扩展功能
文章信息
- 杜心恬, 宋馨, 刘欣欣, 夏永军, 艾连中, 熊智强
- DU Xintian, SONG Xin, LIU Xinxin, XIA Yongjun, AI Lianzhong, XIONG Zhiqiang
- 细菌胞外多糖生物合成转录调控因子研究进展
- Advances in transcription regulators of bacterial exopolysaccharides biosynthesis
- 微生物学通报, 2021, 48(2): 573-581
- Microbiology China, 2021, 48(2): 573-581
- DOI: 10.13344/j.microbiol.china.200169
-
文章历史
- 收稿日期: 2020-03-01
- 接受日期: 2020-05-15
- 网络首发日期: 2020-08-10
细菌胞外多糖(Exopolysaccharide,EPS)是细菌自身合成并分泌到细胞外的糖类化合物,包括粘液多糖和荚膜多糖。根瘤菌、乳酸菌和假单胞菌等细菌是常见的EPS来源菌。作为一种备受青睐的天然食品添加剂[1],细菌EPS可作为增稠剂和稳定剂用于改变产品的质构[2-3]。细菌EPS也是安全的“粘性”发酵剂[4],能在发酵过程中显著改善乳制品的风味和品质[5]。其中,黄原胶、结冷胶和热凝胶等细菌EPS由于产量高、成本低等优点在食品工业中广泛应用。
根据单糖组成,细菌EPS可分为同型多糖和异型多糖。同型多糖(如热凝胶、葡聚糖和果聚糖)在胞外直接合成,无须脂载体参与,主要通过对应的糖基转移酶完成EPS的合成;而异型多糖(如黄原胶和结冷胶)都是脂载体依赖型胞内合成,需要糖核苷酸前体、酶催化系统、酰基供体、脂载体和糖基受体,其合成包括糖核苷酸前体的胞内合成、重复单元的合成和多糖的延伸、聚合及输出[6]。细菌EPS生物合成由eps基因簇控制,包括调控基因、链长决定基因、重复单元合成基因、聚合和输出基因,而且不同细菌中eps基因簇的数量和种类有所差异[7-8]。Song等[9]发现干酪乳杆菌(Lactobacillus casei) eps基因簇中3个关键基因对EPS生物合成至关重要。Xiong等[10-11]完成了嗜热链球菌(Streptococcus thermophilus) S-3的EPS及其前体生物合成基因簇和表型分析。Kong等[12]利用强组成型启动子同时表达了嗜热链球菌eps基因簇中2个关键基因,显著提高了EPS的合成。细菌eps基因簇的深入研究为探索EPS生物合成的转录调控奠定了基础。
转录调控因子是一类具有转录调节活性的蛋白质,参与调控细菌内糖、脂和碳代谢等重要生理活动[13]。调控因子与eps基因簇中靶基因位点结合,启动eps基因的转录及控制其转录效率,从而调控细菌EPS的生物合成[14]。但目前仅少数细菌EPS调控因子被鉴定,且调控机制尚未阐明,尚有大量未知的EPS调控因子亟待研究。本文主要从细菌EPS调控因子的研究方法和调控机制展开综述,为深入研究细菌EPS转录调控提供参考。
1 细菌EPS生物合成转录调控因子的信号传导调控机制转录因子调控EPS生物合成的过程中,调控蛋白对靶基因的识别和结合是调控机制的核心。调控蛋白含有特殊构象的结构域,能特异性识别并结合靶基因中对应的结合位点(保守短序列片段,Motif)[14],调控靶基因表达。许多调控蛋白家族有相似的结合结构域,与EPS生物合成密切相关(表 1)。通过信号分子传导,调控因子影响RNA聚合酶识别特定短序列片段,对下游编码基因的转录表达进行正/负调控。EPS转录因子调控机制中,存在3类经典的信号传导机制:群体感应系统、双组分系统和第二信使分子传导系统。
| Family | Action | Binding domain conformation |
Regulation function | References |
| LuxR | + | HTH | Quorum sensing, biofilm formation, and metabolism | [15-17] |
| TetR | - | HTH | Antibiotic synthesis, biofilm formation, and osmotic stress | [18-19] |
| GntR | - | HTH | General metabolism, CPS synthesis, and virulence | [20-21] |
| LacI | - | HTH | Carbon source utilization, virulence, cell activity, and carbohydrate metabolism | [22-24] |
| DeoR | - | HTH | Antibiotic synthesis and carbohydrate metabolism | [25-27] |
| ArsR | - | HTH | Metal and acid resistance | [28-30] |
| LysR | +/- | HTH | Carbon and nitrogen metabolism, biofilm formation, and flagellar movement | [31-33] |
| OmpR | + | Winged helix | Heavy metal, CPS synthesis, and virulence | [34-35] |
群体感应(Quorum Sensing)是细菌因自身种群密度变化,利用信号分子调控基因表达的机制,参与调控EPS生物合成、抗生素合成和生物膜发育等代谢过程。群体感应系统中,细菌合成的信号分子及其感应机制可分为3类:G-细菌的酰基高丝氨酸内酯类信号分子(N-Acyl-Homoserine Lactones,AHL)系统、G+细菌的寡肽类信号分子(Autoinducing Peptides,AIP)系统和细菌通用信号分子AI-2系统。Licciardello等[17]在波纹假单胞菌(Pseudomonas corrugata)中发现,信号系统PcoI/AHL和LuxR家族调控因子PcoR组成群体感应系统,调节海藻酸钠生物合成。在苜蓿根瘤菌(Rhizobium meliloti)中,信号系统SinI/AHL与调控因子ExpR组成的群体感应系统,调控细菌内多种EPS的生物合成[36]。此外,一些细菌中同时存在多种不同的信号分子调节机制。例如,霍乱弧菌(Vibrio cholerae)中存在3种参与调控因子LuxO调控细菌EPS合成和生物膜形成的群体感应系统:CqsA/CAI-1 (AHL类信号分子)、LuxS/AI-2和一种未知的信号系统[37]。
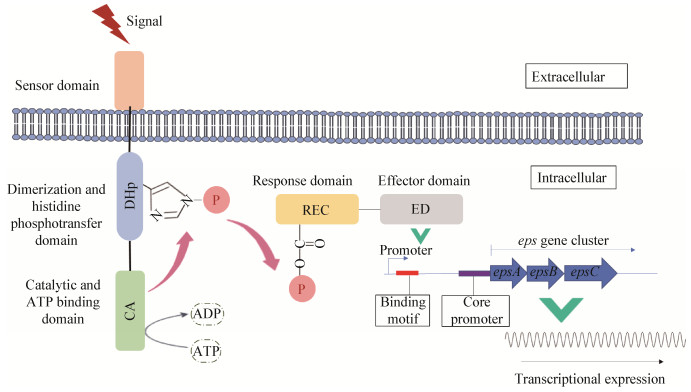
1.2 双组分调控系统双组分系统是细菌适应体内外环境变化的重要信号传导系统,一般由组氨酸激酶与应答调控蛋白组成,通过双组分蛋白磷酸化传递信号,调控胞内基因表达(图 1)[38-39]。Black等[40]在黄色粘球菌(Myxococcus xanthus)中发现途径特异性EPS调控因子EpsW和组氨酸激酶DifE,EpsW被DifE激活后调控EPS合成。转录调控因子RcsB与激酶RcsC组成双组分系统,协同LuxR家族调控因子RcsA共同激活大肠杆菌(Escherichia coli)荚膜多糖基因簇的表达[41]。淀粉液化芽孢杆菌(Bacillus amyloliquefaciens) SQR9中存在双组分调控系统ResDE,ResD被激酶ResE激活后促进生物膜形成,潜在调控EPS[42]。VicRK (调控因子VicR和激酶VicK)、ComDE (调控因子ComE和激酶ComD)和CiaRH (调控因子CiaR和激酶CiaH)等双组分系统在乳酸菌中调控多糖合成等多种代谢过程[43]。此外,细菌中也存在非典型的EPS双组分调控系统,如Minic等[44]发现嗜热链球菌eps基因簇上的酪氨酸激酶EpsD可激活调控蛋白EpsE,调控EPS合成。本课题组过表达嗜热链球菌eps基因簇上关键基因epsA和epsE,能显著提高EPS的合成,其中epsA编码途径特异性调控因子EpsA[12]。此外,我们利用构建的CRISPR-Cas9基因编辑系统解析了干酪乳杆菌LC2W中EPS合成关键基因[45],也发现基因簇上存在途径特异性调控因子LC2W_2170[9]。
