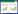扩展功能
文章信息
- 高汉峰, 刘晓芳, 程亮, 朱海霞, 杨琼英, 张纲, 刘玉玲, 郭青云
- GAO Hanfeng, LIU Xiaofang, CHENG Liang, ZHU Haixia, YANG Qiongying, ZHANG Gang, LIU Yuling, GUO Qingyun
- 出芽短梗霉菌株PA-2全基因组测序及分析
- Complete genome sequencing and analysis of Aureobasidium pullulans strain PA-2
- 微生物学通报, 2021, 48(9): 3002-3012
- Microbiology China, 2021, 48(9): 3002-3012
- DOI: 10.13344/j.microbiol.china.200906
-
文章历史
- 收稿日期: 2020-09-10
- 接受日期: 2021-07-08
- 网络首发日期: 2021-07-30
2. 海东市乐都区农业技术推广中心 青海 海东 810799
2. Ledu District Agricultural Technology Extension Center of Haidong City, Haidong, Qinghai 810799, China
出芽短梗霉菌(Aureobasidium pullulans)是广泛存在于自然界、类似于酵母的真菌,因其产生黑色素又称黑酵母[1]。该菌可产生多种代谢物质,如Pullulan、Beta-1, 3-D-Glucan、Aureobasidin A、Exophilin A和Liamocins等,这些代谢物质种类繁多、抑菌活性强、不易产生抗药性,是近年来农产品防腐保鲜[2]、医药开发[3]和植物病草害生物防治[4-8]等领域研究的热点。近年来,有研究者采用2-丁酮提取了出芽短梗霉菌发酵液中的代谢产物,发现含有多元醇脂类物质(Liamocins),该物质对具有较高耐药性的链球菌具有很好的抗菌活性[9]。对出芽短梗霉株CCTCC M2012223的基因组序列及比较基因组学分析发现,该菌株含有多个次级代谢产物合成基因簇,编码聚苹果酸、普鲁兰多糖和黑色素等生物活性物质[10]。Di Francesco等从出芽短梗霉菌株L1和L8发酵产物分离出可抑制大豆立枯丝核菌的非挥发性物质[11]。Wang等[12]从A. pullulans NH6.2发酵产物中分离的嗜铁素(Siderophore)对致病性鳗弧菌(Vibrio anguillarum)具有较强的抑菌效果。赵红霞等[13]从出芽短梗霉菌株NR106发酵产物中分离到了Aureobasidin A——一种对真菌和原生动物具有广谱抗性的环状九肽类抗生素。由于微生物产生的次级代谢产物结构新异、靶点独特、广谱性等特点,因此从微生物中获得具有活性的次级代谢产物合成基因簇及活性成分的研究受到了学者的关注。全基因组测序可将微生物完整的基因组测序出来,进而预测该菌株可能的功能及活性化合物编码基因簇,这为生防菌活性化合物的挖掘和生防机理的研究提供了契机。
课题组前期从青海省海东市平安区患病的杨树上筛选到一株出芽短梗霉菌PA-2菌株,该菌株对藜和猪殃殃等阔叶杂草具有较高的防效,但对青海省主栽作物青稞、油菜等安全[8, 14];岳登高等[15]从该菌株内分离出的环状脂肽类物质对大肠杆菌、樱桃球腔菌和大麦网斑菌等病原菌具有较好的抑菌活性,这说明该菌株具有开发成为生物农药的潜力。利用传统实验技术很难全面地挖掘该菌株的活性物质和解析生防机理。为了深入阐明该菌株内在生防功能,本研究基于Illumina HiSeq平台对该菌株进行全基因组测序分析并预测了其次级代谢产物及碳水化合物活性酶,并对该菌株碳水化合物酶的能力进行了验证,以期为挖掘活性次级代谢产物基因簇及其生防机理研究提供遗传信息基础。
1 材料与方法 1.1 材料 1.1.1 菌种出芽短梗霉菌株PA-2为青海省农业有害生物综合治理重点实验室保存并提供。
1.1.2 培养基和主要试剂发酵培养基(g/L):葡萄糖60.0,(NH4)2SO4 2.0,KH2PO4 0.1,MgSO4 0.1,ZnSO4 0.1,KCl 0.5;PDA培养基(g/L):葡萄糖20.0,马铃薯200.0,琼脂18.0;纤维素培养基(g/L)参考文献[16]并稍做改进,即微晶纤维素5.0,(NH4)2SO4 2.0,K2HPO4 1.0,MgSO4·7H2O 0.5,NaCl 0.5,FeSO4·7H2O 0.1,琼脂粉20.0,pH 7.0−7.2;果胶培养基是将改进后的纤维素培养基中的纤维素改为果胶;0.2%刚果红染液:刚果红2.0 g,蒸馏水1 L;NaCl溶液1.0 mol/L。
1.2 方法 1.2.1 菌体收集将实验室保存的出芽短梗霉菌株PA-2转接到PDA固体培养基上28 ℃培养5 d,用打孔器在菌落边缘取菌块(Φ=8 mm)接种于盛有50 mL发酵培养基的250 mL三角瓶中,25 ℃、180 r/min培养96 h后过滤收集菌丝体,用无菌水冲洗2−3次,液氮中保存备用。
1.2.2 菌株总DNA的提取基因组提取采用Sangon的真菌基因组DNA快速抽提试剂盒,按照试剂盒的操作流程进行提取,提取到的基因组通过1%琼脂糖凝胶电泳检测并进行Qubit®2.0荧光计检测。
1.2.3 全基因组测序和组装委托生工生物工程(上海)股份有限公司完成测序。采用HiSeq 2500高通量测序平台进行基因组DNA测序。Illumina HiSeq得到的原始图像数据文件经CASAVA碱基识别、分析转化为原始测序序列,通过FastQC软件[17]对原始序列进行质量评估,使用Trimmomatic V0.36软件[18]对测序数据进行序列质量剪切,获得质量较高的序列数据。使用SPAdes V3.5.0软件[19]对二代测序数据的序列进行拼接。随后运用GapFiller V1.11软件[20]对拼接得到的Contig进行补洞,最后采用PrInSeS-G V1.0.0软件[21]进行序列矫正。
