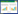扩展功能
文章信息
- 孙剑秋, 张楷, 宋福行, 王龙
- SUN Jianqiu, ZHANG Kai, SONG Fuhang, WANG Long
- 篮状菌属二个中国新记录种
- Two species of Talaromyces new to China
- 微生物学通报, 2022, 49(10): 4080-4089
- Microbiology China, 2022, 49(10): 4080-4089
- DOI: 10.13344/j.microbiol.china.220120
-
文章历史
- 收稿日期: 2022-01-30
- 接受日期: 2022-04-13
- 网络首发日期: 2022-05-19
2. 北京工商大学轻工科学技术学院, 北京 100048;
3. 中国科学院微生物研究所真菌学国家重点实验室, 北京 100101
2. School of Light Industry, Beijing Technology and Business University, Beijing 100048, China;
3. State Key Laboratory of Mycology, Institute of Microbiology, Chinese Academy of Sciences, Beijing 100101, China
相较于陆地环境,滩涂环境可培养真菌物种多样性的研究较少,而滩涂环境可能栖息着大量陆地环境罕见的物种或新物种,栖息于该环境的某些物种可能产生新颖次级代谢产物[1]。我国海岸线长约32 000 km,沿海滩涂面积约2.17万km2[2],为了了解我国沿海滩涂的可培养真菌物种多样性、储备物种资源,科技部启动了科技基础资源调查专项计划的“滩涂丝状真菌资源及多样性调查”项目。在该项目执行过程中,我们从我国福建省漳州市和江苏省盐城市沿海滩涂分离得到了多种青霉(Penicillium Link)和篮状菌(Talaromyces C. R. Benjam.)。篮状菌属最初只包括产生裸囊壳(gymnothecium)和青霉状帚状枝无性繁殖结构的物种,因此,该属作为青霉的一个有性属被放在青霉的分类框架中来研究[3-5]。然而形态学、生态学及分子生物学特征显示,篮状菌及其相关的青霉双轮对称组(sect. Biverticillata-Symmetrica Thom)[4]或双轮亚属(Subgen. Biverticillium Diercks)[5]的物种与青霉其他物种有相当大的差别[6-11]。因此,2012年墨尔本法规认可了Talaromyces作为上述篮状菌及其相关青霉物种的合法属名[12]。篮状菌属目前已发现约175种,可分为8个组[13-14]。其中篮状菌组(sect. Talaromyces Stolk and Samson)是该属物种最多的组,全球已报道约81种[13-17],我国已报道该组44种[14, 17-29];糙刺孢篮状菌组(sect. Trachyspermi Yaguchi & Udagawa)全球已发现29种[13-16, 30-34],我国已发现10种[14, 19, 22-23, 26, 35];螺旋篮状菌组(sect. Helici Yilmaz, Frisvad & Samson)全球已报道13种[13, 22, 36-37],我国只报道了3种[19, 22]。