|
| 图 1 经典双组分调控体系 Figure 1 Classical two-component regulation system |
|
|
第二信使分子与细胞表面的受体结合后,通过受体信号的转导形成信号通路,调控胞外蛋白和EPS分泌。其中,环二鸟苷酸(c-di-GMP)和环磷酸二腺苷(c-di-AMP)在细菌EPS调控中最为常见。Schäper等[46]在草木樨中华根瘤菌(Sinorhizobium meliloti)中发现,AraC家族调控因子CuxR的二聚体在c-di-GMP的作用下调控细菌EPS。恶臭假单胞菌(P. putida) KT2440中,c-di-GMP抑制调控因子FleQ与EPS合成基因bcs启动子的结合[47]。铜绿假单胞菌(P. aeruginosa)中,c-di-GMP抑制FleQ负调控EPS基因簇上pel在内的多个基因[48]。Fazli等[49]发现c-di-GMP在新生伯克霍尔德菌(Burkholderia cepacia)中通过转录调控因子级联调节EPS合成:c-di-GMP首先激活BerB-RpoN体系控制berA编码调控因子BerA,BerA再与c-di-GMP结合,促进EPS合成基因bep转录表达。与已鉴定出数百种结合蛋白的c-di-GMP不同,迄今为止在细菌中只发现了少数与c-di-AMP作用的EPS调控因子。在变形链球菌中,c-di-AMP与受体蛋白CabPA相互作用,影响转录因子VicR调控EPS合成中关键基因gtfB (编码葡萄糖基转移酶)的表达[50]。同样在变形链球菌(S. mutans)中,Cheng等[51]和Rismondo等[52]发现调控因子CdaR调控单二磷酸环化酶CdaA的合成,并通过c-di-AMP信号网络调控氧化反应和EPS生物合成。金黄色葡萄球菌KdpDE双组分系统中的组氨酸激酶KdpD也是c-di-AMP受体蛋白,参与调控荚膜多糖生物合成与生物膜形成[53]。
2 细菌EPS生物合成转录调控因子的研究方法在基因调控过程中,转录调控因子作用的本质是蛋白质-DNA相互作用。研究调控因子常见的方法及其优缺点见表 2。目前染色质免疫沉淀、DNA亲和层析技术和凝胶阻滞实验在细菌EPS调控因子的研究中应用最为广泛。
| 研究方法 Methods |
优点 Advantages |
缺点 Disadvantages |
| 凝胶阻滞实验 EMSA |
体外快速研究DNA与蛋白质相互作用,特异性强 Rapid detection in vitro, strong specificity |
不能真实反映体内情况,不能确定靶序列,结论单一 Cannot reflect the situation in vivo and identify the target motif |
| DNA微阵列技术 DNA microarray |
有效确定下游靶基因 Effective determination of downstream target genes |
不能直观体现作用机制,昂贵且分析要求高 Cannot directly reflect the mechanism of action, expensive and high analysis requirements |
| 酵母单杂交技术 Yeast one hybrid |
确定DNA-蛋白质相互作用,提供蛋白前体的折叠和修饰 Identification of DNA-protein interaction with folding and modification of protein precursors |
非酵母体系中准确率低 Low accuracy in the non-yeast system |
| 染色质免疫沉淀技术 ChIP |
显示DNA-蛋白质在体内动态作用情况,确定靶基因结合位点 Determination of dynamic action between DNA and protein in vivo and identification of the binding sites of target genes |
难以获得特异性蛋白质抗体;调控蛋白的基因限制在特定来源 Difficult to obtain specific protein antibody and limited sources of specific gene for regulator |
| DNA亲和层析技术 DNA affinity chromatography |
活性物质纯度高,性质稳定,步骤简单有效 High purity of active substances, property stability, and simple operation |
配基要求高,不可避免非特异性结合 High requirements for ligand and inevitable non-specific binding |
| RNA-seq | 全基因组水平的基因表达差异研究,定量准确、可重复性高、检测范围广 Gene expression differences at the genome level, quantitative accuracy, high repeatability, and wide-range detection |
存在核糖体RNA影响,不能直观体现相互作用 Ribosomal RNA interference and cannot directly reflect the interaction |
| 噬菌体展示技术 Phage display |
大量快速检测、蛋白结构和活性稳定 Fast and simultaneous test, stability of protein structure and activity |
噬菌体文库的容量和遗传多样性有限制 Limited capacity and genetic diversity of phage library |
| 荧光素酶实验 Luciferase assay |
靶动子和调控因子作用过程光信号强、信噪比高,同时分析多个信号转导通路 Strong light signal and SNR for promoter-regulator interaction, multiple analysis of signal transduction pathways |
仅适用于转录激活检测,无法检测转录抑制 Only suitable for transcriptional activation detection, inapplicable for transcriptional inhibition |
| 生物膜干涉技术 Biolayer interferometry |
利用光干涉原理检测小分子间相互作用,灵敏度高,样本容量大 Detection of molecular interaction based on light interference, large sample size |
样本纯度要求严格,易受非特异性结合影响 Limited sample purity, and easily affected by non-specific binding |
| 扫描探针显微技术 Scanning probe microscope |
原子级分辨率检测调控因子与靶基因作用,准确率高 Detection of DNA-regulator by atomic resolution, high accuracy |
检测环境和样本纯度要求高,不适用于大样本筛选 High requirements on test environment and sample purity, and unsuitable for large sample screening |
| 凝胶阻滞实验 EMSA |
体外快速研究DNA与蛋白质相互作用,特异性强 Rapid detection in vitro, strong specificity |