1.2.4 基因的预测与注释利用Prokka V1.10软件[22]预测tRNA、rRNA等基因元件,使用AUGUSTUS V2.5.5 (http://bioinf.unigreifswald.de/augustus/)软件对Repeat-Masked的基因组做基因预测,利用NCBI BLAST+V2.2.28软件[23]将预测基因的蛋白序列分别与NR、SwissProt数据库[24]、TrEMBL数据库、COG[25]、PFAM (http://pfam.xfam.org/)数据库、CDD (https://www.ncbi.nlm.nih.gov/cdd/)数据库进行比对以获得蛋白功能注释。利用SwissProt、TrEMBL数据库注释结果得到GO (Gene Ontology)[26]功能注释,使用KAAS V2.21软件[27]获得KEGG (Kyoto Encyclopedia of Genes and Genomes)[28]注释信息。
1.2.5 次级代谢产物合成基因簇分析利用antiSMASH 3.0 (https://fungismash.secondarymetabolites.org)在线工具对菌株PA-2中的次级代谢产物合成基因簇进行预测。
1.2.6 CAZymes酶类注释及验证利用在线工具CAZy注释功能(http://www.omicshare.com/tools)对菌株PA-2的碳水化合物活性酶进行预测。
将经过活化的PA-2菌饼(Ф=0.5 cm)分别接入纤维素固体培养基和果胶固体培养基,每个培养基接种1块菌饼,以不接菌饼的培养基作为对照,每种培养基做5个重复。在27 ℃的恒温培养箱里培养10 d,每天观察菌落长势,每5 d测量菌落直径,10 d后将纤维素培养基用0.2%的刚果红染液平板染色15 min,然后用1 mol/L的NaCl溶液清洗2次,一次10 min,十字交叉法测量水解圈直径(D,cm)和菌落直径(d,cm),再计算出纤维素酶相对活性HC值(D/d)。
2 结果与分析 2.1 基因组概述及组分分析采用Illumina HiSeq测序平台,对真菌菌株PA-2进行全基因组测序。菌株PA-2全基因组大小为28 932 793 bp,平均GC含量为50%,包含322个Contigs,总平均长度为1 437.33 bp。由基因GC含量分布图 1A可知GC含量在50%−55%区间内,说明GC含量无明显偏向性。由基因长度分布图 1B可得预测基因长度在800−1 000 bp的基因的数量最多,达1 330个;长度在0−200 bp的基因数量最少,仅仅为73个。基因元件预测结果显示:共有10 839个蛋白编码基因,蛋白编码率为53.85%,27 854个CDS、173个tRNA、56个rRNA (表 1)。将菌株PA-2基因组测序结果提交至NCBI,获得登录号为:PRJNA636834。
|
| 图 1 菌株PA-2全基因组基因GC含量分布(A)和长度分布(B) Figure 1 GC content distribution (A) and length distribution (B) of the whole genome of strain PA-2 |
|
|
| 类别 Class |
数量 Number |
| Size (base) | 28 932 793 |
| Protein coding genes | 10 839 |
| Min length (base) | 120 |
| Max length (base) | 14 895 |
| Average length (base) | 1 437.33 |
| Total coding gene (base) | 15 579 193 |
| Coding ratio (%) | 53.85 |
| tRNA | 173 |
| rRNA | 56 |
| Exon | 27 854 |
| CDS | 27 854 |
| Start codon | 10 801 |
| Stop codon | 10 766 |
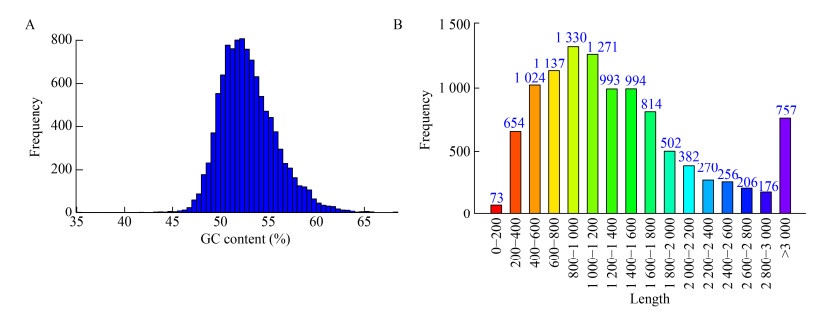
对菌株PA-2的基因组进行NR比对注释、KOG功能分析、GO注释及KEGG通路分析。NR数据库比对结果显示,菌株PA-2基因比对到A. pullulans EXF-150菌株基因数最多(9 350,88.12%),其次是A. subglaciale EXF-2481菌株(487,4.50%)。KOG功能分析显示,菌株PA-2共注释到5 372个基因,注释率为49.56%,分为25种功能组。一般功能预测的基因数量最多(986,18.35%),其余的基因主要集中在翻译后修饰、蛋白质更新及分子伴侣(447,8.32%)和信号转导机制(368,6.85%)等相关基因。GO注释结果显示,该菌株基因组内所有注释基因比对到3大类的54个条目中,总计有6 633条基因序列分别分类到生物学过程、细胞成分、分子功能3大类中,占菌株基因总数的61.2%。在GO生物学过程分类中,细胞过程和单一生物体过程注释到的基因较多,分别为4 643 (70.0%)条和2 683 (40.4%)条;在细胞成分分类中,细胞和细胞部分注释到的基因较多,分别为5 094 (76.8%)条和5 091 (76.7%)条;在分子功能分类中,催化活性和连接功能注释到的基因较多,分别为3 599 (54.3%)条和3 328 (50.2%)条。KEGG通路结果显示,总共有3 890个基因归类到32条通路分类条目中,占菌株基因总数的34.04%,其中碳水化合物代谢、氨基酸代谢以及翻译是最主要的3种代谢通路,分别有440 (11.3%)、394 (10.1%)和343 (8.8%)个基因注释结果(图 2)。
|
| 图 2 A. pullulans strain PA-2 KEGG功能分类 Figure 2 KEGG functional classification of A. pullulans strain PA-2 |
|
|
利用antiSMASH (V5.1.2)对菌株PA-2基因组的次级代谢产物合酶基因簇进行了预测及分析,结果表明该菌株基因组中预测到了23个次级代谢产物合成相关基因簇,主要是I型的聚酮合酶基因簇(T1PKS)和非核糖体多肽合成酶相似基因簇(NRPS-Like)。具体的预测分析结果见表 2,在该菌株预测得到的基因簇中,T1PKS和NRPS及其相似基因簇共有18个,占了预测总基因簇的78.3%,有7个T1PKS、3个NRPS、9个NRPS相似基因簇、2个Betalactone基因簇和4个萜烯类合成途径基因簇。将菌株PA-2所有基因簇与已知的次级代谢产物基因簇做BLAST比对后发现,Betalactone类型的基因簇与黑色素合成基因簇的相似性达到了100%。T1PKS和NRPS这2个基因簇与UNII-YC2Q1O94PT (ACR-Toxin I)合成基因簇相似性达到100%。还有一个T1PKS基因簇与Burnettramic Acid A合成基因簇相似性为22%,另一个T1PKS基因簇与Abscisic Acid合成基因簇相似性为50%。比对后发现,还有19种功能未知的合成基因簇,其中T1PKS类4种、萜烯类4种、NRPS-Like类9种、NRPS类1种、NRPS和Betalactone复合类1种。这说明菌株PA-2中可能存在新的活性物质合成基因簇。