篮状菌物种若存在有性阶段则通常产生壁具小刺的椭球形子囊孢子,如黄色篮状菌[T. flavus (Klöcker) Stolk & Samson]、藤本篮状菌[T. liani (Kamyschko) N. Yilmaz et al.]和橙黄篮状菌(T. aureolinus L. Wang),但也有些物种产生壁具脊状纹饰的椭球形子囊孢子,如柄篮状菌[T. stipitatus (Thom) C.R. Benj.]和鲜绿篮状菌[T. viridis (Stolk & G.F. Orr) Arx][4-5, 15, 17]。本文报道了从我国福建省漳州市和江苏省盐城市滩涂土壤分离到的3种产子囊孢子的篮状菌,即篮状菌组的阿根廷篮状菌(T. argentinensis)、螺旋篮状菌组的巴塞篮状菌(T. barcinensis)和糙刺孢篮状菌组的乌克兰篮状菌(T. ucrainicus),其中,阿根廷篮状菌和巴塞篮状菌为我国新记录种。
1 材料与方法 1.1 样品来源和分离土壤样品采自我国福建省漳州市九龙口(23°55′39.36″N, 117°25′33.6″E)和江苏省盐城市(33°37′28″N, 120°29′1″E)滩涂土壤。菌株分离采用倍比稀释涂布平皿法[38-39],分离到多种青霉和篮状菌,其中有3株产子囊孢子的篮状菌经鉴定后保存于中国普通微生物菌种保藏中心(CGMCC,ZZ2-7-1h=CGMCC 3.16171,ZZ2-1-1= CGMCC 3.16172,JS11-5=CGMCC 3.16173)。
1.2 形态学研究方法总体生理学性状的观测使用查氏酵母精琼脂(Czapek yeast autolysate agar,CYA)和5%麦芽精琼脂(malt extract agar,MEA)培养基。培养基配方、培养条件、分生孢子、菌丝体、渗出液和可溶性色素的描述分别参考Raper等[4]、Pitt[5]、Ridgway[40]和Samson等[41]。显微性状的研究挑取在MEA上25 ℃培养7 d菌落的产孢结构,浸入在85%乳酸水溶液中做普通光学显微镜载片观测[41]。
1.3 分子系统学研究方法基因组DNA的提取、β-微管蛋白基因(β-tubulin gene,BenA)和rDNA ITS1-5.8S-ITS2的PCR引物及扩增方法分别参考Wang等[42]、Glass等[43]、White等[44],PCR产物由诺赛生物技术有限公司进行双向直通测序。原始序列用软件BioEdit 7.0.9[45]校对编辑并提交到GenBank (ZZ2-7-1h=CGMCC 3.16171:BenA=OK104788,ITS=OK087304,Rpb2=OK104792;ZZ2-1-1= CGMCC 3.16172:BenA=OK104789,ITS= OK087305;JS11-5=CGMCC 3.16173:BenA= OK104791,ITS=OK087307,Rpb2=OK104794)。
由于GenBank无T. barcinensis模式菌株CBS 649.95的钙调蛋白基因(calmodulin gene,CaM)和RNA多聚酶第二大亚基基因(DNA- dependent RNA polymerase Ⅱ second largest subunit gene,Rpb2)序列,本文作者经多次尝试也未扩增出我国菌株CGMCC 3.16171的这2个基因序列。因此,分子系统学分析只能依据BenA和ITS序列,将其连接成BenA-ITS组合序列进行统一分析。选择篮状菌组18个种、螺旋篮状菌组5个种和糙刺孢篮状菌19个种的模式菌株和本研究中的3个菌株,以岛蓝状菌组[sect. Islandici (Pitt) Yilmaz, Frisvad & Samson]的鸽色篮状菌(T. columbinus S.W. Peterson & Jurjević)作为外群采用最大似然法(maximum likelihood,ML)分析,并进行1 000次自展(bootstrap)计算各演化支的支持率[46-47]。
