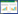扩展功能
文章信息
- 叶晗, 李啸, 张小龙, 肖泽涛, 许超群, 黄聪
- YE Han, LI Xiao, ZHANG Xiaolong, XIAO Zetao, XU Chaoqun, HUANG Cong
- 基于转录组学分析的丙酸钙对酿酒酵母的抑菌机制
- Antimicrobial mechanism of calcium propionate on Saccharomyces cerevisiae based on transcriptomics analysis
- 微生物学通报, 2021, 48(2): 437-448
- Microbiology China, 2021, 48(2): 437-448
- DOI: 10.13344/j.microbiol.china.200244
-
文章历史
- 收稿日期: 2020-03-16
- 接受日期: 2020-04-02
- 网络首发日期: 2020-06-01
2. 湖北省酵母功能重点实验室 安琪酵母股份有限公司 湖北 宜昌 443003
2. Hubei Provincial Key Laboratory of Yeast Function, Angel Yeast Company Limited, Yichang, Hubei 443003, China
耐高糖酵母是一种能够在高浓度糖含量条件下生长并进行发酵作用的酿酒酵母,被广泛应用在烘焙等食品生产领域。丙酸钙作为一种安全的防腐剂常被用于面包等食品中以延长其保质期,而丙酸钙的添加除了起到抑制霉菌等杂菌生长的作用,对酵母的增殖也造成了一定的影响[1-2]。有研究报道,丙酸钙主要是通过在酸性条件下产生游离丙酸,丙酸活性分子穿过霉菌等的细胞壁,抑制胞内酶活性,起到抗菌作用[3]。细胞周期即细胞的增殖过程,细胞周期延滞则细胞增殖被抑制,乙酸毒性能导致酵母细胞内DNA损伤,而DNA损伤会诱导一系列信号转导触发细胞周期延滞[4-5];丙酸钙对洋葱根尖细胞分裂有抑制作用,能改变有丝分裂期的频率,降低细胞内核DNA含量[6]。然而,对于丙酸及丙酸盐等防腐剂对酿酒酵母作用的研究大多聚焦在细胞水平、生理生化水平或群体水平,缺乏对其更深层次的研究。因此,从分子层面上研究丙酸钙的抑菌机制,可为构建及优化耐丙酸钙菌株提供科学依据和理论指导。
转录组,即特定细胞在某一状态下所能转录的所有RNA的总和,包括信使RNA和非编码RNA,转录组学分析已被广泛用于揭示特定生物学过程的分子机理[7]。本文利用高通量测序技术对耐高糖酵母BH1进行转录组测序分析,并用实时荧光定量PCR (qRT-PCR)对结果进行验证,以期在分子水平上探究丙酸钙抑菌作用的机制。转录组测序原始数据保存在NCBI的Sequence Read Archive (SRA)数据库中,序列号为SRP225499。
1 材料与方法 1.1 菌种及培养基耐高糖酵母BH1,由安琪酵母股份有限公司研发中心菌种与分子生物技术研究室提供。菌种活化及发酵液体培养基(g/L):蔗糖100.0,酵母浸粉20.0,KH2PO4 1.0,MgSO4·7H2O 1.0。
1.2 主要试剂和仪器总RNA提取试剂盒,生工生物工程(上海)股份有限公司;PrimeScriptTM RT-PCR试剂盒,宝生物工程(大连)有限公司;SYBR Green RT-PCR试剂盒,南京诺唯赞生物科技有限公司。紫外可见分光光度计,上海光谱仪器有限公司;FUS-50 L (A)发酵罐,上海国强生化工程装备有限公司;HiSeq X Ten高通量测序仪,Illumina公司;实时荧光PCR检测系统,Bio-Rad公司。
1.3 种子活化和批次发酵将斜面中保存的耐高糖酵母BH1接种到初始pH 4.8的液体培养基中,30 ℃、180 r/min培养24 h,随后按照接种量10%、30 ℃、200 r/min、0.035 kPa、通气量400 mL/h条件下培养18 h得到二级种子。50 L发酵罐中批次发酵条件:接种量10%、30 ℃、200 r/min、0.035 kPa、通气量2.4 L/h、初始pH 4.8,在对数期(4 h)向实验组加入丙酸钙后(实验组丙酸钙质量分数为0.25%),控制实验组和对照组pH维持在4.35。用紫外可见分光光度计测定600 nm波长下菌液的吸光值(OD600),并进行活细胞计数。实验重复3次,文中数据为3次的平均值。
1.4 丙酸钙胁迫条件下酵母转录组数据测序及分析在50 L发酵罐中培养耐高糖酵母BH1,取对照组6 h、实验组4 h及6 h时的菌液1.5 mL,在液氮中速冻10 min,-80 ℃保存备用。实验重复3次,共计9个样品用于测序。转录组测序工作委托生工生物工程(上海)股份有限公司高通量测序部完成。质量合格的总RNA和mRNA用于后续建库测序,测序结果的原始数据经进一步过滤后得到Clean Reads,将Clean Reads比对到参考序列酿酒酵母模式菌株S288c的基因组序列上,并进行统计分析得到样品间差异表达基因等数据。
1.5 实时荧光定量PCR (qRT-PCR)验证β-Actin作为内参基因[8]。采用PrimeScriptTM RT-PCR试剂盒对RNA进行反转录合成cDNA,进而采用实时荧光定量PCR检测系统和SYBR Green RT-PCR试剂盒进行后续实验,此过程具体反应条件及相对定量结果分析参考文献[9]。随机选取9个基因,其引物序列见表 1。
| Primers name | Primers sequence (5′→3′) |
| SPO24-F | ACTTCTGACGTTTCTCAACCT |
| SPO24-R | GAGTGTGAGCGGCTTGAAG |
| DAK2-F | GGAAACATCGTTACTCCCTACC |
| DAK2-R | CACCTCCAGAAACCAATGAAAC |
| PGI1-F | TGTCTGGTCGGCTATTGGTT |
| PGI1-R | TAGGCTGGGAATCTGTGCAA |
| TDH1-F | CAACCGTCGATGTTTCCGTT |
| TDH1-R | AATCAGAGGAGACAACGGCA |
| ENO1-F | TTCTACCGGTGTCCACGAAG |
| ENO1-R | AAGCAGCCAAAGAAACACCC |
| CWP2-F | CGTTGCTTTCGTCGCTTTG |
| CWP2-R | TTCGGTGCTGGATGGAGAAA |
| PIR3-F | CTATGCTCCAAAGGACCCGT |
| PIR3-R | CAGTAGTAGTGGCAGCCTGT |
| OPT2-F | AGGTACTGTTGATTACGCCG |
| OPT2-R | GAGTGCGATTTTCATAACCCAG |
| HSP10-F | ACCGTGTCCTTGTCCAAAG |
| HSP10-R | ACTTCTCCACGTTCTTTTCAGG |
| β-actin-F | ACTTTCAACGTTCCAGCCTTC |
| β-actin-R | CGTAAATTGGAACGACGACGTGAGTA |
在摇瓶实验中,向已发酵4 h的酵母菌液中加入不等量的丙酸钙,通过比对不同丙酸钙质量分数下耐高糖酵母BH1的生长情况,发现丙酸钙对耐高糖酵母的生长繁殖具有一定的抑制作用;随着丙酸钙质量分数的提高,其抑制作用略微增强,结果见图 1A。丙酸钙的添加也会引起培养基中渗透压的升高,由图 1B可知,丙酸钙质量分数为0.25%时培养基中的渗透压为0.604 Osm/kg,综合考虑,最终选择0.25%作为丙酸钙质量分数进行后续实验。
|
| 图 1 摇瓶实验中不同丙酸钙质量分数下耐高糖酵母BH1的生长曲线(A)及对应培养基中的渗透压(B) Figure 1 Growth curve of high-glucose resistant yeast BH1 under different mass fractions of calcium propionate in shake flask experiments (A) and osmotic pressure in corresponding medium (B) 注:CP (Calcium Propionate)表示丙酸钙;4 h时向摇瓶中加入丙酸钙,使酵母处于不同浓度的丙酸钙环境中 Note: CP means calcium propionate; At 4 h, add calcium propionate to the flask to keep the yeast in the environment of calcium propionate with different concentrations |