| 基因簇编号 Cluster ID |
基因簇类型 Gene cluster type |
基因大小 Gene size (bp) |
最相似的基因簇 Most similar gene cluster |
相似性 Similarity (%) |
| Cluster 1 | Betalactone | 29 797 | Melanin | 100 |
| Cluster 2 | T1PKS | 46 512 | ||
| Cluster 3 | Terpene | 21 715 | ||
| Cluster 4 | NRPS, betalactone | 54 761 | ||
| Cluster 5 | NRPS-like | 45 073 | ||
| Cluster 6 | NRPS-like | 35 112 | ||
| Cluster 7 | NRPS-like | 42 942 | ||
| Cluster 8 | T1PKS | 47 565 | Burnettramic acid A | 22 |
| Cluster 9 | T1PKS | 46 779 | ||
| Cluster 10 | NRPS-like | 43 621 | ||
| Cluster 11 | NRPS | 54 836 | ||
| Cluster 12 | NRPS-like | 43 127 | ||
| Cluster 13 | Terpene | 21 160 | ||
| Cluster 14 | T1PKS | 45 962 | ||
| Cluster 15 | T1PKS, NRPS | 57 624 | UNII-YC2Q1O94PT (ACR-toxin I) | 100 |
| Cluster 16 | NRPS-like | 45 371 | ||
| Cluster 17 | Terpene | 24 224 | ||
| Cluster 18 | NRPS-like | 43 377 | ||
| Cluster 19 | NRPS-like | 36 965 | ||
| Cluster 20 | Terpene | 23 247 | ||
| Cluster 21 | T1PKS | 42 671 | Abscisic acid | 50 |
| Cluster 22 | T1PKS | 46 168 | ||
| Cluster 23 | NRPS-like | 43 818 |
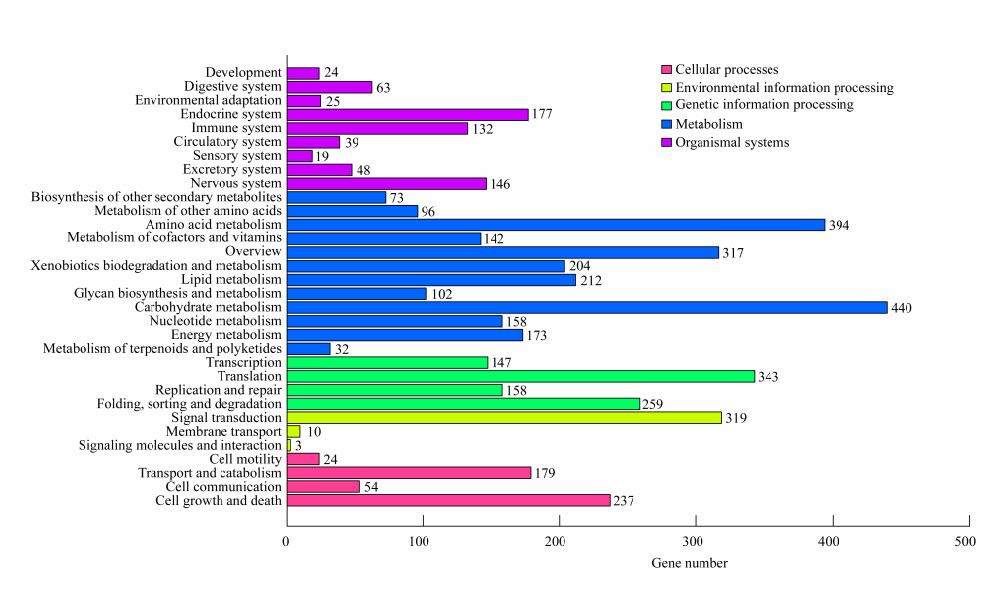
将预测编码蛋白与CAZy数据库进行比对,结果发现在菌株PA-2的基因组内共有1 098个基因编码蛋白属于碳水化合物活性酶家族,该家族成员占菌株所有编码蛋白的10.13%。这些注释到的碳水化合物活性酶家族主要包括555个糖苷水解酶(Glycoside Hydrolases,GH)、332个糖苷转移酶(Glycosyl Transferases,GT)、270个碳水化合物结合模块(Carbohydrate-Binding Modules,CBM)和102个碳水化合物酯酶(Carbohydrate Esterases,CE)等酶类,表明菌株PA-2可能含有较多的碳水化合物活性酶(图 3)。为了进一步确定菌株PA-2基因组的碳水化合物水解酶是否表达,利用纤维素培养基结合刚果红染色法和果胶培养基培养法确定了该菌株水解酶表达情况。结果表明:该菌株能在以纤维素为唯一碳源的培养基上生长,10 d时利用刚果红染色发现形成的HC值达到了1.91。我们还发现该菌株也能在以果胶为唯一碳源的培养基上生长,上述结果表明该菌株能有效利用纤维素和果胶(图 4)。
|
| 图 3 A. pullulans PA-2菌株的CAZy注释 Figure 3 CAZy annotation of A. pullulans PA-2 strain Note: AA: Auxiliary activities; CBM: Carbohydrate-binding modules; CE: Carbohydrate esterases; GH: Glycoside hydrolases; GT: Glycosyl transferases; PL: Polysaccharide lyases |
|
|
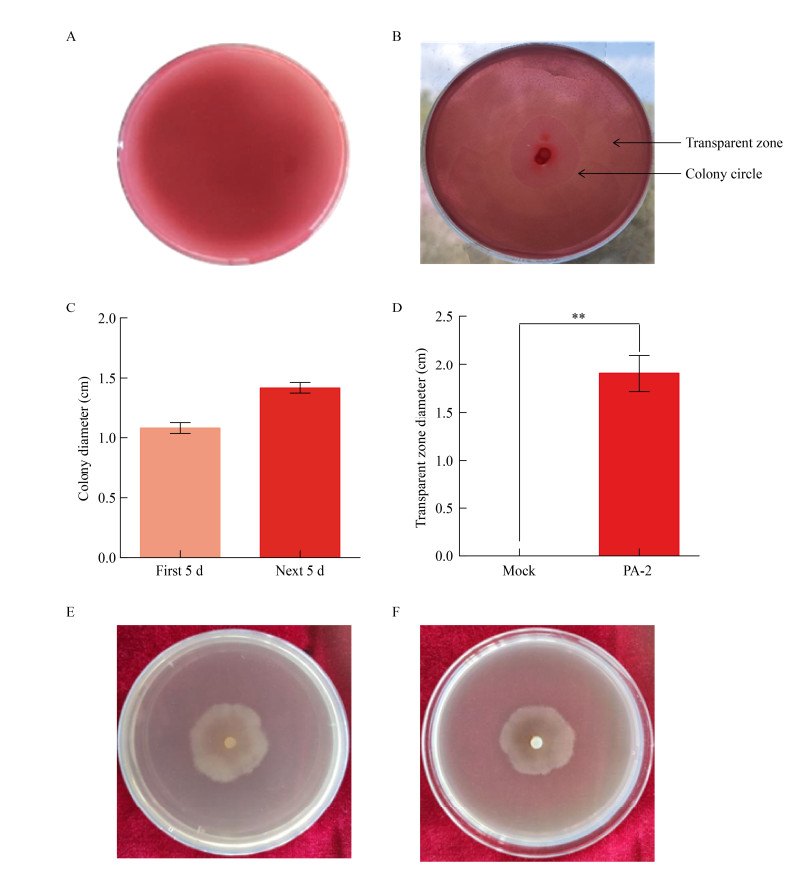
|
| 图 4 A. pullulans strain PA-2对纤维素和果胶的利用(27 ℃,10 d) Figure 4 Utilization of A. pullulans strain PA-2 for cellulose and pectin (27 ℃, 10 d) 注:A:不接菌的纤维素培养基刚果红染色;B:接种菌株PA-2的纤维素培养基刚果红染色;C:菌株PA-2生长在纤维素培养基上10 d的菌落直径;D:菌株PA-2生长在纤维素培养基上的透明圈大小;E:菌株PA-2在果胶培养基上的正面;F:菌株PA-2在果胶培养基上的反面 Note: A: Congo red dyeing of cellulose medium without inoculation; B: Congo red staining of cellulose medium inoculated with strain PA-2; C: Colony diameter of strain PA-2 grown on cellulose medium for 10 days; D: The size of the transparent circle of strain PA-2 grown on the cellulose medium; E: The front of strain PA-2 grown on pectin medium; F: The back of strain PA-2 grown on pectin medium |
|
|
