-804. DOI:10.1046/j.1365-2958.2000.02170.x |
| [22] |
ALKHALAF LM, BARRY SM, REA DA, GALLO A, GRIFFITHS D, LEWANDOWSKI JR, FULOP V, CHALLIS GL. Binding of distinct substrate conformations enables hydroxylation of remote sites in thaxtomin D by cytochrome P450 TxtC[J]. Journal of the American Chemical Society, 2019, 141(1): 216-222. DOI:10.1021/jacs.8b08864 |
| [23] |
KERS JA, WACH MJ, KRASNOFF SB, WIDOM J, CAMERON KD, BUKHALID RA, GIBSON DM, CRANE BR, LORIA R. Nitration of a peptide phytotoxin by bacterial nitric oxide synthase[J]. Nature, 2004, 429(6987): 79-82. DOI:10.1038/nature02504 |
| [24] |
BARRY SM, KERS JA, JOHNSON EG, SONG LJ, ASTON PR, PATEL B, KRASNOFF SB, CRANE BR, GIBSON DM, LORIA R, CHALLIS G. Cytochrome P450–catalyzed L-tryptophan nitration in thaxtomin phytotoxin biosynthesis[J]. Nature Chemical Biology, 2012, 8(10): 814-816. DOI:10.1038/nchembio.1048 |
| [25] |
TOMITA H, KATSUYAMA Y, MINAMI H, OHNISHI Y. Identification and characterization of a bacterial cytochrome P450 monooxygenase catalyzing the 3-nitration of tyrosine in rufomycin biosynthesis[J]. The Journal of Biological Chemistry, 2017, 292(38): 15859-15869. DOI:10.1074/jbc.M117.791269 |
| [26] |
LI YT, LIU JY, ADEKUNLE D, BOWN L, TAHLAN K, BIGNELL DRD. TxtH is a key component of the thaxtomin biosynthetic machinery in the potato common scab pathogen Streptomyces scabies[J]. Molecular Plant Pathology, 2019, 20(10): 1379-1393. DOI:10.1111/mpp.12843 |
| [27] |
JOHNSON EG, KRASNOFF SB, BIGNELL DRD, CHUNG WC, TAO T, PARRY RJ, LORIA R, GIBSON DM. 4-nitrotryptophan is a substrate for the non-ribosomal peptide synthetase TxtB in the thaxtomin A biosynthetic pathway[J]. Molecular Microbiology, 2009, 73(3): 409-418. DOI:10.1111/j.1365-2958.2009.06780.x |
| [28] |
JIANG GD, ZHANG Y, POWELL MM, HYLTON SM, HILLER NW, LORIA R, DING YS. A promiscuous cytochrome P450 hydroxylates aliphatic and aromatic C-H bonds of aromatic 2, 5-diketopiperazines[J]. Chembiochem: a European Journal of Chemical Biology, 2019, 20(8): 1068-1077. DOI:10.1002/cbic.201800736 |
| [29] |
JIANG GD, ZUO R, ZHANG Y, POWELL MM, ZHANG PL, HYLTON SM, LORIA R, DING YS. One-pot biocombinatorial synthesis of herbicidal thaxtomins[J]. ACS Catalysis, 2018, 8(11): 10761-10768. DOI:10.1021/acscatal.8b03317 |
| [30] |
LI YT, TAHLAN K, BIGNELL DRD. Functional cross-talk of MbtH-like proteins during thaxtomin biosynthesis in the potato common scab pathogen Streptomyces scabiei[J]. Frontiers in Microbiology, 2020, 11: 585456. DOI:10.3389/fmicb.2020.585456 |
| [31] |
JIANG GD, ZHANG YC, POWELL MM, ZHANG PL, ZUO R, ZHANG Y, KALLIFIDAS D, TIEU AM, LUESCH H, LORIA R, DING YS. High-yield production of herbicidal thaxtomins and thaxtomin analogs in a nonpathogenic Streptomyces strain[J]. Applied and Environmental Microbiology, 2018, 84(11): e00164-e00118. |
| [32] |
YANG J, TAUSCHEK M, ROBINS-BROWNE RM. Control of bacterial virulence by AraC-like regulators that respond to chemical signals[J]. Trends in Microbiology, 2011, 19(3): 128-135. DOI:10.1016/j.tim.2010.12.001 |
| [33] |
MCCORMICK JR, FLÄRDH K. Signals and regulators that govern Streptomyces development[J]. FEMS Microbiology Reviews, 2012, 36(1): 206-231. DOI:10.1111/j.1574-6976.2011.00317.x |
| [34] |
CHATER KF, CHANDRA G. The evolution of development in Streptomyces analysed by genome comparisons[J]. FEMS Microbiology Reviews, 2006, 30(5): 651-672. DOI:10.1111/j.1574-6976.2006.00033.x |
| [35] |
BIGNELL DRD, FRANCIS IM, FYANS JK, LORIA R. Thaxtomin A production and virulence are controlled by several bld gene global regulators in Streptomyces scabies[J]. Molecular Plant-Microbe Interactions: MPMI, 2014, 27(8): 875-885. DOI:10.1094/MPMI-02-14-0037-R |
| [36] |
ECCLESTON M, ALI RA, SEYLER R, WESTPHELING J, NODWELL J. Structural and genetic analysis of the BldB protein of Streptomyces coelicolor[J]. Journal of Bacteriology, 2002, 184(15): 4270-4276. DOI:10.1128/JB.184.15.4270-4276.2002 |
| [37] |
HUNT AC, SERVÍN-GONZÁLEZ L, KELEMEN GH, BUTTNER MJ. The bldC developmental locus of Streptomyces coelicolor encodes a member of a family of small DNA-binding proteins related to the DNA-binding domains of the MerR family[J]. Journal of Bacteriology, 2005, 187(2): 716-728. DOI:10.1128/JB.187.2.716-728.2005 |