|
|
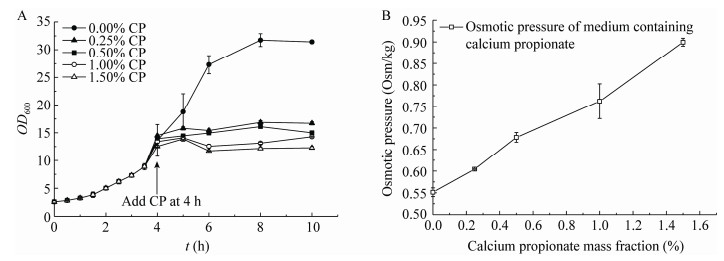
实验中耐高糖酵母BH1在50 L发酵罐中的生长曲线如图 2A所示,在空白对照组(Control Group)中,酵母在10 h左右达到稳定期,此前生物量一直保持较快增长;在4 h前,对照组和实验组在生物量上基本持平,4 h时实验组(CP Group)中添加了丙酸钙,可以看到实验组中OD600 (细胞数)的增长明显放缓,达到稳定时的生物量几乎仅为对照组的一半,丙酸钙对酵母增殖具有明显的抑制作用。
|
| 图 2 50 L发酵罐中耐高糖酵母BH1的生长曲线及细胞死亡率 Figure 2 Growth curve and cell mortality of high-glucose resistant yeast BH1 in 50 L fermentation tank 注:A:耐高糖酵母BH1在50 L发酵罐中的生长曲线(OD600和细胞数);Control group:对照组;CP group (calcium propionate group):4 h时添加丙酸钙的实验组(丙酸钙质量分数为0.25%)。B:细胞死亡率 Note: A: Growth curve (OD600 and cell counts) of high-sugar resistant yeast BH1 in a 50 L fermenter, and the CP group (calcium propionate group) is the experimental group with 0.25% calcium propionate added at 4 h; B: Cell mortality |
|
|
不过,尽管丙酸钙抑制了酵母的增殖,但其并未导致酵母细胞的死亡,对照组和实验组中细胞死亡率并未表现出明显差异,可见丙酸钙对酵母的毒害作用有限,或者说酵母细胞能够通过自身的调节适应丙酸钙带来的胁迫,但其生长繁殖却一直受到抑制。为了探究丙酸钙对酵母的抑菌机制,将经过丙酸钙处理2 h的实验组酵母与未经过处理的对照组酵母和经过丙酸钙处理0 h的实验组酵母进行转录组测序,比较各组间的基因表达差异。
2.2 差异表达基因的筛选6 h对照组(Control Group,CG)、4 h实验组(Calcium Propionate 0 h Group,CP0G)和6 h实验组(Calcium Propionate 2 h Group,CP2G) 3组中均含有3个重复,共计9组转录组测序数据。由表 2样本间相关性分析统计表可知,样本间重复性良好,CP2G与其他2组差异相对较大。
| Samples | CG1 | CG2 | CG3 | CP0G1 | CP0G2 | CP0G3 | CP2G1 | CP2G2 | CP2G3 |
| CG1 | 1.000 | 0.891 | 0.976 | 0.882 | 0.881 | 0.861 | 0.763 | 0.836 | 0.708 |
| CG2 | 0.891 | 1.000 | 0.930 | 0.965 | 0.926 | 0.971 | 0.638 | 0.781 | 0.581 |
| CG3 | 0.976 | 0.932 | 1.000 | 0.914 | 0.904 | 0.904 | 0.736 | 0.827 | 0.681 |
| CP0G1 | 0.882 | 0.965 | 0.914 | 1.000 | 0.981 | 0.987 | 0.734 | 0.832 | 0.683 |
| CP0G2 | 0.881 | 0.926 | 0.904 | 0.981 | 1.000 | 0.954 | 0.816 | 0.871 | 0.776 |
| CP0G3 | 0.861 | 0.971 | 0.904 | 0.987 | 0.954 | 1.000 | 0.682 | 0.810 | 0.625 |
| CP2G1 | 0.763 | 0.638 | 0.736 | 0.734 | 0.816 | 0.682 | 1.000 | 0.942 | 0.986 |
| CP2G2 | 0.836 | 0.781 | 0.827 | 0.832 | 0.871 | 0.810 | 0.942 | 1.000 | 0.897 |
| CP2G3 | 0.708 | 0.581 | 0.681 | 0.683 | 0.776 | 0.625 | 0.986 | 0.897 | 1.000 |
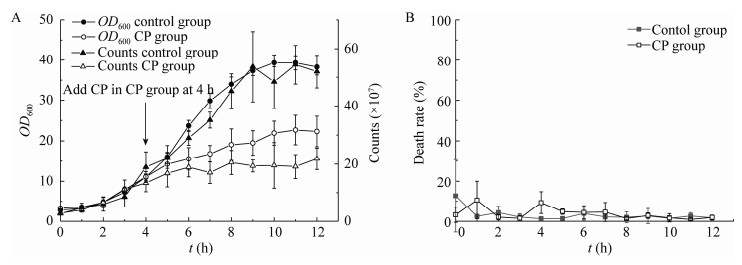
为了得到显著差异的基因,对样本采用DESeq进行分析,筛选条件设为:Q Value < 0.05且基因差异表达量倍数在2倍以上(|Log2(Fold Change)|>1)。如图 3所示,与CG相比,CP2G中有1 438个差异表达基因(Differentially Expressed Genes,DEGs),其中643个基因上调,795个基因下调。然而与CP0G相比,CP2G中共有1 921个DEGs,其中1 438个基因上调,483个基因下调。
|
| 图 3 差异基因韦恩图 Figure 3 Venn diagram of differentially expressed genes |
|
|
通过KEGG功能富集分析发现,丙酸钙会使细胞内MAPK信号途径发生显著变化,即丙酸钙胁迫条件下,细胞内一系列信号被激活转导,如表 3所示。酿酒酵母细胞中至少存在5个MAPK级联系统,它们由5种MAPK蛋白激酶控制,分别与孢子分化、菌丝形成和浸入生长、高渗透压甘油形成、细胞壁完整性及细胞结合过程有关[10]。
| Gene ID | Result | MeanTPM (CP2G) | MeanTPM (CG) | Log2(Fold change) | Q value |
| FUS3 | Up | 6.281 74 | 1.645 03 | 1.933 05 | 2.87×10-8 |
| BNI1 | Up | 33.243 86 | 8.964 49 | 1.890 80 | 1.57×10-9 |
| MTL1 | Up | 160.752 16 | 45.878 96 | 1.808 93 | 3.15×10-6 |
| ROM2 | Up | 22.383 99 | 8.691 38 | 1.364 81 | 1.78×10-10 |
| MIH1 | Up | 28.311 19 | 12.031 53 | 1.234 55 | 3.48×10-10 |
| MID2 | Up | 86.036 61 | 36.767 45 | 1.226 52 | 5.66×10-5 |
| SKM1 | Up | 34.022 86 | 15.142 27 | 1.1679 23 | 4.49×10-8 |
| KDX1 | Down | 5.892 33 | 15.296 11 | -1.376 26 | 1.15×10-5 |
| STE4 | Down | 3.520 30 | 9.768 31 | -1.472 41 | 3.87×10-9 |
| CLB5 | Down | 3.959 05 | 12.456 83 | -1.653 71 | 8.15×10-8 |
| CLB2 | Down | 8.659 02 | 31.639 47 | -1.869 45 | 1.56×10-9 |
| CLB1 | Down | 8.280 04 | 39.216 71 | -2.243 76 | 4.51×10-6 |
| CLB6 | Down | 1.520 54 | 7.860 30 | -2.370 00 | 4.29×10-4 |
| MSB2 | Down | 7.848 16 | 43.478 07 | -2.469 86 | 1.62×10-8 |
| CLN1 | Down | 7.229 71 | 48.032 25 | -2.731 99 | 1.91×10-14 |
| CLN2 | Down | 7.366 67 | 58.052 74 | -2.978 28 | 2.53×10-9 |