2 结果与分析PCR扩增的BenA序列约400 bp,ITS序列约560 bp。连接后的BenA-ITS组合序列矩阵去掉空格共约687个位点(site),ML最适替代模型为K2+GI。基于该组合序列矩阵的分子系统学分析显示,菌株ZZ2-7-1h=CGMCC 3.16171与阿根廷篮状菌的模式菌株NRRL 28750聚在一起,bootstrap值为100%;菌株ZZ2-1-1= CGMCC 3.16172与巴塞篮状菌的模式菌株CBS 649.95聚在一个分支,bootstrap值为88%;菌株JS11-5=CGMCC 3.16173与乌克兰篮状菌的模式菌株CBS 162.67聚在一起,bootstrap值为100%。菌株CGMCC 3.16173的BenA序列在糙刺孢篮状菌组的分析中与包括模式菌株在内的4株乌克兰篮状菌同处一个演化支,bootstrap支持率为100%。通过分子系统学和形态学相结合的分析,支持这3个菌株的鉴定,参考我国已报道的篮状菌组、糙刺孢篮状菌组和螺旋蓝状菌组物种,确定阿根廷篮状菌和巴塞篮状菌为我国新记录种(图 1–3)[14, 17-29]。
|
| 图 1 基于BenA-ITS序列构建的ML系统发育树 Figure 1 The ML tree based on the BenA-ITS sequences. Bootstrap值≥70%的分支标注在分支处;模式菌株用“T”标记;新记录种用粗体标记;菌株BenA和ITS序列的GenBank登录号放在括号内;物种所属的3个组标注在分支处并用粗体显示;T. columbinus作为外群。标尺0.05为每核苷酸位点替代率 Bootstrap percentages ≥70% are indicated at the nodes; ex-type strains are indicated with T; new record species are indicated in boldface; the GenBank accession numbers of BenA and ITS sequences of each strains are in parentheses; the three sections the species belonging to are indicated at each nodes in boldface; T. columbinus is as the outgroup. Bar=0.05 substitutions per nucleotide position. |
|
|
|
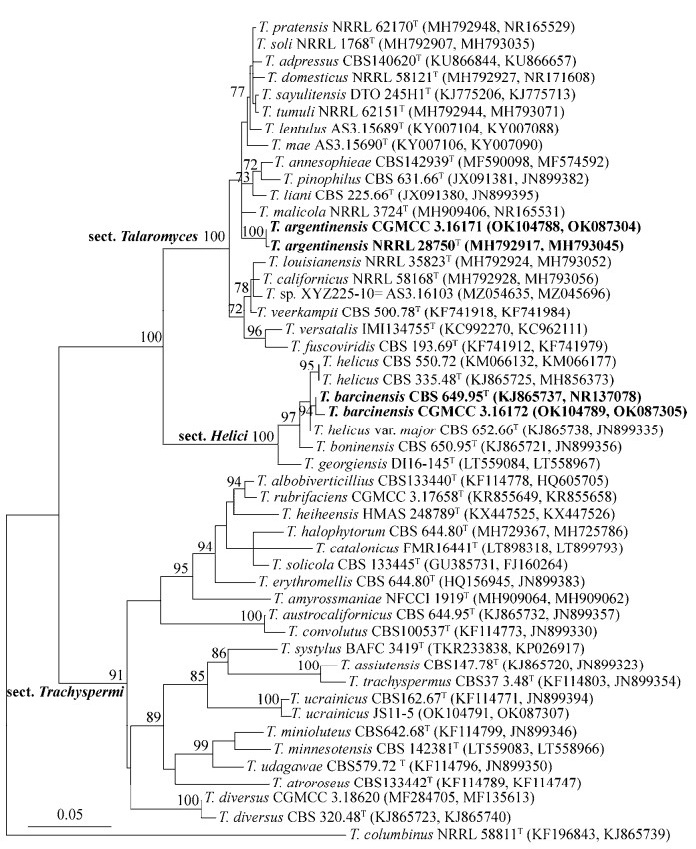
