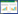扩展功能
文章信息
- 琚慧敏, 杨键, 张偲, 李洁
- JU Huimin, YANG Jian, ZHANG Si, LI Jie
- 荧光标记珊瑚组织来源细菌及其与虫黄藻相互作用的显微观察
- Labeling a coral tissue-derived bacterium with stable fluorescence and microscopic observation of the interaction between the labeled strain and Symbiodiniaceae
- 微生物学通报, 2021, 48(2): 351-361
- Microbiology China, 2021, 48(2): 351-361
- DOI: 10.13344/j.microbiol.china.200118
-
文章历史
- 收稿日期: 2020-02-19
- 接受日期: 2020-03-30
- 网络首发日期: 2020-06-30
2. 中国科学院大学 北京 100049
2. University of Chinese Academy of Sciences, Beijing 100049, China
珊瑚与虫黄藻(Symbiodiniaceae)、细菌、古菌、真菌、病毒等微生物共同构成珊瑚共生体(Coral Holobiont)[1]。珊瑚共生体成员之间的相互作用直接影响珊瑚健康状态[1-2]。研究发现,珊瑚与虫黄藻互利共生,珊瑚为虫黄藻提供栖息环境和营养代谢物[3];虫黄藻通过光合作用,将合成的有机物提供给珊瑚[4]。珊瑚和其他共附生微生物的关系更加复杂微妙。珊瑚为微生物抵挡紫外线使其免受伤害,微生物通过光合作用、固氮作用为珊瑚提供营养,产生抗菌物质防止珊瑚感染疾病等[5-6]。但在胁迫条件下,某些微生物会导致珊瑚发生白化或感染疾病[7-11]。关于虫黄藻和细菌的相互作用研究较少[12]。导致珊瑚白化的施罗氏弧菌(Vibrio shilonii)分泌一种富含脯氨酸的肽类——毒素P,会抑制虫黄藻的光合作用[13]。
利用显微技术针对目标菌进行原位观察,有助于揭示宿主-细菌之间复杂、动态的相互作用。虫黄藻叶绿体含有叶绿素a,用蓝光激发时,可产生波长更长的红色荧光信号。目前大多数虫黄藻的离体培养还未达到无菌纯培养的状态[14],而且无菌可能会影响虫黄藻的生长[15]。因此,为了避免虫黄藻自发荧光的干扰,并且区别于虫黄藻培养液中原有的细菌,对目标细菌进行标记,可利用标记信号通过显微技术研究虫黄藻与目标细菌的原位相互作用。
荧光蛋白被广泛应用于细胞标记[16]。通常采用自然转化、化学转化、电穿孔、接合转移的方法[17],将携带荧光蛋白基因的外源DNA插入目标菌。目前,海洋细菌遗传操作体系的研究有限,仅针对部分海洋细菌建立了成熟的遗传操作系统[17]。例如,霍乱弧菌(Vibrio cholerae)[18]、创伤弧菌(Vibrio vulnificus)[19]、需钠弧菌(Vibrio natriegens)[20]等海洋弧菌具有吸收细胞外遗传物质的天然能力。截至目前,海洋细菌只有V. natriegens能通过化学方法转化[21]。另外,一些海洋菌已通过电穿孔法成功转化,如Bacillus marinus B-9987[22]、Fischerella[23]、Chlorogloeopsis strains[23]、Halomonas sp. O-1[24]、Marinobacter adhaerens HP15[25]、Pseudoalteromonas sp. BSi20429[26]、Roseobacter clade sp.[27]、Synechococcus sp. CC931[28]、Vibrio vulnificus[29]、Vibrio parahaemolyticus[30]等。接合转移通过细菌间的直接接触交换遗传物质,是细菌间水平基因转移的主要方式,驱动微生物环境适应性演化。在基因工程领域,接合转移也是向微生物细胞内导入异源质粒的重要手段。已有的研究表明,接合转移过程需要4个要素协同完成:转移起始序列oriT、弛豫酶(Relaxase)、IV型偶联蛋白T4CP及IV型分泌系统[31]。接合转移最常用的是相对较小、具有自主复制作用的遗传元件——质粒。由于许多宿主携带的限制修饰系统会降解双链质粒DNA[32-33],因此,比起电穿孔转移的双链DNA,通过接合转移方法转移的单链相对更稳定[32]。另外,对于那些难以通过电穿孔和化学方法转化的菌株,接合转移是最有效的转基因方法[17]。目前,只有Dinoroseobacter shibae DFL12[27]、Prochlorococcus sp. MIT9313[34]、Marinobacter adhaerens HP15[25]、Pseudoalteromonas sp. SM9913[35]、Roseobacter clade sp.[27]、Vibrio harveyi ORM4[36]、Vibrio natriegens[37]、Vibrio sp.[38]等海洋细菌已成功通过接合转移将外源遗传物质转入其细胞内。