不能真实反映体内情况,不能确定靶序列,结论单一 Cannot reflect the situation in vivo and identify the target motif |
染色质免疫沉淀技术(Chromatin Immunoprecipitation Assay,ChIP)在体内研究调控因子-靶DNA相互作用,与微阵列芯片(ChIP-on-chip)或高通量测序(ChIP-seq)结合,能准确分析细菌内调控因子的靶基因位点,构建基因表达调控网络。Partridge等[54]利用ChIP-on-chip分析得到大肠杆菌调控因子NsrR的结合位点(AANATGCATTT),该位点存在于细胞膜发育基因mqsR-ygiT的启动子区域,潜在调控EPS合成。ChIP-seq识别伤寒沙门氏菌(Salmonella typhi)与渗透压响应调控因子OmpR相互作用的靶点,其中包括gltA (柠檬酸合酶基因)、sdhC (琥珀酸脱氢酶基因)和tviA (Vi多糖生物合成蛋白基因)等多个与EPS生物合成相关基因[55]。在假单胞菌中,ChIP-seq证明全局调节因子AlgR不仅直接调控海藻酸钠合成和毒力因子表达,还能与c-di-GMP相互作用间接调控EPS生物合成[56]。
2.2 DNA亲和层析技术DNA亲和层析能分离与特定DNA序列作用的蛋白质,利用细菌eps基因启动子片段可寻找未知的EPS调控因子[57]。Wu等[58]在肺炎链球菌(S. pneumoniae) D39中用5′生物素化的荚膜多糖基因簇启动子cpsp亲和层析,筛选得到6个候选调控因子,其中CpsR通过结合cps基因簇阻遏荚膜多糖的生物合成。在金黄色葡萄球菌(Staphylococcus aureus)中,生物素化的psm (酚溶性模块蛋白基因,参与细胞膜形成)操纵子亲和层析筛选出调控蛋白MgrA,MgrA通过抑制psm表达阻遏生物膜形成,潜在调控EPS合成[59]。在铜绿假单胞菌中,细胞分裂基因ftsZ启动子区域亲和层析得到调控因子LexA,并通过DNase I足迹法分析出其DNA结合位点(LexA Box),该位点存在于多个EPS合成基因中[60]。
2.3 凝胶阻滞实验凝胶阻滞实验(Electrophoretic Mobility Shift Assay,EMSA)可以直观显示调控因子和eps基因的相互作用,灵敏度高且特异性强。Zhou等[61]通过EMSA证明调控因子OpaR和AphA与副溶血弧菌(V. parahaemolyticus)荚膜多糖基因cpsQ启动子区域有特异性结合。利用EMSA证明全局调控因子CcpA能与肺炎链球菌cps启动子区域特异性结合,参与调控细菌EPS[62]。转录抑制因子NigR经EMSA表明可特异性结合变形链球菌中糖转运和代谢相关基因,潜在调控细菌EPS[63]。
3 问题和展望目前细菌EPS转录调控研究仍有较大的局限性,已报道的调控因子多集中在肺炎链球菌、金黄色葡萄球菌和铜绿假单胞菌等毒力强的致病菌中[33, 64-66],而在乳酸菌、地衣芽孢杆菌(B. licheniformis)和枯草芽孢杆菌(B. subtilis)等益生菌中鲜有报道[67-69],而且鉴定的多为荚膜多糖调控因子,粘液多糖调控因子的报道较少。此外,细菌中还存在大量EPS潜在或未知的调控因子亟待研究。例如,在乳杆菌中发现的蔗糖代谢调节因子ScrR[70]和中枢糖酵解基因调节因子CggR[71]可能与EPS生物合成相关,但并未实验证实。我们在干酪乳杆菌和嗜热链球菌EPS转录调控研究中,利用其eps启动子区域DNA亲和层析发现多个不同类型的潜在调控因子,包含未知调控因子和尚未证明与EPS合成相关的调控因子,因此还需利用EMSA和DNAase I Footprinting等体内和体外技术方法进一步证实和研究其调控机制。将荧光素酶实验、生物膜干涉技术和扫描探针显微技术等分子生物学方法应用于EPS调控因子研究,有望鉴定出更多新颖的细菌EPS调控因子,为深入解析EPS生物合成调控网络奠定基础。
| [1] |
Anadón A, Martínez-Larrañaga MR, Arés I, Martínez MA. Prebiotics and probiotics: an assessment of their safety and health benefits[A]//Watson RR, Preedy VR. Probiotics, Prebiotics, and Synbiotics[M]. London: Academic Press, 2016: 3-23
|
| [2] |
Dong YQ, Tuo YF, Mu GQ, Jiang SJ, Qian F, Song YL. Screening of exopolysaccharide-producing Lactobacillus strains and study of exopolysaccharide properties[J]. Modern Food Science & Technology, 2017, 33(2): 61-68. (in Chinese) 董阳勤, 妥彦峰, 牟光庆, 姜淑娟, 钱方, 宋莹龙. 产胞外多糖乳杆菌的筛选及多糖功能的研究[J]. 现代食品科技, 2017, 33(2): 61-68. |
| [3] |
Zivkovic M, Miljkovic M, Ruas-Madiedo P, Strahinic I, Tolinacki M, Golic N, Kojic M. Exopolysaccharide production and ropy phenotype are determined by two gene clusters in putative probiotic strain Lactobacillus paraplantarum BGCG11[J]. Applied and Environmental Microbiology, 2015, 81(4): 1387-1396. DOI:10.1128/AEM.03028-14 |
| [4] |
Ale EC, Perezlindo MJ, Pavón Y, Peralta GH, Costa S, Sabbag N, Bergamini C, Reinheimer JA, Binetti AG. Technological, rheological and sensory characterizations of a yogurt containing an exopolysaccharide extract from Lactobacillus fermentum Lf2, a new food additive[J]. Food Research International, 2016, 90: 259-267. DOI:10.1016/j.foodres.2016.10.045 |
| [5] |
Zhang H, Ren W, Guo QB, Xiong ZQ, Wang GQ, Xia YJ, Lai P, Yin BX, Ai LZ. Characterization of a yogurt-quality improving exopolysaccharide from Streptococcus thermophilus AR333[J]. Food Hydrocolloids, 2018, 81: 220-228. DOI:10.1016/j.foodhyd.2017.12.017 |
| [6] |
Kong LH, Zhao LS, Xia YJ, Zhang H, Ai LZ, Xiong ZQ. Research advance in the exopolysaccharide biosynthesis of Streptococcus thermophilus[J]. Journal of Food Safety and Quality, 2019, 10(2): 284-290. (in Chinese) 孔令慧, 赵林森, 夏永军, 张汇, 艾连中, 熊智强. 嗜热链球菌胞外多糖生物合成的研究进展[J]. 食品安全质量检测学报, 2019, 10(2): 284-290. DOI:10.3969/j.issn.2095-0381.2019.02.002 |