| 图 2 阿根廷篮状菌(Talaromyces argentinensis) CGMCC 3.16171的形态特征 Figure 2 Morphological characters of Talaromyces argentinensis CGMCC 3.16171. A、B:在CYA和MEA上25 ℃培养7 d的菌落;C:成熟子囊;D:未成熟子囊;E、F:子囊孢子。标尺:10 μm A, B: Colonies on CYA and MEA at 25 ℃ after 7 d; C: Mature asci; D: Immature asci; E, F: Ascospores. Scale bar: 10 μm. |
|
|
|
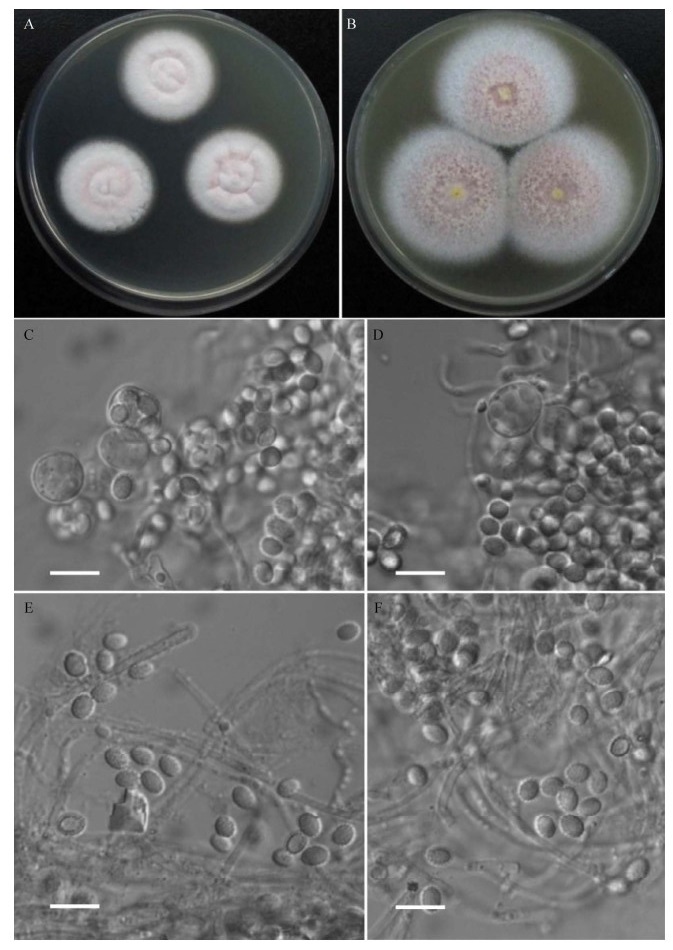
| 图 3 巴塞篮状菌(Talaromyces barcinensis) CGMCC 3.16172的形态特征 Figure 3 Morphological characters of Talaromyces barcinensis CGMCC 3.16172. A、B:在CYA和MEA上25 ℃培养7 d的菌落;C:成熟子囊;D:原基和正在发育的子囊,E:原基和未成熟子囊;F:子囊孢子。标尺:10 μm A, B: Colonies on CYA and MEA at 25 ℃ after 7 d; C: Mature asci; D: Initials and the developing asci; E: Initials and immature asci; F: Ascospores. Scale bar: 10 μm. |
|
|
Talaromyces argentinensis (Jurjevic & S.W. Peterson, Fungal Biology 123: 745–762, 2019[48].)
在CYA上25培养7 d,菌落直径30–33 mm,薄,表面具少量辐射状和同心环状皱纹,边缘整齐;绒状;无分生孢子;在中部可见少量未成熟裸囊壳,皮黄色(buff yellow,R. Pl. Ⅳ);菌丝体白色,在中部呈海贝粉色(seashell pink,R. Pl. ⅩⅣ);无渗出液和可溶性色素;菌落背面呈浅皮黄色至浅赭皮黄色(light buff to light ochraceous-buff,R. Pl. ⅩⅤ)。
在MEA上25培养7 d,菌落直径45–48 mm,稍厚,表面平坦,边缘流苏状;短絮状兼绳状;无分生孢子;中央可见少量帝国黄色(empire yellow)裸囊壳(R. Pl. Ⅳ);菌丝体在边缘为白色,其余为绣球花粉色(hydrangea pink,R. Pl. ⅩⅩⅦ);无渗出液和可溶性色素;菌落背面肉桂红色(cinnamon rufous,R. Pl. ⅩⅣ)。
在CYA上37培养7 d,菌落直径5–8 mm,全部为白色夹杂粉色菌丝体。
在CYA上5培养7 d,不生长。
裸囊壳球形,250–400 μm,帝国黄色(empire yellow,R. Pl. Ⅳ),21 d后成熟;子囊球形至近球形,10–12 μm,内含8个子囊孢子;子囊孢子椭球形,(4–6)×(3.5–5) μm,壁具小刺。
主要特征:生长适度,在MEA上形成短絮状兼绳状菌落,裸囊壳呈帝国黄色,菌丝体白色夹杂粉色;子囊孢子椭球形,壁具小刺。
分布和基物:福建省漳州市九龙口滩涂土壤(ZZ2-7-1h=CGMCC 3.16171)。
2.2 巴塞篮状菌(图 3)Talaromyces barcinensis (Yaguchi & Udagawa, Transactions of Mycological Society of Japan 34: 15–19, 1993[36].)