在MAPK信号途径中,与细胞结合过程相关的STE4、SKM1、FUS3和BNI1等基因的表达发生显著变化。其中,编码G蛋白β亚基的STE4下调表达,该亚基与Ste18p形成二聚体激活交配信号通路,与Gpa1p和Ste18p形成异源三聚体抑制信号传导,而且其可能在交配期间将Rho1p募集到极化的生长部位,在趋化性中起关键作用[11-12]。此外,SKM1、FUS3和BNI1基因均上调表达,Skm1p作为PAK丝氨酸/苏氨酸蛋白激酶家族成员,与Ste20p类似,能够通过磷酸化激活Ste11p进行信号传导,Ste11p会进一步激活Ste7p,Fus3p会被Ste7p磷酸激活,FUS3编码促分裂原活化蛋白激酶,介导信息素诱导的信号转导级联反应中的转录和G1阻滞,被激活的Fus3p能够激活Bni1p以促进极化和细胞融合,Bni1p是线状肌动蛋白丝形成的关键,参与需要极化肌动蛋白簇的细胞过程,如出芽和有丝分裂纺锤体定向[13-16]。
Rho1p同样能够激活Bni1p,其生成量并无显著差异,而Rom2p作为Rho1p和Rho2p的鸟嘌呤核苷酸交换因子能激活Rho1p[17-18],ROM2、MID2及MTL1均上调表达。MID2编码的O-糖基化质膜蛋白充当细胞壁完整性信号传导的传感器并激活该途径,且与Rom2p以及Zeo1p (细胞完整性途径蛋白)相互作用[19-20]。Mtl1p与Mid2p具有相似的结构和功能,与细胞壁完整性信号及胁迫(饥饿胁迫和氧化胁迫)响应相关[21-22],推断丙酸钙对细胞壁造成了一定影响。在细胞壁完整性过程中,下调表达的KDX1基因编码的蛋白激酶能够与许多成分相互作用,其与Rlm1相互作用还能激活酿酒酵母中响应胁迫反应的RCK1基因的表达[23-24],不过其下调表达似乎并没有影响到编码调控1, 3-β-葡聚糖合酶的FKS1、GSC2和FKS3等基因的表达。
酵母细胞壁的主要成分为1, 3-β-葡聚糖、甘露糖蛋白和几丁质,由表 4可知与细胞壁相关的CCW12、CWP2和TOS6等基因下调表达,YPS3、PIR3和YGP1等上调表达,其中CCW12编码细胞壁甘露糖蛋白,在维持新合成的细胞壁区域中发挥作用,定位于小芽的周围和大芽的隔膜区域[25-26];CWP2同样编码共价连接的细胞壁甘露糖蛋白,在稳定细胞壁方面发挥作用,还参与抵御低pH环境[27-28];TOS6编码糖基磷脂酰肌醇依赖性细胞壁蛋白,该基因表达量是周期性变化的,并且在麦角固醇扰动或进入稳定期时减少,消耗Tos6p能为细胞提供应对乳酸的抵抗力[29-32]。YPS3编码的天冬氨酸蛋白酶,通过糖基磷脂酰肌醇连接到质膜上,参与细胞壁的生长和维持[33-34];PIR3编码的O-糖基化共价结合的细胞壁蛋白,是维持细胞壁稳定性所需,其表达受细胞周期调节,并且还受到细胞完整性途径的调节[35-36];YGP1编码细胞壁相关的分泌糖蛋白,在酵母生长停滞时会被诱导表达,可能参与了细胞进入稳定期之前的适应[37-39]。值得注意的是,CCW12和CWP2基因在CP2G比CG间及CP2G比CP0G间均极显著下调表达,这2个基因或在丙酸钙抑菌作用中起重要作用。结合细胞壁完整性途径中的差异基因表达情况,推断在丙酸钙胁迫条件下,细胞壁完整性受到了一定程度的威胁甚至破坏,Mtl1p与Mid2p感受到此信号并将信号往后传导,合成甘露糖蛋白的基因表达受到严重抑制,细胞增殖受到极大抑制,而YGP1等细胞壁蛋白基因则参与维持细胞壁的稳定,保证细胞的正常生命活动不受到严重影响。
| Gene ID | Result | MeanTPM (CP2G) | MeanTPM (CG) | Log2 (Fold change) | Q value |
| CCW12 | Down | 7 643.055 | 19 654.644 | -1.362 65 | 1.64×10-2 |
| CWP2 | Down | 1 179.574 | 6 537.521 | -2.470 48 | 4.49×10-4 |
| TOS6 | Down | 22.048 | 443.400 | -4.329 84 | 4.08×10-14 |
| YPS3 | Up | 213.484 | 48.211 | 2.146 67 | 1.90×10-9 |
| PIR3 | Up | 402.357 | 32.945 | 3.610 321 | 1.86×10-17 |
| YGP1 | Up | 10 700.351 | 1 860.335 | 2.524 024 | 1.92×10-4 |
在高渗透性甘油促分裂原活化蛋白激酶(HOG-MAPK)途径中,信号粘蛋白MSB2基因下调表达,Msb2p在HOG途径中起渗透传感器的作用[40-42],推测丙酸钙并未引起高渗透压胁迫来影响细胞的正常生命活动。
2.4 涉及细胞周期途径的差异表达基因分析从MAPK信号途径中也可以发现细胞周期蛋白CLN2、CLB1和CLB2等基因下调表达。细胞周期蛋白是一类呈细胞周期特异性或时相性表达、累积与分解的蛋白质,其与细胞周期蛋白依赖性激酶共同影响细胞周期的运行。由图 4可见,与对照组CG相比,丙酸钙处理2 h的CP2G中大多细胞周期相关基因下调表达,如PCL1、CLN1、CLN2、CDC5、CDC6、CLB1、CLB5、CLB6、MCD1、IRR1、SMC3、BUB2、PDS1等。PCL1编码参与细胞周期调控的G1/S期特异性周期蛋白,与细胞周期蛋白依赖性激酶Pho85p相互作用,参与细胞生长过程中的极化生长及形态发生和进展的调控,其下调表达意味着细胞由G1期向S期转换的过程受到抑制[43-44]。细胞周期蛋白CLN1和CLN2基因的下调表达意味着其激活Cdc28p激酶,促进G1到S期转变的作用减弱;此外,Clb5p和Clb6p能够激活Cdc28p以促进DNA合成启动,在与Clb3p和Clb4p一起形成有丝分裂纺锤体中起作用,Clb1p (细胞周期蛋白)能够激活Cdc28p以促进从G2到M相的转变[45-46]。CDC5在有丝分裂期间调节细胞核的形状和核膜的扩张,而且与线粒体完整性及细胞活力有关[47-49],有丝分裂细胞中姐妹染色单体凝聚所需的粘着蛋白复合物(由MCD1、IRR1等基因编码)及多蛋白凝聚素复合物(由SMC3编码)的合成均减少[50-52]。另外,细胞周期阻止蛋白(由BUB2编码)参与形成的由GTPase激活的Bfa1p-Bub2p复合物在有丝分裂后期前可响应纺锤体和动粒体损伤,结合Tem1p和纺锤体阻断细胞周期进程[53],分离酶抑制蛋白(由PDS1基因编码)通过结合分离蛋白Esp1p能够抑制有丝分裂后期行为,形成细胞周期阻滞[54],BUB2和PDS1二者的下调表达似乎意味着进入有丝分裂中后期的细胞能够顺利甚至更快速地进入细胞周期下一阶段,参与后期核分裂的Ser/Thr激酶(由DBF20基因编码)的合成增多[55]或许也支持这一观点。值得注意的是,作为细胞周期检查点蛋白基因的MEC1上调表达,Mec1p作为细胞周期停滞和对受损或未复制DNA转录反应所需的信号转导子,其上调表达常常意味着DNA损伤[56]。
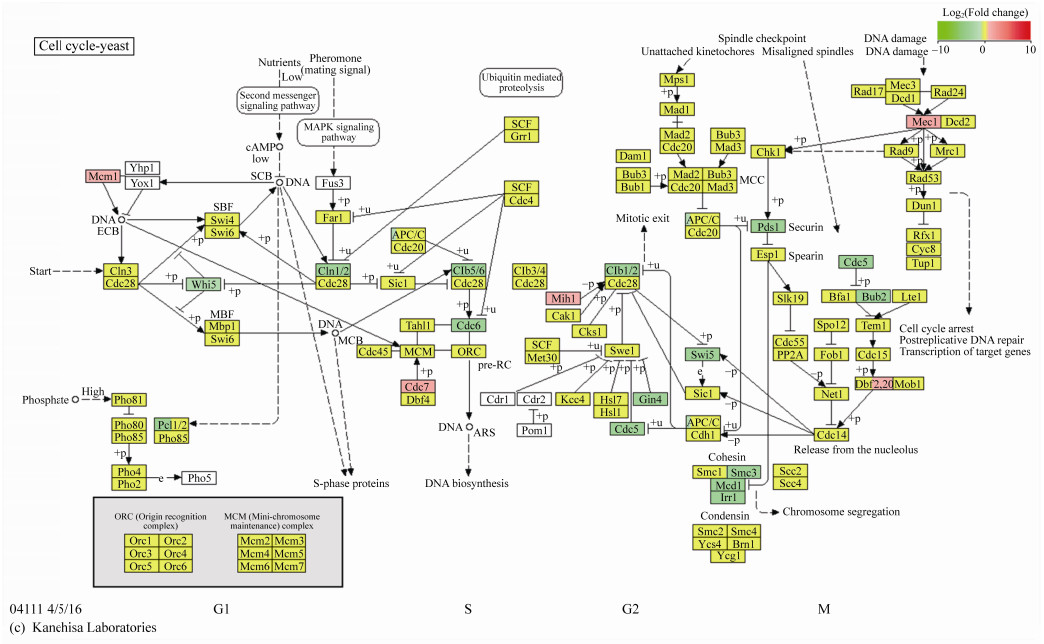
|
| 图 4 丙酸钙对酵母细胞周期过程的影响 Figure 4 Effeet of calcium propionate on yeast cell cycle |
|
|
通过比较CP2G与CP0G的细胞周期相关基因表达情况,发现仅CLB1、CLB2、CLN2及MCD1下调表达,大部分基因为上调表达。结合图 1A中酵母生长曲线判断,在丙酸钙胁迫刚出现时,细胞迅速反应,与细胞增殖相关基因显著下调表达,尤其是细胞周期中起到相间转换关键作用的细胞周期蛋白,而在后续过程中,细胞会逐步适应丙酸钙胁迫,细胞周期相关基因的表达量有所提升,但CLB1、CLB2、CLN2及MCD1等基因的下调表达还是限制了细胞增殖的过程。
2.5 涉及减数分裂途径的差异表达基因分析减数分裂作为一种特殊的细胞增殖方式,其对细胞增殖也有一定影响,与细胞周期共用一些基因,但也有其特异性的基因。MATALPHA2和HMLALPHA2均编码同源盒结构域蛋白,该蛋白能够抑制单倍体特异性基因的转录表达,HMRA2能够抑制HO内切核酸酶的表达以抑制酵母细胞交配型转化,MATALPHA2、HMLALPHA2及HMRA2均上调表达,这可能意味着单倍体孢子的合成及转化过程受到抑制[57-59]。MEK1编码减数分裂特异性丝氨酸/苏氨酸蛋白激酶,在减数分裂检查点起作用,该蛋白激酶可通过抑制姐妹染色单体之间的双链断裂修复而促进同源染色体之间的重组,也可稳定Hop1-Thr318磷酸化以促进减数分裂过程中的同源重组和检查点反应,HOP1编码染色体联会所需的减数分裂特异性蛋白[60]。MND1编码重组和减数分裂核分裂所需的蛋白质[61],与Hop2p形成复合物,后者参与染色体配对和减数分裂双链断裂的修复[62]。减数分裂特异性调节亚基(由GIP1编码)合成增多,孢子壁和隔膜蛋白的形成受到调节[63]。由SPO24编码的小蛋白质参与孢子形成,定位于孢子膜,在减数分裂过程中被磷酸化[64]。这些基因均上调表达。不过,SPS4编码不需要孢子形成但能在孢子形成过程中被诱导表达的蛋白质,其在大肠杆菌中异源表达会引起SOS反应(对DNA损伤的响应)[65],SSP1合成参与减数分裂核分裂控制的蛋白质,参与减数分裂与孢子形成的协调[66],SPS4和SSP1略微下调表达。在这些差异表达基因中,最值得注意的是SPO24,其表达量变化极显著。综合分析减数分裂途径中的差异表达基因,推断在丙酸钙条件下,减数分裂检查点作用增强,细胞内的联会同源重组过程略有增强,孢子壁的形成或有所增强。