| [7] |
Schmid J. Recent insights in microbial exopolysaccharide biosynthesis and engineering strategies[J]. Current Opinion in Biotechnology, 2018, 53: 130-136. DOI:10.1016/j.copbio.2018.01.005 |
| [8] |
Lebellenger L, Verrez-bagnis V, Passerini D, Delbarre-Ladrat C. Comparative genomics reveals a widespread distribution of an exopolysaccharide biosynthesis gene cluster among Vibrionaceae[J]. BMC Research Notes, 2018, 11(1): 102. DOI:10.1186/s13104-018-3214-z |
| [9] |
Song X, Xiong ZQ, Kong LH, Wang GQ, Ai LZ. Relationship between putative eps genes and production of exopolysaccharide in Lactobacillus casei LC2W[J]. Frontiers in Microbiology, 2018, 9: 1882. DOI:10.3389/fmicb.2018.01882 |
| [10] |
Xiong ZQ, Kong LH, Meng HL, Cui JM, Xia YJ, Wang SJ, Ai LZ. Comparison of gal-lac operons in wild-type galactose-positive and -negative Streptococcus thermophilus by genomics and transcription analysis[J]. Journal of Industrial Microbiology & Biotechnology, 2019, 46(5): 751-758. |
| [11] |
Xiong ZQ, Kong LH, Lai PFH, Xia YJ, Liu JC, Li QY, Ai LZ. Genomic and phenotypic analyses of exopolysaccharide biosynthesis in Streptococcus thermophilus S-3[J]. Journal of Dairy Science, 2019, 102(6): 4925-4934. DOI:10.3168/jds.2018-15572 |
| [12] |
Kong LH, Xiong ZQ, Song X, Xia YJ, Zhang N, Ai LZ. Characterization of a panel of strong constitutive promoters from Streptococcus thermophilus for fine-tuning gene expression[J]. ACS Synthetic Biology, 2019, 8(6): 1469-1472. DOI:10.1021/acssynbio.9b00045 |
| [13] |
Ikawa Y, Tsuge S. The quantitative regulation of the hrp regulator HrpX is involved in sugar-source-dependent hrp gene expression in Xanthomonas oryzae pv. oryzae[J]. FEMS Microbiology Letters, 2016, 363(10): fnw071. DOI:10.1093/femsle/fnw071 |
| [14] |
Elmas A, Wang XD, Samoilov MS. Reconstruction of novel transcription factor regulons through inference of their binding sites[J]. BMC Bioinformatics, 2015, 16: 299. DOI:10.1186/s12859-015-0685-y |
| [15] |
Subhadra B, Kim J, Kim DH, Woo K, Oh MH, Choi CH. Local repressor AcrR regulates AcrAB efflux pump required for biofilm formation and virulence in Acinetobacter nosocomialis[J]. Frontiers in Cellular and Infection Microbiology, 2018, 8: 270. DOI:10.3389/fcimb.2018.00270 |
| [16] |
Kleinman CL, Sycz G, Bonomi HR, Rodríguez RM, Zorreguieta A, Sieira R. ChIP-Seq analysis of the LuxR-type regulator VjbR reveals novel insights into the Brucella virulence gene expression network[J]. Nucleic Acids Research, 2017, 45(10): 5757-5769. DOI:10.1093/nar/gkx165 |
| [17] |
Licciardello G, Caruso A, Bella P, Gheleri R, Strano CP, Anzalone A, Trantas EA, Sarris PF, Almeida NF, Catara V. The LuxR regulators PcoR and RfiA co-regulate antimicrobial peptide and alginate production in Pseudomonas corrugata[J]. Frontiers in Microbiology, 2018, 9: 521. DOI:10.3389/fmicb.2018.00521 |
| [18] |
Liu JL, Stone VN, Ge XC, Tang M, Elrami F, Xu P. TetR family regulator BrpT modulates biofilm formation in Streptococcus sanguinis[J]. PLoS One, 2017, 12(1): e0169301. DOI:10.1371/journal.pone.0169301 |
| [19] |
Taylor DL, Ante VM, Bina XR, Howard MF, Bina JE. Substrate-dependent activation of the Vibrio cholerae vexAB RND efflux system requires vexR[J]. PLoS One, 2015, 10(2): e0117890. DOI:10.1371/journal.pone.0117890 |
| [20] |
Taw MN, Lee HI, Lee SH, Chang WS. Characterization of MocR, a GntR-like transcriptional regulator, in Bradyrhizobium japonicum: its impact on motility, biofilm formation, and soybean nodulation[J]. Journal of Microbiology, 2015, 53(8): 518-525. DOI:10.1007/s12275-015-5313-z |