在CYA上25培养7 d,菌落直径33–34 mm,薄,表面平坦,边缘整齐;绒状;无分生孢子;在中部可见少量未成熟裸囊壳,皮黄色(buff yellow,R. Pl. Ⅳ);菌丝体在边缘为白色,其余为皮黄色至玉米黄色(buff yellow to maize yellow,R. Pl. Ⅳ);无渗出液和可溶性色素;菌落背面中部呈橙红色(orange rufous,R. Pl. Ⅱ),外围浅至旱金莲黄色(capucine yellow,R. Pl. Ⅲ)。
在MEA上25培养7 d,菌落直径37–40 mm,较薄,表面平坦,边缘流苏状;质地绒状兼短絮状;无分生孢子;菌丝体在边缘呈白色,其余呈萘黄色(naphthalene yellow,R. Pl. ⅩⅣ);无渗出液和可溶性色素;菌落背面呈浅黄色。
在CYA上37培养7 d,不生长。
在CYA上5培养7 d,不生长。
裸囊壳球形,120–240 μm,皮黄色,21 d后成熟;子囊球形至椭球形,5–8 μm,内含8个子囊孢子;子囊孢子椭球形,(2.5–3.5)×(2–2.5) μm,壁近光滑至稍粗糙。
主要特征:生长适度,形成绒状菌落,菌丝体白色夹杂萘黄色;裸囊壳较小,呈皮黄色,21 d后成熟;子囊孢子椭球形,较小,壁近光滑至稍粗糙。
分布和基物:福建省漳州市九龙口滩涂土壤(ZZ2-1-1=CGMCC 3.16172)。
3 讨论与结论阿根廷篮状菌此前只发现于阿根廷和加纳,而且GenBank只记录了Peterson等[48]论文中包括模式菌株的2株菌的序列。我国菌株CGMCC 3.16171与这2株菌在生长速度和菌落形态上几乎无差别,它们在MEA上均形成絮状兼绳状菌落,产生少量黄色裸囊壳;其子囊和子囊孢子的形态和大小无差别,但国外的这2株菌在25培养7 d时产生少量分生孢子,而我国菌株则未产生分生孢子。CGMCC 3.16171与模式菌株NRRL 28750在BenA序列上无差别,与另外一个菌株NRRL 28758有2个核苷酸的差别(OK104788 vs. MH792917和MH792918),在ITS序列上与模式菌株NRRL 28750只有1个核苷酸的差别,而与NRRL 28758有5个核苷酸的差别(OK087304 vs. MH793045和MH793046),在Rpb2序列上与模式菌株NRRL 28750只有1个核苷酸的差别,与NRRL 28758有6个核苷酸的差别(OK087304 vs. MH793108和MH793109)。因此,我国菌株CGMCC 3.16171与模式菌株亲缘关系更近。
Yilmaz等[15]把T. barcinensis作为T. helicus (Raper & Fennell) C.R. Benj.的异名,但T. barcinensis与T. helicus在子囊发育过程中的原基(initials)有着本质不同,即T. barcinensis的原基由不规则膨大的细胞构成,而T. helicus的原基则是由产囊体(ascogonium)和螺旋缠绕其上的雄器(antheridium)构成。因此,Yilmaz等[15]将T. barcinensis作为T. helicus的异名是不正确的。巴塞篮状菌此前只发现于西班牙、日本和尼泊尔。我国菌株CGMCC 3.16172的菌落形态、子囊和子囊孢子大小及形状均与模式菌株CBS 649.95的无明显差别,但模式菌株在25培养7 d时产生稀疏分生孢子,而CGMCC 3.16172则未产生分生孢子。另外模式菌株的子囊孢子壁稀疏粗糙,但CGMCC 3.16172的分生孢子近于光滑或只是稍粗糙[36]。CGMCC 3.16172与模式菌株的BenA序列无差别(OK104789 vs. KJ865737),ITS序列有3个碱基的差别(OK087305 vs. MH862547)。GenBank中未记录其他巴塞篮状菌菌株的BenA序列,但有3个菌株的ITS序列(MT510027、KJ942582、KC800596),由于ITS无法区分巴塞篮状菌及其近缘种螺旋篮状菌(T. helices C.R. Benj.),因此这3个菌株不能确定是巴塞篮状菌。Yilmaz等[15]的研究表明,BenA序列基本可以区分目前发现的所有篮状菌物种,通过上述BenA序列的比较,确定菌株CGMCC 3.16171和CGMCC 3.16172鉴定无误。
| [1] |
Nicoletti R, Trincone A. Bioactive compounds produced by strains of Penicillium and Talaromyces of marine origin[J]. Marine Drugs, 2016, 14(2): 37. DOI:10.3390/md14020037 |
| [2] |
Long XH, Liu LP, Shao TY, Shao HB, Liu ZP. Developing and sustainably utilize the coastal mudflat areas in China[J]. Science of the Total Environment, 2016, 569/570: 1077-1086. DOI:10.1016/j.scitotenv.2016.06.170 |
| [3] |
Benjamin CR. Ascocarps of Aspergillus and Penicillium[J]. Mycologia, 1955, 47(5): 669-687. DOI:10.1080/00275514.1955.12024485 |
| [4] |
Raper KB, Thom C. Manual of the penicillia[J]. Soil Science, 1949, 68(5): 415. |
| [5] |
Pitt J. The genus Penicillium and its teleomorphic states Eupenicillium and Talaromyces[M]. London, UK: Academic Press, 1981.