2.6 实时荧光定量PCR (qRT-PCR)验证结果随机选取的9个差异显著的基因中,包括6个上调表达基因和3个下调表达基因。Ct (β-Actin,对照)=19.214 006±0.052 520 083,Ct (β-Actin,实验)=19.716 068 860±0.108 304 549,内参基因表达相对稳定。差异表达基因的qRT-PCR结果如图 5所示,与RNA-Seq的结果在基因表达幅度上有一定差异,但表达趋势是一致的,说明转录组测序的结果是可信的。
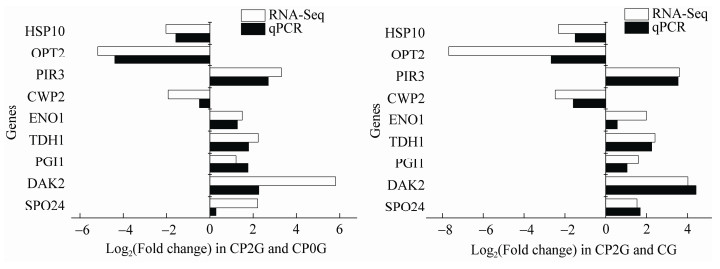
|
| 图 5 实时荧光定量PCR (qRT-PCR)验证 Figure 5 Real-time quantitative PCR (qRT-PCR) verification |
|
|
食品防腐剂的作用机理主要分3种:对细胞壁和细胞膜系统起作用;对遗传物质或遗传微粒结构起作用;对酶或功能蛋白起作用。然而酸型防腐剂主要靠未电离形式的分子聚集在细胞膜表面或进入细胞抑制胞内酶活使微生物正常代谢受阻,目前普遍认为丙酸钙对微生物的抑菌作用主要是通过丙酸抑制胞内酶活性[3]。丙酸解离常数(pKa)为4.87,在低pH条件下,丙酸进入细胞会造成胞内酸化抑制细胞生长。pH 4.0-5.0范围为酵母适宜生长条件,4 h后实验组与对照组培养基中pH均维持在4.35左右,丙酸钙抑制酵母生长的主要原因或不在于胞内酸化。转录组结果显示,在丙酸钙胁迫条件下,耐高糖酵母BH1细胞内一系列基因的表达发生变化,差异基因较多集中在信号转导机制、细胞周期和减数分裂等方面。丙酸钙对DNA合成具有抑制作用,而且能够改变有丝分裂的时相[6],这与我们的结果一致。我们发现细胞周期蛋白基因CLB1、CLB2及CLN2等的下调表达使细胞周期更多地延滞在细胞间期(G1、S和G2期),尤其是G1期,G1到S期转变过程受阻使得DNA复制也受到极大抑制,这些变化均限制了细胞的增殖,起到了一定的抑菌作用;除此之外,合成细胞壁主要成分甘露糖蛋白的基因CCW12、CWP2等的表达受到严重抑制,CCW12和CWP2或在丙酸钙抑制酵母增殖中起到重要作用,细胞壁完整性传感器Mtl1p与Mid2p感受到内外界环境刺激并进行信号转导,YGP1等细胞壁蛋白基因上调表达以维持细胞壁的稳定,保证细胞的正常生命活动不受到严重影响。除细胞壁完整性途径外,其他信号转导相关基因的差异表达也调控着细胞内的代谢过程,例如碳水化合物转运代谢、氨基酸运输代谢等,细胞内酶活性也受到影响,这或许意味着丙酸钙可能作为信号分子调控酵母的增殖过程,而对于丙酸钙的抑菌作用的更深入探究,还有待进一步分析挖掘转录组数据及开展对关键基因作用的验证。
| [1] |
O'Connell CA, Dollimore D. A study of the decomposition of calcium propionate, using simultaneous TG-DTA[J]. Thermochimica Acta, 2000, 357-358: 79-87. DOI:10.1016/S0040-6031(00)00371-3 |
| [2] |
Olson Jr JC, Macy H. Propionic acid, sodium propionate and calcium propionate as inhibitors of mold growth. I. Observations on the use of pro-pionate-treated parchment in inhibiting mold growth on the surface of butter[J]. Journal of Dairy Science, 1945, 28(9): 701-710. DOI:10.3168/jds.S0022-0302(45)95224-6 |
| [3] |
Wang JB, Gao XM, Li P, Wang XL. Inhibitory effect of calcium propionate on mold growth in foods[J]. Food and Fermentation Industries, 1984(6): 6-13. (in Chinese) 汪锦邦, 高学敏, 李平, 王秀玲. 丙酸钙对食品中霉菌生长的抑制作用[J]. 食品与发酵工业, 1984(6): 6-13. |
| [4] |
Ribeiro GF, Côrte-Real M, Johansson B. Characterization of DNA damage in yeast apoptosis induced by hydrogen peroxide, acetic acid, and hyperosmotic shock[J]. Molecular Biology of the Cell, 2006, 17(10): 4584-4591. DOI:10.1091/mbc.e06-05-0475 |
| [5] |
Amoussouvi A, Teufel L, Reis M, Seeger M, Schlichting JK, Schreiber G, Herrmann A, Klipp E. Transcriptional timing and noise of yeast cell cycle regulators: a single cell and single molecule approach[J]. npj Systems Biology and Applications, 2018, 4: 17. DOI:10.1038/s41540-018-0053-4 |
| [6] |
Türkoğlu Ş. Evaluation of genotoxic effects of sodium propionate, calcium propionate and potassium propionate on the root meristem cells of Allium cepa[J]. Food and Chemical Toxicology, 2008, 46(6): 2035-2041. DOI:10.1016/j.fct.2008.01.043 |
| [7] |
Costa V, Angelini C, De Feis I, Ciccodicola A. Uncovering the complexity of transcriptomes with RNA-Seq[J]. Journal of Biomedicine and Biotechnology, 2010, 2010: 853916. |
| [8] |
Bian J, Guo YF, Zhang Z, Zhang SH, Zhao XJ. Selection of reference genes in Saccharomyces cerevisiae under nickel stress[J]. Journal of Inner Mongolia University of Science and Technology, 2018, 37(1): 11-14, 75. (in Chinese) 边金, 郭艳飞, 张治, 张淑慧, 赵秀娟. 重金属镍胁迫下酿酒酵母实时荧光定量PCR内参基因的筛选[J]. 内蒙古科技大学学报, 2018, 37(1): 11-14, 75. |
| [9] |