| [21] |
Tsypik O, Makitrynskyy R, Bera A, Song LJ, Wohlleben W, Fedorenko V, Ostash B. Role of GntR family regulatory gene SCO1678 in gluconate metabolism in Streptomyces coelicolor M145[J]. BioMed Research International, 2017, 2017: 9529501. |
| [22] |
Fillenberg SB, Grau FC, Seidel G, Muller YA. Structural insight into operator dre-sites recognition and effector binding in the GntR/HutC transcription regulator NagR[J]. Nucleic Acids Research, 2015, 43(2): 1283-1296. DOI:10.1093/nar/gku1374 |
| [23] |
Kuge T, Teramoto H, Inui M, Zhulin IB. AraR, an L-arabinose-responsive transcriptional regulator in Corynebacterium glutamicum ATCC 31831, exerts different degrees of repression depending on the location of its binding sites within the three target promoter regions[J]. Journal of Bacteriology, 2015, 197(24): 3788-3796. DOI:10.1128/JB.00314-15 |
| [24] |
Wilson CM, Klingeman DM, Schlachter C, Syed MH, Wu CW, Guss AM, Brown SD, Parales RE. LacI transcriptional regulatory networks in Clostridium thermocellum DSM1313[J]. Applied and Environmental Microbiology, 2016, 83(5): e02751-16. |
| [25] |
Kaznadzey A, Shelyakin P, Belousova E, Aleksandra E, Shvyreva U, Bykova D, Emelianenko V, Korosteleva A, Tutukina M, Gelfand MS. The genes of the sulphoquinovose catabolism in Escherichia coli are also associated with a previously unknown pathway of lactose degradation[J]. Scientific Reports, 2018, 8(1): 3177. DOI:10.1038/s41598-018-21534-3 |
| [26] |
Hirooka K, Kodoi Y, Satomura T, Fujita Y. Regulation of the rhaEWRBMA operon involved in L-rhamnose catabolism through two transcriptional factors, RhaR and CcpA, in Bacillus subtilis[J]. Journal of Bacteriology, 2016, 198(5): 830-845. DOI:10.1128/JB.00856-15 |
| [27] |
Turner SE, Pang YY, O'Malley MR, Weisberg AJ, Fraser VN, Yan Q, Chang JH, Anderson JC. A DeoR-Type transcription regulator is required for sugar-induced expression of type Ⅲ secretion-encoding genes in Pseudomonas syringae pv. tomato DC3000[J]. Molecular Plant-Microbe Interactions, 2020, 33(3): 509-518. DOI:10.1094/MPMI-10-19-0290-R |
| [28] |
Sai R, Li QM, Xie LX, Xie JP. Molecular mechanisms underlying the function diversity of ArsR family metalloregulator[J]. Critical Reviews in Eukaryotic Gene Expression, 2017, 27(1): 19-35. DOI:10.1615/CritRevEukaryotGeneExpr.2016018476 |
| [29] |
Servetas SL, Carpenter BM, Haley KP, Gilbreath JJ, Gaddy JA, Merrell DS, Silhavy TJ. Characterization of key Helicobacter pylori regulators identifies a role for ArsRS in biofilm formation[J]. Journal of Bacteriology, 2016, 198(18): 2536-2548. DOI:10.1128/JB.00324-16 |
| [30] |
Li QM, Li CY, Xie LX, Zhang CH, Feng YH, Xie JP. Characterization of a putative ArsR transcriptional regulator encoded by Rv2642 from Mycobacterium tuberculosis[J]. Journal of Biomolecular Structure and Dynamics, 2017, 35(9): 2031-2039. DOI:10.1080/07391102.2016.1206037 |
| [31] |
Mao DN, Bushin LB, Moon K, Wu YH, Seyedsayamdost MR. Discovery of ScmR as a global regulator of secondary metabolism and virulence in Burkholderia thailandensis E264[J]. Proceedings of the National Academy of Sciences of the United States of America, 2017, 114(14): E2920-E2928. DOI:10.1073/pnas.1619529114 |
| [32] |
Park H, Do E, Kim M, Park HJ, Lee J, Han SW. A LysR-type transcriptional regulator LcrX is involved in virulence, biofilm formation, swimming motility, siderophore secretion, and growth in sugar sources in Xanthomonas axonopodis pv. glycines[J]. Frontiers in Plant Science, 2019, 10: 1657. |