|
| [6] |
Malloch D. The Trichocomaceae: relationships with other Ascomycetes[A]//Samson RA, Pitt JI (Eds). Advances in Penicillium and Aspergillus Systematics[M]. New York, USA: Plenum Press, 1986
|
| [7] |
LoBuglio KF, Pitt JI, Taylor JW. Phylogenetic analysis of two ribosomal DNA regions indicates multiple independent losses of a sexual Talaromyces state among asexual Penicillium species in subgenus Biverticillium[J]. Mycologia, 1993, 85(4): 592-604. DOI:10.1080/00275514.1993.12026313 |
| [8] |
Berbee ML, Yoshimura A, Sugiyama J, Taylor JW. Is Penicillium monophyletic? An evaluation of phylogeny in the family Trichocomaceae from 18S, 5.8S and ITS ribosomal DNA sequence data[J]. Mycologia, 1995, 87(2): 210. DOI:10.1080/00275514.1995.12026523 |
| [9] |
Wang L, Zhuang WY. Phylogenetic analyses of penicillia based on partial calmodulin gene sequences[J]. Biosystems, 2007, 88(1/2): 113-126. |
| [10] |
Houbraken J, Samson RA. Phylogeny of Penicillium and the segregation of Trichocomaceae into three families[J]. Studies in Mycology, 2011, 70: 1-51. DOI:10.3114/sim.2011.70.01 |
| [11] |
Samson RA, Yilmaz N, Houbraken J, Spierenburg H, Seifert KA, Peterson SW, Varga J, Frisvad JC. Phylogeny and nomenclature of the genus Talaromyces and taxa accommodated in Penicillium subgenus Biverticillium[J]. Studies in Mycology, 2011, 70: 159-183. DOI:10.3114/sim.2011.70.04 |
| [12] |
McNeill J, Barrie FR, Buck WR, Demoulin V, Greuter W, Hawksworth DL, Herendeen PS, Knapp S, Marhold K, Prado J, et al. International Code of Nomenclature for Algae, Fungi, and Plants (Melbourne Code) Regnum Vegetabile 154[M]. Koenigstein: Koeltz Scientific Books, 2012: 1-257.
|
| [13] |
Houbraken J, Kocsubé S, Visagie CM, Yilmaz N, Wang XC, Meijer M, Kraak B, Hubka V, Bensch K, Samson RA, et al. Classification of Aspergillus, Penicillium, Talaromyces and related genera (Eurotiales): an overview of families, genera, subgenera, sections, series and species[J]. Studies in Mycology, 2020, 95: 5-169. DOI:10.1016/j.simyco.2020.05.002 |
| [14] |
Sun BD, Chen AJ, Houbraken J, Frisvad JC, Wu WP, Wei HL, Zhou YG, Jiang XZ, Samson RA. New section and species in Talaromyces[J]. MycoKeys, 2020, 68: 75-113. DOI:10.3897/mycokeys.68.52092 |
| [15] |
Yilmaz N, Visagie CM, Houbraken J, Frisvad JC, Samson RA. Polyphasic taxonomy of the genus Talaromyces[J]. Studies in Mycology, 2014, 78: 175-341. DOI:10.1016/j.simyco.2014.08.001 |
| [16] |
Guevara-Suarez M, García D, Cano-Lira JF, Guarro J, Gené J. Species diversity in Penicillium and Talaromyces from herbivore dung, and the proposal of two new genera of Penicillium-like fungi in Aspergillaceae[J]. Fungal Systematics and Evolution, 2020, 5: 39-75. DOI:10.3114/fuse.2020.05.03 |
| [17] |
Wei SZ, Xu XL, Wang L. Four new species of Talaromyces section Talaromyces discovered in China[J]. Mycologia, 2021, 113(2): 492-508. DOI:10.1080/00275514.2020.1853457 |
| [18] |
Tzean SS, Chiu SC, Chen JL, Hseu SH, Lin GH. Penicillium and Related Teleomorphs from Taiwan[M]. Hsinchu, China: Food Industry Research and Development Institute, 1994: 1-158.