Wei XW, Ma C, Xiong L, Zhang MM, Zhao XQ, Bai FW. Effect of vacuolar proteinase B on high temperature ethanol fermentation of Saccharomyces cerevisiae[J]. Microbiology China, 2015, 42(10): 1841-1846. (in Chinese) 魏小文, 马翠, 熊亮, 张明明, 赵心清, 白凤武. 液泡蛋白酶B对酿酒酵母高温乙醇发酵效率的影响[J]. 微生物学通报, 2015, 42(10): 1841-1846. |
| [10] |
Chen RE, Thorner J. Function and regulation in MAPK signaling pathways: lessons learned from the yeast Saccharomyces cerevisiae[J]. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research, 2007, 1773(8): 1311-1340. DOI:10.1016/j.bbamcr.2007.05.003 |
| [11] |
Whiteway M, Hougan L, Dignard D, Thomas DY, Bell L, Saari GC, Grant FJ, O'Hara P, MacKay VL. The STE4 and STE18 genes of yeast encode potential β and γ subunits of the mating factor receptor-coupled G protein[J]. Cell, 1989, 56(3): 467-477. DOI:10.1016/0092-8674(89)90249-3 |
| [12] |
Guo M, Aston C, Burchett SA, Dyke C, Fields S, Rajarao SJR, Uetz P, Wang YQ, Young K, Dohlman HG. The yeast G protein α subunit Gpa1 transmits a signal through an RNA binding effector protein Scp160[J]. Molecular Cell, 2003, 12(2): 517-524. DOI:10.1016/S1097-2765(03)00307-1 |
| [13] |
Elion EA, Satterberg B, Kranz JE. FUS3 phosphorylates multiple components of the mating signal transduction cascade: evidence for STE12 and FAR1[J]. Molecular Biology of the Cell, 1993, 4(5): 495-510. DOI:10.1091/mbc.4.5.495 |
| [14] |
Matheos D, Metodiev M, Muller E, Stone D, Rose MD. Pheromone-induced polarization is dependent on the Fus3p MAPK acting through the formin Bni1p[J]. Journal of Cell Biology, 2004, 165(1): 99-109. DOI:10.1083/jcb.200309089 |
| [15] |
Yu L, Qi MS, Sheff MA, Elion EA. Counteractive control of polarized morphogenesis during mating by mitogen- activated protein kinase Fus3 and G1 cyclin-dependent kinase[J]. Molecular Biology of the Cell, 2008, 19(4): 1739-1752. DOI:10.1091/mbc.e07-08-0757 |
| [16] |
Kusari AB, Molina DM, Sabbagh W, Lau CS, Bardwell L. A conserved protein interaction network involving the yeast MAP kinases Fus3 and Kss1[J]. The Journal of Cell Biology, 2004, 164(2): 267-277. DOI:10.1083/jcb.200310021 |
| [17] |
Ozaki K, Tanaka K, Imamura H, Hihara T, Kameyama T, Nonaka H, Hirano H, Matsuura Y, Takai Y. Rom1p and Rom2p are GDP/GTP exchange proteins (GEPs) for the Rho1p small GTP binding protein in Saccharomyces cerevisiae[J]. The EMBO Journal, 1996, 15(9): 2196-2207. DOI:10.1002/j.1460-2075.1996.tb00573.x |
| [18] |
Krause SA, Cundell MJ, Poon PP, McGhie J, Johnston GC, Price C, Gray JV. Functional specialisation of yeast Rho1 GTP exchange factors[J]. Journal of Cell Science, 2012, 125(11): 2721-2731. DOI:10.1242/jcs.100685 |
| [19] |
Philip B, Levin DE. Wsc1 and Mid2 are cell surface sensors for cell wall integrity signaling that act through Rom2, a guanine nucleotide exchange factor for Rho1[J]. Molecular and Cellular Biology, 2001, 21(1): 271-280. DOI:10.1128/MCB.21.1.271-280.2001 |
| [20] |
Green R, Lesage G, Sdicu AM, Ménard P, Bussey H. A synthetic analysis of the Saccharomyces cerevisiae stress sensor Mid2p, and identification of a Mid2p-interacting protein, Zeo1p, that modulates the PKC1-MPK1 cell integrity pathway[J]. Microbiology, 2003, 149(9): 2487-2499. DOI:10.1099/mic.0.26471-0 |
| [21] |
Vilella F, Herrero E, Torres J, De La Torre-Ruiz MA. Pkc1 and the upstream elements of the cell integrity pathway in Saccharomyces cerevisiae, Rom2 and Mtl1, are required for cellular responses to oxidative stress[J]. Journal of Biological Chemistry, 2005, 280(10): 9149-9159. DOI:10.1074/jbc.M411062200 |
| [22] |
Rajavel M, Philip B, Buehrer BM, Errede B, Levin DE. Mid2 is a putative sensor for cell integrity signaling in Saccharomyces cerevisiae[J]. Molecular and Cellular Biology, 1999, 19(6): 3969-3976. DOI:10.1128/MCB.19.6.3969 |
| [23] |