本团队前期从青海省海东市平安区患病的杨树叶片上分离了一株出芽短梗霉菌株PA-2,通过致病性和安全性试验发现菌株PA-2对多种阔叶杂草具有较高除草活性,但对作物安全性较高,后期也发现从该菌株内可以分离出脂肽类抑菌物质[8, 14-15]。为了深入了解该菌,挖掘具有除草活性及抑菌活性基因簇,从而解析其生防机制。本研究利用全基因组测序的方法获得了菌株PA-2的全基因组序列,确定了菌株基因组大小及编码基因数目,同时将菌株PA-2全基因组数据在NR、KOG、GO、KEGG等数据库完成比对分析,并在分子生物学层面探讨了菌株PA-2在生防中可能涉及的基因簇。
利用antiSMASH工具预测了菌株PA-2的次级代谢产物合成基因簇,发现该菌株有一个与黑色素合成基因簇相似性达100%的基因簇,而且该菌株在PDA培养基上生长至后期也会产生黑色,这与预测结果一致。研究表明UNII-YC2Q1O94PT的本质是ACR-Toxin I,其是由19碳多元醇中带有一个α-二氢吡喃酮环的聚酮组成的一种植物毒素[29]。先前的研究显示,由一种链格孢菌(Alternaria alternata)产生的寄主选择性的植物毒素ACR-Toxin可以引起粗皮柠檬(Citrus jambhiri)的叶斑病且该毒素作用位点是寄主线粒体[29-30]。本研究发现PA-2菌株也有一个与ACR-Toxin I的合成基因簇相似性为100%的基因簇,推测该菌株在侵染寄主植物后可能分泌该种植物毒素引起寄主死亡。先前的研究显示,病原菌侵染寄主时会分泌一些脱落酸的确会增加寄主植物体内的脱落酸含量,而且高浓度的脱落酸会抑制植物的生长[31-34]。菌株PA-2中存在脱落酸合成基因簇,尽管相似度仅为50%,推测可能合成了脱落酸或其类似物抑制寄主生长为其除草机理之一。也有报道证明从灰葡萄孢菌(Botrytis cinerea)或瓜果腐霉[Pythium aphanidermatum (Edson) Fitzp.]等菌的代谢物中分离并鉴定的脱落酸及其衍生物也有较强的除草活性[35]。Burnettramic Acid A是一种吡咯烷二酮类物质,还是一种新类型的抗生素[36-37]。菌株PA-2也有与Burnettramic Acid A合成基因簇相似的基因簇,相似性仅为22%,这说明该基因簇可能合成了其他类型的抑菌物质。尽管在PA-2菌株内预测出了23个次级代谢物合成基因簇,但只有4个已知功能的基因簇,还有19个功能未知的合成基因簇,这说明菌株PA-2可能还编码其他类型的活性物质,需要今后进一步研究。事实上,岳登高等[15]从菌株PA-2发酵液中分离出了具有较好活性的环状脂肽类抗生素,这也证明了该菌株内可能还有其他活性物质的推测。
碳水化合物活性酶(CAZymes)作为植物病原菌中的一类重要酶,在病原菌的生长和发育中发挥重要的作用,尤其在病原菌侵染植物或拮抗微生物的过程中[38-39]。先前的研究显示,能与寄主保持共生关系的病原菌分泌组的纤维素酶和果胶酶数量较少可能与其不会降解植物组织并引起植物死亡的特点相符[40]。相反的是,致病性病原菌含有较多的编码水解酶活性的基因,其与病原菌在侵染过程中降解植物组织并导致寄主死亡密切相关[41]。除了在病原菌侵染寄主的过程中发挥作用外,研究表明CAZymes酶类也与抑菌作用相关[42]。本研究将菌株PA-2碳水化合物活性酶基因进行了预测,结果表明共预测到1 098个碳水化合物活性酶(CAZymes)家族。这与田李等对引起棉花黄萎病的大丽轮枝菌(Verticillium dahliae)基因组内预测的158个CAZymes酶相比多的多[39]。这表明菌株PA-2可能具有较好的细胞壁水解活性。植物细胞壁的主要成分是纤维素和果胶,我们对菌株PA-2水解纤维素和果胶能力进行了测试,发现该菌株有较好的纤维素水解能力(HC=1.91)。该菌株也能在以果胶为唯一碳源的培养基上生长,说明具有水解和利用果胶的能力。事实上,已经有很多的报道表明出芽短梗霉菌能产生纤维素酶和果胶酶[43-46]。Rong等的研究表明,筛选自海洋中的A. pullulans 98可以生产高产量的纤维素酶[47]。Merín等首次从A. pullulans GM-R-22中发现在低温下也有较好活性的果胶酶存在[48]。上述研究与本研究结果相符,推测水解寄主细胞壁为菌株PA-2除草及抑菌的机理之一。本研究弥补了该菌株在遗传信息方面的空白,也为进一步解析该菌株生防机制提供基因背景信息。
| [1] |
Liu J, Liu ZQ, Chi ZM, Zhang L, Zhang DC. Intraspecific diversity of Aureobasidium pullulans strains from different marine environments[J]. Journal of Ocean University of China, 2009, 8(3): 241-246. DOI:10.1007/s11802-009-0241-3 |
| [2] |
Cheng KC, Demirci A, Catchmark JM. Pullulan: biosynthesis, production, and applications[J]. Applied Microbiology and Biotechnology, 2011, 92(1): 29-44. DOI:10.1007/s00253-011-3477-y |
| [3] |
Lu XD. Screening of pullulan strains by UV mutagensis and optimization of the fermentation conditions[D]. Tianjin: Master's Thesis of Tianjin University of Science & Technology, 2013 (in Chinese) 卢星达. 产普鲁兰多糖出芽短梗霉的菌种筛选及发酵条件优化[D]. 天津: 天津科技大学硕士学位论文, 2013 |
| [4] |
Zhu GL, Tong QY. Research progress on microbial polysaccharide[J]. Science and Technology of Food Industry, 2012, 33(6): 444-448. (in Chinese) 朱桂兰, 童群义. 微生物多糖的研究进展[J]. 食品工业科技, 2012, 33(6): 444-448. |
| [5] |