细菌SCSIO 12696分离自鹿角杯形珊瑚(Pocillopora damicornis)组织,需在添加海水的培养基中生长。基于16S rRNA基因序列分析,发现其与Porticoccus litoralis IMCC2115T (Gammaproteobacteria,Cellvibrionales,Porticoccaceae)的相似性最高(91.83%),是港口球菌科(Porticoccaceae)的潜在新属。据报道,Cellvibrionales clade BD1-7是柳珊瑚“核心菌群”的成员,并且很可能在珊瑚共生体中发挥有益作用[39]。港口球菌科的菌株能够利用藻类产生的代谢物促进生长[40]。在珊瑚微生物学领域,对珊瑚-致病菌互作的研究相对较多,例如利用绿色荧光蛋白标记珊瑚条件致病菌溶珊瑚弧菌(Vibrio coralliilyticus),进而观察标记菌株与珊瑚宿主的互作,以期揭示其致病机理[41],而关于珊瑚益生菌及其益生机制的研究有限[42]。港口球菌科成员SCSIO 12696有望作为模式菌株研究珊瑚-共生藻-益生菌三者相互作用模式与机理。荧光标记可有效将目标细菌从复杂的原位环境中区分开,是研究生物相互作用的重要技术手段。本研究以菌株SCSIO 12696为受体菌,以带有性菌毛的Escherichia coli WM3064为供体菌[43],采用接合转移的方法构建绿色荧光蛋白标记菌株。我们希望通过首次建立纤维弧菌目成员SCSIO 12696的遗传操作与荧光蛋白标记技术体系,以期为原位环境虫黄藻-细菌相互作用的研究提供材料,为进一步深入研究该潜在有益菌的生态功能奠定基础。
1 材料与方法 1.1 菌株和质粒研究所用菌株和质粒的信息详见表 1。
| 菌株和质粒Strains and plasmids | 特征Characteristics | 来源Source |
| SCSIO 12696 | Porticoccaceae, wild type strain is sensitive to ampicillin and chloramphenicol, but resistant to kanamycin, gentamycin, and streptomycin | Isolated from coral tissue of Pocillopora damicornis by Professor LI Jie |
| WM3064 | Escherichia coli, 2, 6-diaminopimelic acid auxotrophic | Given by Professor WANG Xiaoxue |
| pBBR1MCS2-Tac-mCherry plasmid | Red fluorescent protein expression plasmid, a broad host plasmid, 6 195 bp, kanamycin resistance | Given by Associate Professor YANG Jian |
| pET22b-EGFP plasmid | Red fluorescent protein expression plasmid, expression host: E. coli BL21(DE3), 6 177 bp, ampicillin resistance | Purchase from Miaoling plasmid |
卡那霉素(Kanamycin Sulfate,Kana)、氨苄青霉素(Ampicillin Sodium Salt,Amp)、氯霉素(Chloramphenicol,Chl)、链霉素(Streptomycin Sulfate,Str)、庆大霉素(Gentamycin Sulfate,Gen),生工生物工程(上海)股份有限公司;二氨基庚二酸(Diaminopimelic Acid,DAP),Sigma公司;柠檬酸铁,Aladdin公司。PCR仪、电转杯,Bio-Rad公司;Taq酶、Pfu酶,北京全式金生物技术有限公司;琼脂糖凝胶DNA回收试剂盒、质粒小量快速抽提试剂盒,Magen公司;荧光显微镜、激光扫描共聚焦显微镜,Leica公司;Isopore聚碳酸酯膜,Millipore公司。
1.3 培养基普通海水培养基(MB):BD Difco™ Marine Broth商品培养基37.4 g/L,纯水1.0 L;普通海水培养基(MA):MB培养基中加琼脂15.0 g/L;LB改良培养基(LBmodified,g/L):胰蛋白胨5.0,酵母粉1.0,柠檬酸铁0.137,人工海水(盐度为3%) 1.0 L;藻-菌共培养所用的培养基为f/2-Si培养基[44]。
1.4 荧光标记接合质粒的构建利用SnapGene软件设计引物,序列信息见表 2。引物由上海美吉生物医药科技有限公司合成。
| 引物名称 Primer |
序列 Sequences (5′→3′) |
| Amp-F | TGAGGATCGTTTCGCATGAGTATTCAACATTTCCG |
| Amp-R | ACCCCAGAGTCCCGCTTACCAATGCTTAATCAGTG |
| pBBR1MCS2-Tac-mCherry-Amp-F (pBBR-Amp-F) | GCGAAACGATCCTCATCCTGTCTCT |