Breitkreutz A, Choi H, Sharom JR, Boucher L, Neduva V, Larsen B, Lin ZY, Breitkreutz BJ, Stark C, Liu GM, et al. A global protein kinase and phosphatase interaction network in yeast[J]. Science, 2010, 328(5981): 1043-1046. DOI:10.1126/science.1176495 |
| [24] |
Watanabe Y, Takaesu G, Hagiwara M, Irie K, Matsumoto K. Characterization of a serum response factor-like protein in Saccharomyces cerevisiae, Rlm1, which has transcriptional activity regulated by the Mpk1 (Slt2) mitogen-activated protein kinase pathway[J]. Molecular and Cellular Biology, 1997, 17(5): 2615-2623. DOI:10.1128/MCB.17.5.2615 |
| [25] |
Mrsa V, Ecker M, Strahl-Bolsinger S, Nimtz M, Lehle L, Tanner W. Deletion of new covalently linked cell wall glycoproteins alters the electrophoretic mobility of phosphorylated wall components of Saccharomyces cerevisiae[J]. Journal of Bacteriology, 1999, 181(10): 3076-3086. DOI:10.1128/JB.181.10.3076-3086.1999 |
| [26] |
Ragni E, Piberger H, Neupert C, García-Cantalejo J, Popolo L, Arroyo J, Aebi M, Strahl S. The genetic interaction network of CCW12, a Saccharomyces cerevisiae gene required for cell wall integrity during budding and formation of mating projections[J]. BMC Genomics, 2011, 12: 107. DOI:10.1186/1471-2164-12-107 |
| [27] |
Van Der Vaart JM, Caro LH, Chapman JW, Klis FM, Verrips CT. Identification of three mannoproteins in the cell wall of Saccharomyces cerevisiae[J]. Journal of Bacteriology, 1995, 177(11): 3104-3110. DOI:10.1128/JB.177.11.3104-3110.1995 |
| [28] |
Skrzypek M, Lester RL, Dickson RC. Suppressor gene analysis reveals an essential role for sphingolipids in transport of glycosylphosphatidylinositol-anchored proteins in Saccharomyces cerevisiae[J]. Journal of Bacteriology, 1997, 179(5): 1513-1520. DOI:10.1128/JB.179.5.1513-1520.1997 |
| [29] |
Hamada K, Fukuchi S, Arisawa M, Baba M, Kitada K. Screening for glycosylphosphatidylinositol (GPI)-dependent cell wall proteins in Saccharomyces cerevisiae[J]. Molecular and General Genetics MGG, 1998, 258(1/2): 53-59. |
| [30] |
Tu BP, Kudlicki A, Rowicka M, McKnight SL. Logic of the yeast metabolic cycle: temporal compartmentalization of cellular processes[J]. Science, 2005, 310(5751): 1152-1158. DOI:10.1126/science.1120499 |
| [31] |
Bammert GF, Fostel JM. Genome-wide expression patterns in Saccharomyces cerevisiae: comparison of drug treatments and genetic alterations affecting biosynthesis of ergosterol[J]. Antimicrobial Agents and Chemotherapy, 2000, 44(5): 1255-1265. DOI:10.1128/AAC.44.5.1255-1265.2000 |
| [32] |
Kawahata M, Masaki K, Fujii T, Iefuji H. Yeast genes involved in response to lactic acid and acetic acid: acidic conditions caused by the organic acids in Saccharomyces cerevisiae cultures induce expression of intracellular metal metabolism genes regulated by Aft1p[J]. FEMS Yeast Research, 2006, 6(6): 924-936. DOI:10.1111/j.1567-1364.2006.00089.x |
| [33] |
Olsen V, Cawley NX, Brandt J, Egel-Mitani M, Loh YP. Identification and characterization of Saccharomyces cerevisiae yapsin 3, a new member of the yapsin family of aspartic proteases encoded by the YPS3 gene[J]. Biochemical Journal, 1999, 339(2): 407-411. DOI:10.1042/bj3390407 |
| [34] |
Krysan DJ, Ting EL, Abeijon C, Kroos L, Fuller RS. Yapsins are a family of aspartyl proteases required for cell wall integrity in Saccharomyces cerevisiae[J]. Eukaryotic Cell, 2005, 4(8): 1364-1374. DOI:10.1128/EC.4.8.1364-1374.2005 |
| [35] |
Mrsă V, Seidl T, Gentzsch M, Tanner W. Specific labelling of cell wall proteins by biotinylation. Identification of four covalently linked O-mannosylated proteins of Saccharomyces cerevisiae[J]. Yeast, 1997, 13(12): 1145-1154. DOI:10.1002/(SICI)1097-0061(19970930)13:12<1145::AID-YEA163>3.0.CO;2-Y |