Liu GJ. Studies on hard capsules material based on pullulan—carrageenan[D]. Wuxi: Master's Thesis of Jiangnan University, 2014 (in Chinese) 刘国军. 普鲁兰多糖—卡拉胶基硬胶囊囊材的研究[D]. 无锡: 江南大学硕士学位论文, 2014 |
| [6] |
Roberti R, Di Francesco A, Innocenti G, Mari M. Potential for biocontrol of Pleurotus ostreatus green mould disease by Aureobasidium pullulans De Bary (Arnaud)[J]. Biological Control, 2019, 135: 9-15. DOI:10.1016/j.biocontrol.2019.04.016 |
| [7] |
Rusin C, Di Francesco A, Di Foggia M, D'Aquino S, Rombolà A, Tugnoli V, Botelho RV, Baraldi E. An emerging problem affecting apple production: Neofusicoccum parvum. Aureobasidium pullulans L1 and L8 strains as an alternative control strategy[J]. Biological Control, 2019, 134: 157-162. DOI:10.1016/j.biocontrol.2019.04.015 |
| [8] |
Guo QY, Cheng L, Zhu HX, Li W, Wei YH, Chen HY, Guo LZ, Weng H, Wang J. Herbicidal activity of Aureobasidium pullulans PA-2 on weeds and optimization of its solid-state fermentation conditions[J]. Journal of Integrative Agriculture, 2020, 19(1): 173-182. DOI:10.1016/S2095-3119(19)62738-3 |
| [9] |
Wu LQ, Yang SL, Qi LM, Liu GJ, Gu HK. Liamocins produced by Aeruobasidium spp.[J]. Acta Microbiologica Sinica, 2021. (in Chinese) 武利勤, 杨素玲, 亓丽梅, 刘桂君, 顾海科. 短梗霉产liamocins[J]. 微生物学报, 2021. DOI:10.13343/j.cnki.wsxb.20200541 |
| [10] |
Wang YK, Song XD, Li XR, Yang ST, Zou X. Complete genome sequencing of polymalic acid-producing strain Aureobasidium pullulans CCTCC M2012223[J]. Acta Microbiologica Sinica, 2017, 57(1): 97-108. (in Chinese) 王永康, 宋晓丹, 李晓荣, 杨尚天, 邹祥. 聚苹果酸生产菌出芽短梗霉CCTCC M2012223的全基因组测序及序列分析[J]. 微生物学报, 2017, 57(1): 97-108. |
| [11] |
Di Francesco A, Di Foggia M, Corbetta M, Baldo D, Ratti C, Baraldi E. Biocontrol activity and plant growth promotion exerted by Aureobasidium pullulans strains[J]. Journal of Plant Growth Regulation, 2021, 40(3): 1233-1244. DOI:10.1007/s00344-020-10184-3 |
| [12] |
Wang WL, Chi Z, Liu GL, Buzdar MA, Chi ZM, Gu QQ. Chemical and biological characterization of siderophore produced by the marine-derived Aureobasidium pullulans HN6.2 and its antibacterial activity[J]. BioMetals, 2009, 22(6): 965-972. DOI:10.1007/s10534-009-9248-x |
| [13] |
Zhao HX, Gou P, Liu XP, Liu SH, Guo XJ. Effects of aureobasidin A on Botrytis cinerea's growth[J]. Plant Protection, 2015, 41(6): 44-48, 59. (in Chinese) 赵红霞, 苟萍, 刘小平, 刘圣红, 郭星军. 短梗霉素A对灰葡萄孢菌生长的影响[J]. 植物保护, 2015, 41(6): 44-48, 59. DOI:10.3969/j.issn.0529-1542.2015.06.007 |
| [14] |
Li YL, Cheng L, Zhu HX, Guo QY. Herbicidal activity of Aureobasidium pullulans PA-2 on weeds and its safety on crops[J]. Chinese Journal of Biological Control, 2014, 30(2): 232-238. (in Chinese) 李永龙, 程亮, 朱海霞, 郭青云. 出芽短梗霉菌菌株PA-2的除草活性及对作物的安全性[J]. 中国生物防治学报, 2014, 30(2): 232-238. |
| [15] |