| pBBR1MCS2-Tac-mCherry-Amp-R (pBBR-Amp-R) | GCGGGACTCTGGGGTTCGAA |
| Green-F | CAGGAAACAGTATTCATGGTGAGCAAGGGCGAGGA |
| Green-R | AGCAGCCTAGGTTAATTACTTGTACAGCTCGTCCA |
| pBBR1MCS2-Tac-mCherry-Green-F (pBBR-Green-F) | TTAACCTAGGCTGCTGCCACCGCTGAGCAA |
| pBBR1MCS2-Tac-mCherry-Green-R (pBBR-Green-R) | GAATACTGTTTCCTGTGTGAAATATCCGCT |
| M13-F | CAGGAAACAGCTATGACC |
| M13-R | TGTAAAACGACGGCCAGT |
因野生型菌株SCSIO 12696对氨苄青霉素敏感而对卡那霉素不敏感,需将广宿主表达质粒中的卡那抗性基因替换成氨苄抗性基因。分别以Amp-F/R为引物,以质粒pET22b-EGFP为模板扩增氨苄抗性基因;以pBBR1MCS2-Tac-mCherry-Amp-F/R为引物,以广宿主质粒pBBR1MCS2-Tac-mCherry为模板反向扩增线性化载体。PCR扩增体系(50 µL):引物Amp-F/R或pBBR1MCS2-Tac-mCherry-Amp- F/R (10 µmol/L)各2 µL,模板pET22b-EGFP (40 µg/mL)或pBBR1MCS2-Tac-mCherry (38 µg/mL)质粒2 µL,2×TransStart® FastPfu PCR SuperMix (−Dye) 25 µL,ddH2O 19 µL。PCR反应条件:95 ℃ 5 min;95 ℃ 30 s,55 ℃ 30 s,72 ℃ 50 s,32个循环;72 ℃ 5 min。按照琼脂糖凝胶DNA回收试剂盒说明书纯化目的片段。按照无缝连接试剂盒ClonExpress®Ⅱ One Step Cloning Kit说明书,将氨苄抗性基因和线性化广宿主载体连接,连接产物转化至大肠杆菌DH5α感受态细胞中,利用添加氨苄抗生素(50 µg/mL)的LB平板37 ℃培养12 h,筛选出单克隆。挑单克隆至含氨苄(50 µg/mL)抗性的LB液体培养基中,37 ℃、200 r/min扩大培养12 h后,进行PCR验证质粒是否成功转化至大肠杆菌感受态细胞(引物用Amp-F/R),获得含氨苄抗性和红色荧光蛋白的广宿主质粒pBBR1MCS2-Tac-mCherry-Amp。参照上述方法,将质粒pBBR1MCS2-Tac-mCherry-Amp中的红色荧光蛋白基因替换成egfp基因,所用引物为Green-F/R和pBBR1MCS2-Tac-mCherry-Green-F/R,构建重组质粒pBBR1MCS2-Tac-EGFP-Amp。
1.5 接合转移实验电击转化(2 500 V电压)质粒pBBR1MCS2-Tac-EGFP-Amp至E. coli WM3064,即得重组供体菌E. coli WM3064/pBBR1MCS2-Tac-EGFP-Amp。取2 mL供体菌(OD600为1.034±0.003),分别取1、2或4 mL受体菌SCSIO 12696 (OD600为0.775±0.002),4 000×g离心5 min收集菌体,分别用100 µL FASW (0.2 µm Filtered Autoclaved Seawater,盐度3%)重悬,供体菌细胞量为2.06×109个,受体菌细胞量分别为5.39×108、1.08×109或2.16×109个,按约4׃1、2׃1、1׃1的供、受体菌细胞比例,将混合菌液滴于添加二氨基庚二酸(0.3 µmol/L)的固体LB改良培养基(LBmodified)上,分别置于25 ℃和30 ℃的培养箱中培养48 h。利用氨苄抗性筛选接合子。
挑取添加氨苄抗生素(50 µg/mL)的改良LB培养基上生长的单克隆,用M13-F/R引物对进行PCR扩增。PCR扩增体系(50 µL):引物M13-F/R (10 µmol/L)各2 µL,模板菌液1 µL,2×Easy Taq® PCR SuperMix (+Dye)酶25 µL,ddH2O 20 µL。PCR反应条件:95 ℃ 10 min;95 ℃ 30 s,55 ℃ 50 s,72 ℃ 50 s,32个循环;72 ℃ 5 min。PCR产物进行琼脂糖凝胶电泳和测序验证。在荧光显微镜下观察荧光信号,判断细菌体内的荧光蛋白基因是否表达。DAPI信号观察条件:激发波长350±50 nm,发射波长460±50 nm;绿色荧光蛋白信号观察条件:激发波长480±40 nm,发射波长527±30 nm。
1.6 测定接合转移效率及质粒稳定性参照1.5描述的方法进行接合转移,将25 ℃和30 ℃培养48 h后的混合菌分别用1 mL FASW从平板上洗脱,收集菌液,稀释101、102、103、104、105、106、107、108倍,取100 µL涂布于添加Amp (50 µg/mL)的LBmodified平板上,分别置于25 ℃和30 ℃培养48 h后,计菌落数,每组3个平行。将野生型受体菌SCSIO 12696用FASW稀释105、106倍,各取100 µL涂布于LBmodified平板,计菌落数,每组3个平行。根据以下公式计算接合转移效率[45-46]:

|
(1) |
将SCSIO 12696/pBBR1MCS2-Tac-EGFP-Amp接合子在不添加Amp的MA培养基上传代5次后,随机挑取100个单菌落接种于添加50 µg/mL Amp的MA培养基上,观察菌株生长情况,以判断质粒的稳定性。
1.7 虫黄藻CCMP 2433与SCSIO 12696/ pBBR1MCS2-Tac-EGFP-Amp的共培养观察虫黄藻CCMP 2433 (Symbiodinium)分离自鹿角杯形珊瑚,由National Center for Marine Algae and Microbiota (NCMA)保藏。菌株SCSIO 12696/pBBR1MCS2-Tac-EGFP-Amp在添加50 µg/mL Amp的MB培养基于25 ℃培养48 h后,用f/2-Si培养基洗涤2次,4 000×g离心5 min收集细菌细胞。随后与CCMP 2433在无菌天然海水配制的f/2-Si培养基中共培养。菌株SCSIO 12696/pBBR1MCS2- Tac-EGFP-Amp和虫黄藻CCMP 2433的初始细胞比例为10׃1。培养条件为25 ℃、光照强度为25 µmol/(m2·s)、光周期为12 L:12 D。2 d后取1 mL培养液用减压抽滤装置过滤到直径25 mm、孔径0.2 µm的聚碳酸酯膜上,在激光扫描共聚焦显微镜下观察。叶绿素a观察通道:激发波长488 nm,发射波长650−690 nm;SCSIO 12696/pBBR1MCS2- Tac-EGFP-Amp观察通道:激发波长488 nm,发射波长495−520 nm。
2 结果与分析 2.1 表达荧光蛋白广宿主接合质粒的构建广宿主质粒pBBR1分离自支气管败血博德特氏杆菌(Bordetella bronchiseptica),含有2个关键蛋白Rep和Mob编码序列。Rep蛋白使该质粒在多个宿主内复制,Mob蛋白与该质粒在不同宿主间的移动密切相关[47-48]。该质粒可在革兰氏阴性细菌如布鲁氏菌(Brucella)、博德特氏菌(Bordetella)、弧菌(Vibrio)、大肠杆菌(Escherichia)、假单胞菌(Pseudomonas)和根瘤菌(Rhizobium)等[49]以及革兰氏阳性细菌如芽孢杆菌(Bacillus)[50]中稳定复制。由于其优良的可移动性和宿主适应性,pBBR1自发现以来被广泛改造应用于异源蛋白在非模式细菌体内表达。鉴于该质粒的成功应用案例,本研究尝试改造该质粒使其应用于珊瑚共附生细菌SCSIO 12696的荧光标记。
结合细菌SCSIO 12696及虫黄藻的特性,首先采用两步Gibson Assembly方法,按图 1所示流程改造pBBR1MCS2-Tac-mCherry,替换该质粒卡那抗性基因为氨苄抗性基因,同时将该质粒原有的红色荧光蛋白基因替换为绿色荧光蛋白基因。经PCR测序验证后,成功构建了pBBR1MCS2-Tac-EGFP- Amp。该质粒图谱如图 1所示,绿色荧光蛋白基因egfp位于Tac启动子下游,同时含有蛋白Rep和Mob编码序列。质粒pBBR1MCS2-Tac-EGFP-Amp用于后续接合转移研究。
|
| 图 1 构建质粒pBBR1MCS2-Tac-EGFP-Amp的流程示意图 Figure 1 Flow chart of constructing plasmid pBBR1MCS2-Tac-EGFP-Amp |
|
|
由于菌株SCSIO 12696仅在海水培养基(如MA)上生长,但是DAP缺陷型的大肠杆菌WM3064可以在不添加DAP的MA培养基上生长(表 3),所以MA+Amp培养基不适合在接合转移实验中用于接合子的筛选。然而供、受体菌株在接合转移过程中的活力是接合转移效率的决定因素之一。因此,选择合适的培养基用于大肠杆菌WM3064和SCSIO 12696接合转移实验十分关键。考虑到DAP缺陷型的大肠杆菌WM3064在未添加DAP的LB培养中不生长(表 3),本研究以LB为基础培养基进行改良。
| 培养基Medium | E. coli WM3064 | E. coli WM3064/pBBR1MCS2-Tac-EGFP-Amp | SCSIO 12696 |
| MA | + | + | + |
| MA+DAP | + | + | + |
| LB | − | − | − |
| LB+DAP | + | + | − |
| LBmodified | − | − | + |
| LBmodified+Amp | − | − | − |
| LBmodified+DAP | + | + | + |
| LBmodified+DAP+Amp | − | + | − |
| 注:+:菌株能生长;−:菌株不能生长 Note: +: The strain can grow; −: The strain cannot grow |
|||
LB改良培养基(LBmodified)既能满足海洋细菌SCSIO 12696的生长需求,同时,WM3064/pBBR1MCS2-Tac-EGFP-Amp在添加DAP的LBmodified培养基上能生长,但不会在缺乏DAP的LBmodified培养基上生长(表 3)。因此,添加DAP的LBmodified培养基可用于接合转移过程中供/受体菌的共培养,而不添加DAP的LBmodified+Amp培养基可用于接合子的筛选。