| [33] |
Yang XJ, Zhang ZQ, Huang ZW, Zhang XX, Li DH, Sun L, You JJ, Pan XW, Yang HJ. A putative LysR-type transcriptional regulator inhibits biofilm synthesis in Pseudomonas aeruginosa[J]. Biofouling, 2019, 35(5): 541-550. DOI:10.1080/08927014.2019.1627337 |
| [34] |
Zhang Y, Xia L, Lin LP, Tang H, Osei-Adjei G, Xu SG, Zhang YQ, Huang XX. Reciprocal regulation of OmpR and Hfq and their regulatory actions on the Vi polysaccharide capsular antigen in Salmonella enterica Serovar Typhi[J]. Current Microbiology, 2018, 75(6): 773-778. DOI:10.1007/s00284-018-1447-7 |
| [35] |
Tipton KA, Rather PN. An ompR-envZ two-component system ortholog regulates phase variation, osmotic tolerance, motility, and virulence in Acinetobacter baumannii strain AB5075[J]. Journal of Bacteriology, 2017, 199(3): e00705-16. |
| [36] |
Charoenpanich P, Meyer S, Becker A, Matthew M. Temporal expression program of quorum sensing-based transcription regulation in Sinorhizobium meliloti[J]. Journal of Bacteriology, 2013, 195(14): 3224-3236. DOI:10.1128/JB.00234-13 |
| [37] |
Solano C, Echeverz M, Iñigo L. Biofilm dispersion and quorum sensing[J]. Current Opinion in Microbiology, 2014, 18: 96-104. DOI:10.1016/j.mib.2014.02.008 |
| [38] |
Haag AF, Bagnoli F. The role of two-component signal transduction systems in Staphylococcus aureus virulence regulation[A]//Bagnoli F, Rappuoli R, Grandi G. Staphylococcus Aureus[M]. Cham: Springer, 2017: 145-198
|
| [39] |
Wei CF, Tsai YH, Tsai SH, Lin CS, Chang CJ, Lu CC, Huang HC, Lai HC. Cross-talk between bacterial two-component systems drives stepwise regulation of flagellar biosynthesis in swarming development[J]. Biochemical and Biophysical Research Communications, 2017, 489(1): 70-75. DOI:10.1016/j.bbrc.2017.05.077 |
| [40] |
Black WP, Wang LL, Davis MY, Yang ZM. The orphan response regulator EpsW is a substrate of the DifE kinase and it regulates exopolysaccharide in Myxococcus xanthus[J]. Scientific Reports, 2016, 5: 17831. DOI:10.1038/srep17831 |
| [41] |
Pannen D, Fabisch M, Gausling L, Karin S. Interaction of the RcsB response regulator with auxiliary transcription regulators in Escherichia coli[J]. The Journal of Biological Chemistry, 2016, 291(5): 2357-2370. DOI:10.1074/jbc.M115.696815 |
| [42] |
Zhou X, Zhang N, Xia LM, Li Q, Shao JH, Shen QR, Zhang RF. ResDE two-component regulatory system mediates oxygen limitation-induced biofilm formation by Bacillus amyloliquefaciens SQR9[J]. Applied and Environmental Microbiology, 2018, 84(8): e02744-17. |
| [43] |
Monedero V, Revilla-Guarinos A, Zúñiga M. Physiological role of two-component signal transduction systems in food-associated lactic acid bacteria[J]. Advances in Applied Microbiology, 2017, 99: 1-51. |
| [44] |
Minic Z, Marie C, Delorme C, Faurie JM, Mercier G, Ehrlich D, Renault P. Control of EpsE, the phosphoglycosyltransferase initiating exopolysaccharide synthesis in Streptococcus thermophilus, by EpsD tyrosine kinase[J]. Journal of Bacteriology, 2007, 189(4): 1351-1357. DOI:10.1128/JB.01122-06 |
| [45] |
Song X, Huang H, Xiong ZQ, Ai LZ, Yang S. CRISPR-Cas9D10A nickase-assisted genome editing in Lactobacillus casei[J]. Applied and Environmental Microbiology, 2017, 83(22): e01259-17. |
| [46] |
Schäper S, Steinchen W, Krol E, Altegoer F, Skotnicka D, Sogaard-Andersen L, Bange G, Becker A. AraC-like transcriptional activator CuxR binds c-di-GMP by a PilZ-like mechanism to regulate extracellular polysaccharide production[J]. Proceedings of the National Academy of Sciences of the United States of America, 2017, 114(24): E4822-E4831. DOI:10.1073/pnas.1702435114 |