Yue DG, Cheng L, Guo QY. Antibacterial activities and optimization of fermentation conditions of lipopeptides produced by Aureobasidium pullulans PA-2[J]. Acta Agriculturae Zhejiangensis, 2021, 33(3): 479-489. (in Chinese) 岳登高, 程亮, 郭青云. 出芽短梗霉菌PA-2脂肽类物质抑菌活性及其发酵条件优化[J]. 浙江农业学报, 2021, 33(3): 479-489. |
| [16] |
Li YB, Zhang Q, Teng LP, Liu ZW. Screening of a cotton Fusarium oxysporum f. sp. vasinfectum and Verticillium dahliae antagonistic strain from cotton decomposing microorganisms[J]. Acta Agriculturae Boreali-occidentalis Sinica, 2016, 25(11): 1730-1735. (in Chinese) 李艳宾, 张琴, 滕立平, 刘占文. 1株拮抗棉花枯、黄萎病菌的纤维素分解菌筛选[J]. 西北农业学报, 2016, 25(11): 1730-1735. DOI:10.7606/j.issn.1004-1389.2016.11.021 |
| [17] |
Brown J, Pirrung M, McCue LA. FQC Dashboard: integrates FastQC results into a web-based, interactive, and extensible FASTQ quality control tool[J]. Bioinformatics, 2017, 33(19): 3137-3139. DOI:10.1093/bioinformatics/btx373 |
| [18] |
Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data[J]. Bioinformatics, 2014, 30(15): 2114-2120. DOI:10.1093/bioinformatics/btu170 |
| [19] |
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing[J]. Journal of Computational Biology, 2012, 19(5): 455-477. DOI:10.1089/cmb.2012.0021 |
| [20] |
Boetzer M, Pirovano W. Toward almost closed genomes with GapFiller[J]. Genome Biology, 2012, 13(6): R56. DOI:10.1186/gb-2012-13-6-r56 |
| [21] |
Massouras A, Hens K, Gubelmann C, Uplekar S, Decouttere F, Rougemont J, Cole ST, Deplancke B. Primer-initiated sequence synthesis to detect and assemble structural variants[J]. Nature Methods, 2010, 7(7): 485-486. DOI:10.1038/nmeth.f.308 |
| [22] |
Seemann T. Prokka: rapid prokaryotic genome annotation[J]. Bioinformatics, 2014, 30(14): 2068-2069. DOI:10.1093/bioinformatics/btu153 |
| [23] |
Altschul SF, Madden TL, Schäffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs[J]. Nucleic Acids Research, 1997, 25(17): 3389-3402. DOI:10.1093/nar/25.17.3389 |
| [24] |
Boutet E, Lieberherr D, Tognolli M, Schneider M, Bansal P, Bridge AJ, Poux S, Bougueleret L, Xenarios I. UniProtKB/Swiss-prot, the manually annotated section of the UniProt KnowledgeBase: how to use the entry view[J]. Methods in Molecular Biology, 2016, 1374: 23-54. |
| [25] |
Tatusov RL, Natale DA, Garkavtsev IV, Tatusova TA, Shankavaram UT, Rao BS, Kiryutin B, Galperin MY, Fedorova ND, Koonin EV. The COG database: new developments in phylogenetic classification of proteins from complete genomes[J]. Nucleic Acids Research, 2001, 29(1): 22-28. DOI:10.1093/nar/29.1.22 |
| [26] |
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Michael Cherry J, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene Ontology: tool for the unification of biology[J]. Nature Genetics, 2000, 25(1): 25-29. DOI:10.1038/75556 |
| [27] |
Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. KAAS: an automatic genome annotation and pathway reconstruction server[J]. Nucleic Acids Research, 2007, 35(Suppl_2): W182-W185. |