2.2.2 接合转移条件优化接合转移效率分析结果表明,在相同温度下,供、受体菌的比例在一定范围内,受体菌的比例越高其接合转移效率越高(表 4);供、受体菌比例一定时,30 ℃的接合转移效率高于25 ℃ (表 4)。
| 温度Temperature (℃) | 供/受体菌细胞比例Donor/receptor cell amount ratio | 接合子Conjugants (×106 CFU/mL) | 受体菌Receptor (CFU/mL) | 接合转移效率Conjugation efficiency (%) |
| 25 | 4׃1 | 2.23±0.16 | 5.39×108 | 0.41±0.02 |
| 2׃1 | 2.57±0.31 | 5.39×108 | 0.48±0.03 | |
| 1׃1 | 2.73±0.53 | 5.39×108 | 0.51±0.01 | |
| 30 | 4׃1 | 4.42±0.51 | 5.39×108 | 0.82±0.05 |
| 2׃1 | 6.72±0.27 | 5.39×108 | 1.25±0.03 | |
| 1׃1 | 10.07±0.63 | 5.39×108 | 1.99±0.06 |
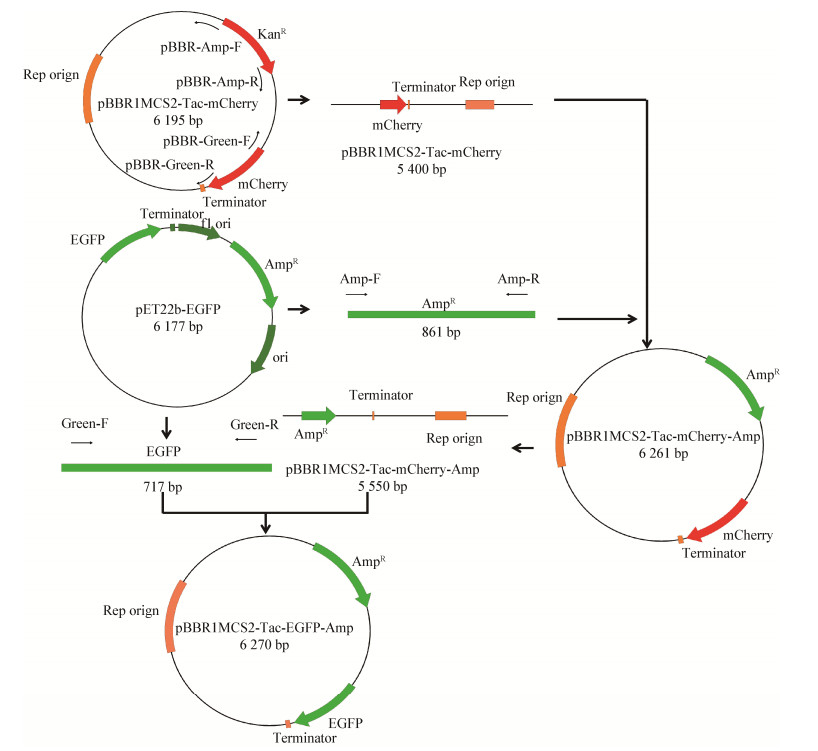
依照上述优化后的接合转移条件,挑取添加氨苄的改良LB培养基上生长的单克隆进行PCR扩增,PCR产物的琼脂糖凝胶电泳结果表明其与目的片段大小吻合。切胶回收目的条带,测序验证结果显示与egfp基因完全匹配。同时,荧光显微镜下可清晰观察到绿色荧光蛋白表达的信号(图 2)。以上结果表明已成功构建EGFP标记菌株SCSIO 12696/pBBR1MCS2-Tac-EGFP-Amp。
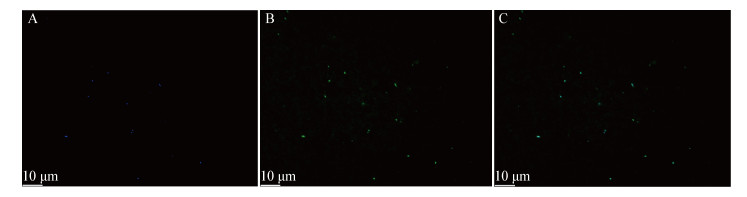
|
| 图 2 egfp基因在SCSIO 12696/pBBR1MCS2-Tac-EGFP-Amp中的表达 Figure 2 Expression of egfp in SCSIO 12696/pBBR1MCS2-Tac-EGFP-Amp 注:A:为SCSIO 12696/pBBR1MCS2-Tac-EGFP-Amp经DAPI染色后的显微观察照片;B:SCSIO 12696/pBBR1MCS2-Tac-EGFP-Amp在激发波长为480±40 nm,发射波长为527±30 nm通道下可见绿色荧光蛋白表达信号;C:DAPI和绿色荧光蛋白信号重叠图 Note: A: The microscopic observation of SCSIO 12696/pBBR1MCS2-Tac-EGFP-Amp stained by DAPI; B: The green fluorescent protein signal of SCSIO 12696/pBBR1MCS2-Tac-EGFP-Amp observed at the excitation wavelength of 480±40 nm and emission wavelength of 527±30 nm; C: The overlapping of DAPI and GFP signals |
|
|
由于在后续共培养体系中不便加入抗生素,因此在无选择压力条件下,检测重组质粒pBBR1MCS2- Tac-EGFP-Amp能否在宿主传代过程中稳定存在和正常表达,才可确保后续实验中观察到荧光信号。SCSIO 12696/pBBR1MCS2-Tac-EGFP-Amp在无抗性的MA培养基上传代5次后,随机挑取100个单菌落接种于添加Amp的MA培养基上,结果显示100个单菌落均能生长,说明质粒pBBR1MCS2-Tac- EGFP-Amp可在SCSIO 12696中稳定存在。
2.4 SCSIO 12696/pBBR1MCS2-Tac-EGFP-Amp和CCMP 2433共培养的显微观察对虫黄藻CCMP 2433和绿色荧光蛋白标记菌SCSIO 12696/pBBR1MCS2-Tac-EGFP-Amp共培养物进行显微观察。激光扫描共聚焦显微镜观察结果显示,在激发波长488 nm和发射波长650−690 nm通道下清晰可见虫黄藻叶绿素a荧光信号;在激发波长488 nm和发射波长495−520 nm通道下清晰可见SCSIO 12696/pBBR1MCS2-Tac-EGFP-Amp绿色荧光信号(图 3)。在共培养条件下,菌株SCSIO 12696可附着在虫黄藻CCMP 2433细胞表面(图 3),是否进入虫黄藻细胞内部仍需进一步实验验证。
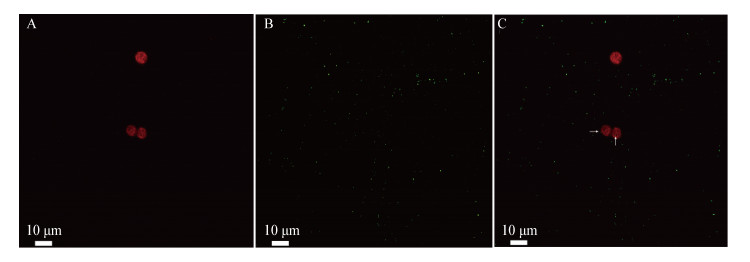
|