| [47] |
Xiao YJ, Nie HL, Liu HZ, Luo XS, Chen WL, Huang QY. c-di-GMP regulates the expression of lapA and bcs operons via FleQ in Pseudomonas putida KT2440[J]. Environmental Microbiology Reports, 2016, 8(5): 659-666. DOI:10.1111/1758-2229.12419 |
| [48] |
Baraquet C, Harwood CS. FleQ DNA binding consensus sequence revealed by studies of FleQ-dependent regulation of biofilm gene expression in Pseudomonas aeruginosa[J]. Journal of Bacteriology, 2015, 198(1): 178-186. |
| [49] |
Fazli M, Rybtke M, Steiner E, Weidel E, Berthelsen J, Groizeleau J, Wu B, Zhi BZ, Zhang YM, Kaever V, et al. Regulation of Burkholderia cenocepacia biofilm formation by RpoN and the c-di-GMP effector BerB[J]. MicrobiologyOpen, 2017, 6(4): e00480. DOI:10.1002/mbo3.480 |
| [50] |
Peng X, Zhang Y, Bai GC, Zhou XD, Wu H. Cyclic di-AMP mediates biofilm formation[J]. Molecular Microbiology, 2016, 99(5): 945-959. DOI:10.1111/mmi.13277 |
| [51] |
Cheng XQ, Zheng X, Zhou XD, Zeng JM, Ren Z, Xu X, Cheng L, Li MY, Li JY, Li YQ. Regulation of oxidative response and extracellular polysaccharide synthesis by a diadenylate cyclase in Streptococcus mutans[J]. Environmental Microbiology, 2016, 18(3): 904-922. DOI:10.1111/1462-2920.13123 |
| [52] |
Rismondo J, Gibhardt J, Rosenberg J, Kaever V, Halbedel S, Commichau FM. Phenotypes associated with the essential diadenylate cyclase CdaA and its potential regulator CdaR in the human pathogen Listeria monocytogenes[J]. Journal of Bacteriology, 2016, 198(3): 416-426. DOI:10.1128/JB.00845-15 |
| [53] |
Moscoso JA, Schramke H, Zhang Y, Tosi T, Dehbi A, Jung K, Gründling A. Binding of cyclic di-AMP to the Staphylococcus aureus sensor kinase KdpD occurs via the universal stress protein domain and downregulates the expression of the Kdp potassium transporter[J]. Journal of Bacteriology, 2016, 198(1): 98-110. DOI:10.1128/JB.00480-15 |
| [54] |
Partridge JD, Bodenmiller DM, Humphrys MS, Spiro S. NsrR targets in the Escherichia coli genome: new insights into DNA sequence requirements for binding and a role for NsrR in the regulation of motility[J]. Molecular Microbiology, 2009, 73(4): 680-694. DOI:10.1111/j.1365-2958.2009.06799.x |
| [55] |
Perkins TT, Davies MR, Klemm EJ, Rowley G, Wileman T, James K, Keane T, Maskell D, Hinton JCD, Dougan G, et al. ChIP-seq and transcriptome analysis of the OmpR regulon of Salmonella enterica serovars Typhi and Typhimurium reveals accessory genes implicated in host colonization[J]. Molecular Microbiology, 2013, 87(3): 526-538. DOI:10.1111/mmi.12111 |
| [56] |
Kong WN, Zhao JR, Kang HP, Zhu M, Zhou TH, Deng X, Liang HH. ChIP-seq reveals the global regulator AlgR mediating cyclic di-GMP synthesis in Pseudomonas aeruginosa[J]. Nucleic Acids Research, 2015, 43(17): 8268-8282. DOI:10.1093/nar/gkv747 |
| [57] |
Zhang QH, Huang Q, Fang Q, Li HT, Tang H, Zou G, Wang D, Li SQ, Bei WC, Chen HC, et al. Identification of genes regulated by the two-component system response regulator NarP of Actinobacillus pleuropneumoniae via DNA-affinity-purified sequencing[J]. Microbiological Research, 2020, 230: 126343. DOI:10.1016/j.micres.2019.126343 |
| [58] |
Wu KF, Xu HM, Zheng YQ, Wang LB, Zhang XM, Yin YB. CpsR, a GntR family regulator, transcriptionally regulates capsular polysaccharide biosynthesis and governs bacterial virulence in Streptococcus pneumoniae[J]. Scientific Reports, 2016, 6: 29255. DOI:10.1038/srep29255 |
| [59] |