|
| [19] |
Kong HZ, Wang L. Flora Fungorum Sinicorum. Vo1. 35. Penicillium et Teleomorphi Cognate[M]. Beijing, China: Science Press, 2007. (in Chinese) 孔华忠, 王龙. 中国真菌志第35卷•青霉属及其相关有性型属[M]. 北京: 科学出版社, 2007. |
| [20] |
Wang QM, Zhang YH, Wang B, Wang L. Talaromyces neofusisporus and T. qii, two new species of section Talaromyces isolated from plant leaves in Tibet, China[J]. Scientific Reports, 2016, 6: 18622. DOI:10.1038/srep18622 |
| [21] |
Wang XC, Chen K, Xia YW, Wang L, Li TH, Zhuang WY. A new species of Talaromyces (Trichocomaceae) from the Xisha Islands, Hainan, China[J]. Phytotaxa, 2016, 267(3): 187. DOI:10.11646/phytotaxa.267.3.2 |
| [22] |
Chen AJ, Sun BD, Houbraken J, Frisvad JC, Yilmaz N, Zhou YG, Samson RA. New Talaromyces species from indoor environments in China[J]. Studies in Mycology, 2016, 84: 119-144. DOI:10.1016/j.simyco.2016.11.003 |
| [23] |
Wang XC, Chen K, Qin WT, Zhuang WY. Talaromyces heiheensis and T. mangshanicus, two new species from China[J]. Mycological Progress, 2017, 16(1): 73-81. DOI:10.1007/s11557-016-1251-3 |
| [24] |
Su L, Niu YC. Multilocus phylogenetic analysis of Talaromyces species isolated from cucurbit plants in China and description of two new species, T. cucurbitiradicus and T. endophyticus[J]. Mycologia, 2018, 110(2): 375-386. DOI:10.1080/00275514.2018.1432221 |
| [25] |
Jiang XZ, Yu ZD, Ruan YM, Wang L. Three new species of Talaromyces sect. Talaromyces discovered from soil in China[J]. Scientific Reports, 2018, 8: 4932. DOI:10.1038/s41598-018-23370-x |
| [26] |
Chen H, Ding G, Sun BD, Zhang Z, Wang L, Chen J. Current taxonomy and three new Chinese records of Talaromyces[J]. Mycosystema, 2021, 40(5): 1200-1215. (in Chinese) 陈晗, 丁刚, 孙炳达, 张争, 王龙, 陈娟. 篮状菌属分类概述及三个中国新记录种[J]. 菌物学报, 2021, 40(5): 1200-1215. DOI:10.13346/j.mycosystema.200288 |
| [27] |
Shan XN, Xu KX, Ruan YM, Wang L. Four species of Talaromyces new to China[J]. Mycosystema, 2021, 40(5): 1216-1231. (in Chinese) 单夏男, 徐可心, 阮永明, 王龙. 篮状菌属的四个中国新记录种[J]. 菌物学报, 2021, 40(5): 1216-1231. DOI:10.13346/j.mycosystema.200323 |
| [28] |
Xu KX, Shan XN, Yu ZH, Deng JX, Wang L. Three species of Talaromyces (Ascomycota, Eurotiales) new to China[J]. Mycosystema, 2021, 40(8): 2181-2190. (in Chinese) 徐可心, 单夏男, 余知和, 邓建新, 王龙. 篮状菌属的三个中国新记录种[J]. 菌物学报, 2021, 40(8): 2181-2190. DOI:10.13346/j.mycosystema.210107 |
| [29] |
Wang L, Jin SY. Two species of Talaromyces sect. Talaromyces new to China[J]. Journal of Liaocheng University: Natural Science Edition, 2022, 35(3): 56-61. (in Chinese) 王龙, 金世宇. 篮状菌属蓝状菌组的两个中国新记录种[J]. 聊城大学学报(自然科学版), 2022, 35(3): 56-61. |
| [30] |
Rodríguez-Andrade E, Stchigel AM, Guarro J, Cano-Lira JF. Fungal diversity of deteriorated sparkling wine and cork stoppers in Catalonia, Spain[J]. Microorganisms, 2019, 8(1): 12. DOI:10.3390/microorganisms8010012 |
| [31] |
Crous PW, Carnegie AJ, Wingfield MJ, Sharma R, Mughini G, Noordeloos ME, Santini A, Shouche YS, Bezerra JDP, Dima B, et al. Fungal planet description sheets: 868-950[J]. Persoonia, 2019, 42: 291-473. DOI:10.3767/persoonia.2019.42.11 |
| [32] |
Crous PW, Cowan DA, Maggs-Kölling G, Yilmaz N, Thangavel R, Wingfield MJ, Noordeloos ME, Dima B, Brandrud TE, Jansen GM, et al. Fungal planet description sheets: 951–1041[J]. Persoonia, 2019, 43: 223-425. DOI:10.3767/persoonia.2019.43.06 |