| 图 3 虫黄藻CCMP 2433与egfp标记的SCSIO 12696的共存方式 Figure 3 Coexistence of Symbiodinium CCMP 2433 and egfp-labeled SCSIO 12696 注:A:在激发波长488 nm和发射波长650−690 nm通道下虫黄藻叶绿素a的荧光信号;B:在激发波长488 nm和发射波长495−520 nm通道下菌株SCSIO 12696/pBBR1MCS2-Tac-EGFP-Amp表达的绿色荧光蛋白信号;C:图A和图B叠加图,箭头指示标记菌株SCSIO 12696 Note: A: The fluorescence signal of chlorophyll a of zooxanthellae under the channel of excitation wavelength 488 nm and emission wavelength 650-690 nm; B: The green fluorescent protein expressed by SCSIO 12696/pBBR1MCS2-Tac-EGFP-Amp observed under the channel of excitation wavelength of 488 nm and emission wavelength of 495-520 nm; C: An overlay of Figure A and B, arrows indicate cells of the eGFP-labeled strain SCSIO 12696 |
|
|
近年来,国内外学者对于海洋细菌遗传操作系统的研究有限[17]。自然转化和化学转化对细菌本身有一定的要求[17];电穿孔虽然是一种快速、高效率的手段,但因为盐分的限制使其不适用于一些需盐或需海水方可生长的海洋细菌。因此,接合转移是最可能被广泛应用于海洋细菌的转化技术[33]。本研究所用的菌株SCSIO 12696是从鹿角杯形珊瑚组织中分离的港口球菌科潜在新属,需在添加海水的培养基中生长。然而,DAP营养缺陷型大肠杆菌WM3064在不含DAP的常用海洋菌培养基MA上也能正常生长,因此MA培养基虽然能满足供、受体菌的生长需求,但不适用于接合子筛选。常见的接合转移方法没有提供一种专性海洋细菌能够正常生长且具有筛选作用的培养基。本研究改良了LB培养基,使其盐度发生变化,使供体菌和专性海洋受体菌都能生长,并且能利用抗生素抗性和营养缺陷进行接合子的筛选,为专性海洋菌的接合转移遗传操作提供了关键培养条件。研究发现,在本系统中接合转移效率与供、受体菌的比例及温度有关,相关结果可为珊瑚共附生细菌的遗传操作提供借鉴。
众所周知,在全球气候变化和人类活动的影响下,世界范围内的珊瑚正面临着大面积的白化和死亡[51]。探究珊瑚共生体各成员之间的相互关系是寻求珊瑚健康维护和恢复手段的关键[42]。本研究成功建立了适合专性海洋细菌的遗传操作体系,为深入研究珊瑚-细菌、虫黄藻-细菌相互作用奠定了基础。构建的egfp基因标记菌株,将为后续从微观角度研究虫黄藻-细菌原位相互作用提供材料。同时,替换不同的荧光蛋白基因可使目标菌株携带其他的荧光蛋白信号,使之更适用于在珊瑚共生体复杂自发荧光背景下的原位观察。此外,基于本研究所建立的遗传操作体系及荧光标记技术,可对相关海洋菌进行遗传操作,以便于进一步深入研究细菌的附着动力学过程和机制、细菌在宿主微环境中的增殖能力和方式以及细菌与宿主或虫黄藻的互作机制。
致谢: 感谢中国科学院南海海洋研究所王晓雪研究员赠予菌株Escherichia coli WM3064,以及李茹在实验操作中给予的指导。| [1] |
Bourne DG, Garren M, Work TM, Rosenberg E, Smith GW, Harvell CD. Microbial disease and the coral holobiont[J]. Trends in Microbiology, 2009, 17(12): 554-562. DOI:10.1016/j.tim.2009.09.004 |
| [2] |
Krediet CJ, Ritchie KB, Paul VJ, Teplitski M. Coral-associated micro-organisms and their roles in promoting coral health and thwarting diseases[J]. Proceedings of the Royal Society B Biological Sciences, 2013, 280(1755): 20122328. DOI:10.1098/rspb.2012.2328 |
| [3] |
Rowan R. Coral bleaching: thermal adaptation in reef coral symbionts[J]. Nature, 2004, 430(7001): 742. DOI:10.1038/430742a |
| [4] |
Dubinsky Z, Jokiel PL. Ratio of energy and nutrient fluxes regulates symbiosis between zooxanthellae and corals[J]. Pacific Science, 1994, 48(3): 313-324. |
| [5] |
Rädecker N, Pogoreutz C, Voolstra CR, Wiedenmann J, Wild C. Nitrogen cycling in corals: the key to understanding holobiont functioning?[J]. Trends in Microbiology, 2015, 23(8): 490-497. DOI:10.1016/j.tim.2015.03.008 |