Jiang Q, Jin ZY, Sun BL. MgrA negatively regulates biofilm formation and detachment by repressing the expression of psm operons in Staphylococcus aureus[J]. Applied and Environmental Microbiology, 2018, 84(16): e01008-18. |
| [60] |
Honda T, Morimoto D, Sako Y, Yoshida T. LexA binds to transcription regulatory site of cell division gene ftsZ in toxic cyanobacterium Microcystis aeruginosa[J]. Marine Biotechnology, 2018, 20(4): 549-556. DOI:10.1007/s10126-018-9826-4 |
| [61] |
Zhou DS, Yan XJ, Qu F, Wang L, Zhang YQ, Hou J, Hu Y, Li J, Xin SJ, Qiu JF, et al. Quorum sensing modulates transcription of cpsQ-mfpABC and mfpABC in Vibrio parahaemolyticus[J]. International Journal of Food Microbiology, 2013, 166(3): 458-463. DOI:10.1016/j.ijfoodmicro.2013.07.008 |
| [62] |
Wang LB, Xu HM, Wu KF, Zheng YQ, Wang JM, Ma F, Zhang XM, Yin YB, Zhang Q. Regulation effect of CcpA protein on the biosynthesis of capsular polysaccharide in Streptococcus pneumoniae[J]. Acta Microbiologica Sinica, 2015, 55(6): 732-738. (in Chinese) 王丽滨, 徐红梅, 吴凯峰, 郑玉强, 王建敏, 马峰, 张雪梅, 尹一兵, 张群. 肺炎链球菌糖代谢蛋白CcpA对荚膜多糖的调控作用[J]. 微生物学报, 2015, 55(6): 732-738. |
| [63] |
Vujanac M, Iyer VS, Sengupta M, Ajdic D. Regulation of Streptococcus mutans PTSBio by the transcriptional repressor NigR[J]. Molecular Oral Microbiology, 2015, 30(4): 280-294. DOI:10.1111/omi.12093 |
| [64] |
Peng D, Li X, Liu P, Zhou XP, Luo M, Su KW, Chen S, Zhang ZS, He Q, Qiu JF, et al. Transcriptional regulation of galF by RcsAB affects capsular polysaccharide formation in Klebsiella pneumoniae NTUH-K2044[J]. Microbiological Research, 2018, 216: 70-78. DOI:10.1016/j.micres.2018.08.010 |
| [65] |
Yang Y, Luo MJ, Zhou HK, Li C, Luk A, Zhao GP, Fung K, IP M. Role of two-component system response regulator BceR in the antimicrobial resistance, virulence, biofilm formation, and stress response of group B Streptococcus[J]. Frontiers in Microbiology, 2019, 10: 10. DOI:10.3389/fmicb.2019.00010 |
| [66] |
Huertas-Rosales O, Romero M, Heeb S, Espinosa-Urgel M, Cámara M, Ramos-González MI. The Pseudomonas putida CsrA/RsmA homologues negatively affect c-di-GMP pools and biofilm formation through the GGDEF/EAL response regulator CfcR[J]. Environmental Microbiology, 2017, 19(9): 3551-3566. DOI:10.1111/1462-2920.13848 |
| [67] |
Vastano V, Perrone F, Marasco R, Sacco M, Muscariello L. Transcriptional analysis of exopolysaccharides biosynthesis gene clusters in Lactobacillus plantarum[J]. Archives of Microbiology, 2016, 198(3): 295-300. DOI:10.1007/s00203-015-1169-1 |
| [68] |
Li BL, Ding XY, Evivie SE, Jin D, Meng YY, Huo GC, Liu F. Short communication: genomic and phenotypic analyses of exopolysaccharides produced by Streptococcus thermophilus KLDS SM[J]. Journal of Dairy Science, 2018, 101(1): 106-112. DOI:10.3168/jds.2017-13534 |
| [69] |
Dertli E, Mayer MJ, Colquhoun IJ, Narbad A. EpsA is an essential gene in exopolysaccharide production in Lactobacillus johnsonii FI9785[J]. Microbial Biotechnology, 2016, 9(4): 496-501. DOI:10.1111/1751-7915.12314 |
| [70] |
Teixeira JS, Abdi R, Su MSW, Schwab C, Gänzle MG. Functional characterization of sucrose phosphorylase and scrR, a regulator of sucrose metabolism in Lactobacillus reuteri[J]. Food Microbiology, 2013, 36(2): 432-439. DOI:10.1016/j.fm.2013.07.011 |
| [71] |
Rud I, Naterstad K, Bongers RS, Molenaar D, Kleerebezem M, Axelsson L. Functional analysis of the role of CggR (central glycolytic gene regulator) in Lactobacillus plantarum by transcriptome analysis[J]. Microbial Biotechnology, 2011, 4(3): 345-356. DOI:10.1111/j.1751-7915.2010.00223.x |
 2021, Vol. 48
2021, Vol. 48