| [38] |
den HENGST CD, TRAN NT, BIBB MJ, CHANDRA G, LESKIW BK, BUTTNER MJ. Genes essential for morphological development and antibiotic production in Streptomyces coelicolor are targets of BldD during vegetative growth[J]. Molecular Microbiology, 2010, 78(2): 361-379. DOI:10.1111/j.1365-2958.2010.07338.x |
| [39] |
SCHUMACHER MA, ZENG WJ, FINDLAY KC, BUTTNER MJ, BRENNAN RG, TSCHOWRI N. The Streptomyces master regulator BldD binds c-di-GMP sequentially to create a functional BldD2-(c-di-GMP)4 complex[J]. Nucleic Acids Research, 2017, 45(11): 6923-6933. DOI:10.1093/nar/gkx287 |
| [40] |
TSCHOWRI N, SCHUMACHER MA, SCHLIMPERT S, CHINNAM NB, FINDLAY KC, BRENNAN RG, BUTTNER MJ. Tetrameric c-di-GMP mediates effective transcription factor dimerization to control Streptomyces development[J]. Cell, 2014, 158(5): 1136-1147. DOI:10.1016/j.cell.2014.07.022 |
| [41] |
TAKANO E, TAO M, LONG F, BIBB MJ, WANG L, LI W, BUTTNER MJ, BIBB MJ, DENG ZX, CHATER KF. A rare leucine codon in adpA is implicated in the morphological defect of bldA mutants of Streptomyces coelicolor[J]. Molecular Microbiology, 2003, 50(2): 475-486. DOI:10.1046/j.1365-2958.2003.03728.x |
| [42] |
SCHLÖSSER A, ALDEKAMP T, SCHREMPF H. Binding characteristics of CebR, the regulator of the ceb operon required for cellobiose/cellotriose uptake in Streptomyces reticuli[J]. FEMS Microbiology Letters, 2000, 190(1): 127-132. DOI:10.1111/j.1574-6968.2000.tb09274.x |
| [43] |
MARUSHIMA K, OHNISHI Y, HORINOUCHI S. CebR as a master regulator for cellulose/cellooligosaccharide catabolism affects morphological development in Streptomyces griseus[J]. Journal of Bacteriology, 2009, 191(19): 5930-5940. DOI:10.1128/JB.00703-09 |
| [44] |
FRANCIS IM, JOURDAN S, FANARA S, LORIA R, RIGALI S. The cellobiose sensor CebR is the gatekeeper of Streptomyces scabies pathogenicity[J]. mBio, 2015, 6(2): e02018. |
| [45] |
WANG X, FU YD, WANG MY, NIU GQ. Synthetic cellobiose-inducible regulatory systems allow tight and dynamic controls of gene expression in Streptomyces[J]. ACS Synthetic Biology, 2021, 10(8): 1956-1965. DOI:10.1021/acssynbio.1c00152 |
| [46] |
JOURDAN S, FRANCIS IM, KIM MJ, SALAZAR JJC, PLANCKAERT S, FRÈRE JM, MATAGNE A, KERFF F, DEVREESE B, LORIA R, RIGALI S. The CebE/MsiK transporter is a doorway to the cello-oligosaccharide-mediated induction of Streptomyces scabies pathogenicity[J]. Scientific Reports, 2016, 6: 27144. DOI:10.1038/srep27144 |
| [47] |
SCHLÖSSER A, JANTOS J, HACKMANN K, SCHREMPF H. Characterization of the binding protein-dependent cellobiose and cellotriose transport system of the cellulose degrader Streptomyces reticuli[J]. Applied and Environmental Microbiology, 1999, 65(6): 2636-2643. DOI:10.1128/AEM.65.6.2636-2643.1999 |
| [48] |
KOS V, FORD RC. The ATP-binding cassette family: a structural perspective[J]. Cellular and Molecular Life Sciences, 2009, 66(19): 3111-3126. DOI:10.1007/s00018-009-0064-9 |
| [49] |
JOURDAN S, FRANCIS IM, DEFLANDRE B, TENCONI E, RILEY J, PLANCKAERT S, TOCQUIN P, MARTINET L, DEVREESE B, LORIA R, RIGALI S. Contribution of the β-glucosidase BglC to the onset of the pathogenic lifestyle of Streptomyces scabies[J]. Molecular Plant Pathology, 2018, 19(6): 1480-1490. DOI:10.1111/mpp.12631 |
| [50] |
van KEULEN G, DYSON PJ. Production of specialized metabolites by Streptomyces coelicolor A3(2)[J]. Advances in Applied Microbiology, 2014, 89: 217-266. |
| [51] |
PROCÓPIO RE, SILVA IR, MARTINS MK, AZEVEDO JL, ARAÚJO JM. Antibiotics produced by Streptomyces[J]. The Brazilian Journal of Infectious Diseases: an Official Publication of the Brazilian Society of Infectious Diseases, 2012, 16(5): 466-471. DOI:10.1016/j.bjid.2012.08.014 |
| [52] |
LEE N, HWANG S, LEE Y, CHO S, PALSSON B, CHO BK. Synthetic biology tools for novel secondary metabolite discovery in Streptomyces[J]. Journal of Microbiology and Biotechnology, 2019, 29(5): 667-686. DOI:10.4014/jmb.1904.04015 |
 2023, Vol. 50
2023, Vol. 50