| [6] |
Shnit-Orland M, Sivan A, Kushmaro A. Antibacterial activity of Pseudoalteromonas in the coral holobiont[J]. Microbial Ecology, 2012, 64(4): 851-859. DOI:10.1007/s00248-012-0086-y |
| [7] |
Thompson JR, Rivera HE, Closek CJ, Medina M. Microbes in the coral holobiont: partners through evolution, development, and ecological interactions[J]. Frontiers Cellular Infection Microbiology, 2015, 4: 176. |
| [8] |
Ben-Haim Y, Banim E, Kushmaro A, Loya Y, Rosenberg E. Inhibition of photosynthesis and bleaching of zooxanthellae by the coral pathogen Vibrio shiloi[J]. Environmental Microbiology, 1999, 1(3): 223-229. DOI:10.1046/j.1462-2920.1999.00027.x |
| [9] |
Ben-Haim Y, Thompson FL, Thompson CC, Cnockaert MC, Hoste B, Swings J, Rosenberg E. Vibrio coralliilyticus sp. nov., a temperature-dependent pathogen of the coral Pocillopora damicornis[J]. International Journal of Systematic and Evolutionary Microbiology, 2003, 53(1): 309-315. DOI:10.1099/ijs.0.02402-0 |
| [10] |
Rosenberg E, Kushmaro A. Microbial diseases of corals: pathology and ecology[A]//Dubinsky Z, Stambler N. Coral Reefs: An Ecosystem in Transition[M]. Dordrecht: Springer, 2011: 451-464
|
| [11] |
Richardson LL, Aronson RB. Infectious diseases of reef corals[A]//Proceedings 9th International Coral Reef Symposium[C]. Bali, Indonesia, 2000, 2: 1225-1230
|
| [12] |
Ritchie KB. Bacterial symbionts of corals and Symbiodinium[A]//Rosenberg E, Gophna U. Beneficial Microorganisms in Multicellular Life Forms[M]. Berlin, Heidelberg: Springer, 2012: 139-150
|
| [13] |
Banin E, Khare SK, Naider F, Rosenberg E. Proline-rich peptide from the coral pathogen Vibrio shiloi that inhibits photosynthesis of zooxanthellae[J]. Applied and Environmental Microbiology, 2001, 67(4): 1536-1541. DOI:10.1128/AEM.67.4.1536-1541.2001 |
| [14] |
Dixon GK, Syrett PJ. The growth of dinoflagellates in laboratory cultures[J]. New Phytologist, 1988, 109(3): 297-302. DOI:10.1111/j.1469-8137.1988.tb04198.x |
| [15] |
Alavi M, Miller T, Erlandson K, Schneider R, Belas R. Bacterial community associated with Pfiesteria-like dinoflagellate cultures[J]. Environmental Microbiology, 2001, 3(6): 380-396. DOI:10.1046/j.1462-2920.2001.00207.x |