| [36] |
Mrša V, Tanner W. Role of NaOH-extractable cell wall proteins Ccw5p, Ccw6p, Ccw7p and Ccw8p (members of the Pir protein family) in stability of the Saccharomyces cerevisiae cell wall[J]. Yeast, 1999, 15(10A): 813-820. DOI:10.1002/(SICI)1097-0061(199907)15:10A<813::AID-YEA421>3.0.CO;2-Y |
| [37] |
Destruelle M, Holzer H, Klionsky DJ. Identification and characterization of a novel yeast gene: the YGP1 gene product is a highly glycosylated secreted protein that is synthesized in response to nutrient limitation[J]. Molecular and Cellular Biology, 1994, 14(4): 2740-2754. DOI:10.1128/MCB.14.4.2740 |
| [38] |
Riou C, Nicaud JM, Barre P, Gaillardin C. Stationary-phase gene expression in Saccharomyces cerevisiae during wine fermentation[J]. Yeast, 1997, 13(10): 903-915. DOI:10.1002/(SICI)1097-0061(199708)13:10<903::AID-YEA145>3.0.CO;2-1 |
| [39] |
Chang YW, Howard SC, Budovskaya YV, Rine J, Herman PK. The rye mutants identify a role for Ssn/Srb proteins of the RNA polymerase Ⅱ holoenzyme during stationary phase entry in Saccharomyces cerevisiae[J]. Genetics, 2001, 157(1): 17-26. |
| [40] |
O'Rourke SM, Herskowitz I. A third osmosensing branch in Saccharomyces cerevisiae requires the Msb2 protein and functions in parallel with the Sho1 branch[J]. Molecular and Cellular Biology, 2002, 22(13): 4739-4749. DOI:10.1128/MCB.22.13.4739-4749.2002 |
| [41] |
Vadaie N, Dionne H, Akajagbor DS, Nickerson SR, Krysan DJ, Cullen PJ. Cleavage of the signaling mucin Msb2 by the aspartyl protease Yps1 is required for MAPK activation in yeast[J]. Journal of Cell Biology, 2008, 181(7): 1073-1081. DOI:10.1083/jcb.200704079 |
| [42] |
Rodríguez-Peña JM, García R, Nombela C, Arroyo J. The high-osmolarity glycerol (HOG) and cell wall integrity (CWI) signalling pathways interplay: a yeast dialogue between MAPK routes[J]. Yeast, 2010, 27(8): 495-502. DOI:10.1002/yea.1792 |
| [43] |
Espinoza FH, Ogas J, Herskowitz I, Morgan DO. Cell cycle control by a complex of the cyclin HCS26 (PCL1) and the kinase PHO85[J]. Science, 1994, 266(5189): 1388-1391. DOI:10.1126/science.7973730 |
| [44] |
Moffat J, Andrews B. Late-G1 cyclin-CDK activity is essential for control of cell morphogenesis in budding yeast[J]. Nature Cell Biology, 2004, 6(1): 59-66. DOI:10.1038/ncb1078 |
| [45] |
Hadwiger JA, Wittenberg C, Richardson HE, De Barros Lopes M, Reed SI. A family of cyclin homologs that control the G1 phase in yeast[J]. Proceedings of the National Academy of Sciences of the United States of America, 1989, 86(16): 6255-6259. DOI:10.1073/pnas.86.16.6255 |
| [46] |
Wittenberg C, Sugimoto K, Reed SI. G1-specific cyclins of S. cerevisiae: cell cycle periodicity, regulation by mating pheromone, and association with the p34CDC28 protein kinase[J]. Cell, 1990, 62(2): 225-237. DOI:10.1016/0092-8674(90)90361-H |
| [47] |
Iacovella MG, Daly CN, Kelly JS, Michielsen AJ, Clyne RK. Analysis of polo-like kinase Cdc5 in the meiosis recombination checkpoint[J]. Cell Cycle, 2010, 9(6): 1182-1193. DOI:10.4161/cc.9.6.11068 |
| [48] |
Walters AD, May CK, Dauster ES, Cinquin BP, Smith EA, Robellet X, D'Amours D, Larabell CA, Cohen-Fix O. The yeast polo kinase Cdc5 regulates the shape of the mitotic nucleus[J]. Current Biology, 2014, 24(23): 2861-2867. DOI:10.1016/j.cub.2014.10.029 |
| [49] |
Mishra PK, Olafsson G, Boeckmann L, Westlake TJ, Jowhar ZM, Dittman LE, Baker RE, D'Amours D, Thorpe PH, Basrai MA. Cell cycle–dependent association of polo kinase Cdc5 with CENP-A contributes to faithful chromosome segregation in budding yeast[J]. Molecular Biology of the Cell, 2019, 30(8): 1020-1036. DOI:10.1091/mbc.E18-09-0584 |
| [50] |