| [33] |
Rajeshkumar KC, Yilmaz N, Marathe SD, Seifert KA. Morphology and multigene phylogeny of Talaromyces amyrossmaniae, a new synnematous species belonging to the section Trachyspermi from India[J]. MycoKeys, 2019, 45: 41-56. DOI:10.3897/mycokeys.45.32549 |
| [34] |
You YH, Aktaruzzaman M, Heo I, Park JM, Hong JW, Hong SB. Talaromyces halophytorum sp. nov. isolated from roots of Limonium tetragonum in Korea[J]. Mycobiology, 2020, 48(2): 133-138. DOI:10.1080/12298093.2020.1723389 |
| [35] |
Luo Y, Lu XH, Bi W, Liu F, Gao WW. Talaromyces rubrifaciens, a new species discovered from heating, ventilation and air conditioning systems in China[J]. Mycologia, 2016, 108(4): 773-779. DOI:10.3852/15-233 |
| [36] |
Yaguchi T, Miyadoh S, Udagawa S. Talaromyces barcinensis, a new soil Ascomycete[J]. Transactions of Mycological Society of Japan, 1993, 34(1): 15-19. |
| [37] |
Varriale S, Houbraken J, Granchi Z, Pepe O, Cerullo G, Ventorino V, Chin-A-Woeng T, Meijer M, Riley R, Grigoriev IV, et al. Talaromyces borbonicus, sp. nov., a novel fungus from biodegraded Arundo donax with potential abilities in lignocellulose conversion[J]. Mycologia, 2018, 110(2): 316-324. DOI:10.1080/00275514.2018.1456835 |
| [38] |
Malloch D. Moulds Their Isolation, Cultivation and Identification[M]. Toronto, Canada: University of Toronto Press, 1981: 1-97.
|
| [39] |
Wang L, Sun JQ, Jin SY. Two new records of Talaromyces section islandici species from China[J]. Journal of Liaocheng University: Natural Science Edition, 2020, 33(4): 78-84. (in Chinese) 王龙, 孙剑秋, 金世宇. 篮状菌属岛篮状菌组的两个中国新记录种[J]. 聊城大学学报(自然科学版), 2020, 33(4): 78-84. |
| [40] |
Ridgway R. Color Standards and Color Nomenclature[M]. Washington, D. C.: Published by the author, 1912.
|
| [41] |
Samson RA, Houbraken J, Thrane U, Frisvad JC, Andersen B. Food and Indoor Fungi[M]. 2nd ed. Utrecht, the Netherlands: CBS-KNAW Fungal Biodiversity Centre, 2010: 1-390.
|
| [42] |
Wang L, Zhuang WY. Designing primer sets for amplification of partial calmodulin genes from penicillia[J]. Mycosystema, 2004, 23(4): 466-473. |
| [43] |
Glass NL, Donaldson GC. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous Ascomycetes[J]. Applied and Environmental Microbiology, 1995, 61(4): 1323-1330. |
| [44] |
White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics[A]//Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds. ) PCR Protocols: A Guide to Methods and Applications[M]. San Diego, USA: Academic Press, 1990: 315-322
|
| [45] |
Hall T. BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT[J]. Nucleic Acids Symposium Series, 1999, 41(41): 95-98. |
| [46] |
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA 6: molecular evolutionary genetics analysis version 6.0[J]. Molecular Biology and Evolution, 2013, 30(12): 2725-2729. |
| [47] |
Hall BG. Building phylogenetic trees from molecular data with MEGA[J]. Molecular Biology and Evolution, 2013, 30(5): 1229-1235. |
| [48] |
Peterson SW, Jurjević Ž. The Talaromyces pinophilus species complex[J]. Fungal Biology, 2019, 123(10): 745-762. |
 2022, Vol. 49
2022, Vol. 49