| [28] |
Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M. From genomics to chemical genomics: new developments in KEGG[J]. Nucleic Acids Research, 2006, 34(Suppl_1): D354-D357. |
| [29] |
Izumi Y, Ohtani K, Miyamoto Y, Masunaka A, Fukumoto T, Gomi K, Tada Y, Ichimura K, Peever TL, Akimitsu K. A polyketide synthase gene, ACRTS2, is responsible for biosynthesis of host-selective ACR-toxin in the rough lemon pathotype of Alternaria alternata[J]. Molecular Plant-Microbe Interactions, 2012, 25(11): 1419-1429. DOI:10.1094/MPMI-06-12-0155-R |
| [30] |
Ohtani K, Yamamoto H, Akimitsu K. Sensitivity to Alternaria alternata toxin in Citrus because of altered mitochondrial RNA processing[J]. Proceedings of the National Academy of Sciences of the United States of America, 2002, 99(4): 2439-2444. DOI:10.1073/pnas.042448499 |
| [31] |
Tuomi T, Ilvesoksa J, Laakso S, Rosenqvist H. Interaction of abscisic acid and indole-3-acetic acid-producing fungi with Salix leaves[J]. Journal of Plant Growth Regulation, 1993, 12(3): 149-156. DOI:10.1007/BF00189646 |
| [32] |
Marumo S, Katayama M, Komori E, Ozaki Y, Natsume M, Kondo S. Microbial production of abscisic acid by Botrytis cinerea[J]. Agricultural and Biological Chemistry, 1982, 46(7): 1967-1968. |
| [33] |
Zhou YY, Zhang Y, Wang XW, Han X, An Y, Lin SW, Shen C, Wen JL, Liu C, Yin WL, et al. Root-specific NF-Y family transcription factor, PdNF-YB21, positively regulates root growth and drought resistance by abscisic acid-mediated indoylacetic acid transport in Populus[J]. New Phytologist, 2020, 227(2): 407-426. DOI:10.1111/nph.16524 |
| [34] |
Rowe JH, Topping JF, Liu JL, Lindsey K. Abscisic acid regulates root growth under osmotic stress conditions via an interacting hormonal network with cytokinin, ethylene and auxin[J]. New Phytologist, 2016, 211(1): 225-239. DOI:10.1111/nph.13882 |
| [35] |
Zhang J, Dong YX, Zhang JL, Dong JG. A fungal strain having high herbicidal activity[J]. Mycosystema, 2008, 27(5): 645-651. (in Chinese) 张剑, 董晔欣, 张金林, 董金皋. 一株具有高除草活性的真菌菌株[J]. 菌物学报, 2008, 27(5): 645-651. |
| [36] |
Li J, Chen MH, Hao XM, Li SS, Li F, Yu LY, Xiao CL, Gan ML. Structural revision and absolute configuration of burnettramic acid A[J]. Organic Letters, 2020, 22(1): 98-101. DOI:10.1021/acs.orglett.9b04008 |
| [37] |
Li H, Gilchrist CLM, Lacey HJ, Crombie A, Vuong D, Pitt JI, Lacey E, Chooi YH, Piggott AM. Discovery and heterologous biosynthesis of the burnettramic acids: rare PKS-NRPS-derived bolaamphiphilic pyrrolizidinediones from an Australian fungus, Aspergillus burnettii[J]. Organic Letters, 2019, 21(5): 1287-1291. DOI:10.1021/acs.orglett.8b04042 |
| [38] |