Guacci V, Koshland D, Strunnikov A. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae[J]. Cell, 1997, 91(1): 47-57. DOI:10.1016/S0092-8674(01)80008-8 |
| [51] |
Tóth A, Ciosk R, Uhlmann F, Gálová M, Schleiffer A, Nasmyth K. Yeast Cohesin complex requires a conserved protein, Eco1p(Ctf7), to establish cohesion between sister chromatids during DNA replication[J]. Genes & Development, 1999, 13(3): 320-333. |
| [52] |
Ciosk R, Zachariae W, Michaelis C, Shevchenko A, Mann M, Nasmyth K. An ESP1/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast[J]. Cell, 1998, 93(6): 1067-1076. DOI:10.1016/S0092-8674(00)81211-8 |
| [53] |
Hu FH, Elledge SJ. Bub2 is a cell cycle regulated phospho-protein controlled by multiple checkpoints[J]. Cell Cycle, 2002, 1(5): 340-344. DOI:10.4161/cc.1.5.154 |
| [54] |
Yamamoto A, Guacci V, Koshland D. Pds1p, an inhibitor of anaphase in budding yeast, plays a critical role in the APC and checkpoint pathway(s)[J]. The Journal of Cell Biology, 1996, 133(1): 99-110. DOI:10.1083/jcb.133.1.99 |
| [55] |
Toyn JH, Johnston LH. The Dbf2 and Dbf20 protein kinases of budding yeast are activated after the metaphase to anaphase cell cycle transition[J]. The EMBO Journal, 1994, 13(5): 1103-1113. DOI:10.1002/j.1460-2075.1994.tb06359.x |
| [56] |
Weinert TA, Kiser GL, Hartwell LH. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair[J]. Genes & Development, 1994, 8(6): 652-665. |
| [57] |
Porter SD, Smith M. Homoeo-domain homology in yeast MATα2 is essential for repressor activity[J]. Nature, 1986, 320(6064): 766-768. DOI:10.1038/320766a0 |
| [58] |
Goutte C, Johnson AD. a1 protein alters the DNA binding specificity of α2 repressor[J]. Cell, 1988, 52(6): 875-882. DOI:10.1016/0092-8674(88)90429-1 |
| [59] |
Jensen R, Sprague Jr GF, Herskowitz I. Regulation of yeast mating-type interconversion: feedback control of HO gene expression by the mating-type locus[J]. Proceedings of the National Academy of Sciences of the United States of America, 1983, 80(10): 3035-3039. DOI:10.1073/pnas.80.10.3035 |
| [60] |
Hollingsworth NM, Byers B. HOP1: a yeast meiotic pairing gene[J]. Genetics, 1989, 121(3): 445-462. DOI:10.1093/genetics/121.3.445 |
| [61] |
Rabitsch KP, Tóth A, Gálová M, Schleiffer A, Schaffner G, Aigner E, Rupp C, Penkner AM, Moreno-Borchart AC, Primig M, et al. A screen for genes required for meiosis and spore formation based on whole-genome expression[J]. Current Biology, 2001, 11(13): 1001-1009. DOI:10.1016/S0960-9822(01)00274-3 |
| [62] |
Tsubouchi H, Roeder GS. The Mnd1 protein forms a complex with Hop2 to promote homologous chromosome pairing and meiotic double-strand break repair[J]. Molecular and Cellular Biology, 2002, 22(9): 3078-3088. DOI:10.1128/MCB.22.9.3078-3088.2002 |
| [63] |
Tachikawa H, Bloecher A, Tatchell K, Neiman AM. A Gip1p-Glc7p phosphatase complex regulates septin organization and spore wall formation[J]. Journal of Cell Biology, 2001, 155(5): 797-808. DOI:10.1083/jcb.200107008 |
| [64] |
Hurtado S, Kim Guisbert KS, Sontheimer EJ. SPO24 is a transcriptionally dynamic, small ORF-encoding locus required for efficient sporulation in Saccharomyces cerevisiae[J]. PLoS One, 2014, 9(8): e105058. DOI:10.1371/journal.pone.0105058 |
| [65] |
Hepworth SR, Ebisuzaki LK, Segall J. A 15-base-pair element activates the SPS4 gene midway through sporulation in Saccharomyces cerevisiae[J]. Molecular and Cellular Biology, 1995, 15(7): 3934-3944. DOI:10.1128/MCB.15.7.3934 |
| [66] |
Nag DK, Koonce MP, Axelrod J. SSP1, a gene necessary for proper completion of meiotic divisions and spore formation in Saccharomyces cerevisiae[J]. Molecular and Cellular Biology, 1997, 17(12): 7029-7039. DOI:10.1128/MCB.17.12.7029 |
 2021, Vol. 48
2021, Vol. 48