Brink J, De Vries RP. Fungal enzyme sets for plant polysaccharide degradation[J]. Applied Microbiology and Biotechnology, 2011, 91(6): 1477-1492. DOI:10.1007/s00253-011-3473-2 |
| [39] |
Tian L, Chen JY, Chen XY, Wang JN, Dai XF. Prediction and analysis of Verticillium dahliae VdLs.17 secretome[J]. Scientia Agricultura Sinica, 2011, 44(15): 3142-3153. (in Chinese) 田李, 陈捷胤, 陈相永, 汪佳妮, 戴小枫. 大丽轮枝菌(Verticillium dahliae VdLs.17)分泌组预测及分析[J]. 中国农业科学, 2011, 44(15): 3142-3153. |
| [40] |
Mueller O, Kahmann R, Aguilar G, Trejo-Aguilar B, Wu A, De Vries RP. The secretome of the maize pathogen Ustilago maydis[J]. Fungal Genetics and Biology, 2008, 45: S63-S70. DOI:10.1016/j.fgb.2008.03.012 |
| [41] |
Fradin EF, Thomma BPHJ. Physiology and molecular aspects of Verticillium wilt diseases caused by V. dahliae and V. albo-atrum[J]. Molecular Plant Pathology, 2006, 7(2): 71-86. DOI:10.1111/j.1364-3703.2006.00323.x |
| [42] |
Zhang DF, Gao YX, Ke XL, Wang YJ, Ren Y, Shi CB. Genomic analysis of Bacillus velezensis LF01 strain and the biocontrol effect of its secondary metabolites[J]. Journal of Fisheries of China, 2021. 张德锋, 高艳侠, 可小丽, 王亚军, 任燕, 石存斌. 贝莱斯芽孢杆菌LF01基因组序列分析及其代谢产物的生防作用[J]. 水产学报, 2021. DOI:10.11964/jfc.20201012442 |
| [43] |
Vieira MM, Kadoguchi E, Segato F, Da Silva SS, Chandel AK. Production of cellulases by Aureobasidium pullulans LB83: optimization, characterization, and hydrolytic potential for the production of cellulosic sugars[J]. Preparative Biochemistry & Biotechnology, 2021, 51(2): 153-163. |
| [44] |
Chi ZM, Wang F, Chi Z, Yue LX, Liu GL, Zhang T. Bioproducts from Aureobasidium pullulans, a biotechnologically important yeast[J]. Applied Microbiology and Biotechnology, 2009, 82(5): 793-804. DOI:10.1007/s00253-009-1882-2 |
| [45] |
Merín MG, Martín MC, Rantsiou K, Cocolin L, De Ambrosini VIM. Characterization of pectinase activity for enology from yeasts occurring in Argentine Bonarda grape[J]. Brazilian Journal of Microbiology, 2015, 46(3): 815-823. DOI:10.1590/S1517-838246320140160 |
| [46] |
Onetto CA, Borneman AR, Schmidt SA. Investigating the effects of Aureobasidium pullulans on grape juice composition and fermentation[J]. Food Microbiology, 2020, 90: 103451. DOI:10.1016/j.fm.2020.103451 |
| [47] |
Rong YJ, Zhang L, Chi ZM, Wang XH. A carboxymethyl cellulase from a marine yeast (Aureobasidium pullulans 98): its purification, characterization, gene cloning and carboxymethyl cellulose digestion[J]. Journal of Ocean University of China, 2015, 14(5): 913-921. DOI:10.1007/s11802-015-2574-4 |
| [48] |
Merín MG, Morata De Ambrosini VI. Kinetic and metabolic behaviour of the pectinolytic strain Aureobasidium pullulans GM-R-22 during pre-fermentative cold maceration and its effect on red wine quality[J]. International Journal of Food Microbiology, 2018, 285: 18-26. DOI:10.1016/j.ijfoodmicro.2018.07.003 |
 2021, Vol. 48
2021, Vol. 48