| [16] |
Balch WE, Der CJ, Hall A. Regulators and effectors of small GTPases: Rho family[J]. Methods in Enzymology, 2006, 406: 1-798. |
| [17] |
Zeaiter Z, Mapelli F, Crotti E, Borin S. Methods for the genetic manipulation of marine bacteria[J]. Electronic Journal of Biotechnology, 2018, 33: 17-28. DOI:10.1016/j.ejbt.2018.03.003 |
| [18] |
Pollack-Berti A, Wollenberg MS, Ruby EG. Natural transformation of Vibrio fischeri requires tfoX and tfoY[J]. Environmental Microbiology, 2010, 12(8): 2302-2311. |
| [19] |
Gulig PA, Tucker MS, Thiaville PC, Joseph JL, Brown RN. USER friendly cloning coupled with chitin-based natural transformation enables rapid mutagenesis of Vibrio vulnificus[J]. Applied and Environmental Microbiology, 2009, 75(15): 4936-4949. DOI:10.1128/AEM.02564-08 |
| [20] |
Dalia TN, Hayes CA, Stolyar S, Marx CJ, Mckinlay JB, Dalia AB. Multiplex genome editing by natural transformation (MuGENT) for synthetic biology in Vibrio natriegens[J]. ACS Synthetic Biology, 2017, 6(9): 1650-1655. DOI:10.1021/acssynbio.7b00116 |
| [21] |
Weinstock MT, Hesek ED, Wilson CM, Gibson DG. Vibrio natriegens as a fast-growing host for molecular biology[J]. Nature Methods, 2016, 13(10): 849-851. DOI:10.1038/nmeth.3970 |
| [22] |
Liu Y, Zheng H, Zhan GH, Qin W, Tian L, Li WL. Establishment of an efficient transformation protocol and its application in marine-derived Bacillus strain[J]. Science China Life Sciences, 2014, 57(6): 627-635. DOI:10.1007/s11427-014-4632-3 |
| [23] |
Stucken K, Ilhan J, Roettger M, Dagan T, Martin WF. Transformation and conjugal transfer of foreign genes into the filamentous multicellular cyanobacteria (subsection V) Fischerella and Chlorogloeopsis[J]. Current Microbiology, 2012, 65(5): 552-560. DOI:10.1007/s00284-012-0193-5 |
| [24] |
Harris JR, Lundgren BR, Grzeskowiak BR, Mizuno K, Nomura CT. A rapid and efficient electroporation method for transformation of Halomonas sp. O-1[J]. Journal of Microbiological Methods, 2016, 129: 127-132. DOI:10.1016/j.mimet.2016.08.009 |
| [25] |
Sonnenschein EC, Gärdes A, Seebah S, Torres-Monroy I, Grossart HP, Ullrich MS. Development of a genetic system for Marinobacter adhaerens HP15 involved in marine aggregate formation by interacting with diatom cells[J]. Journal of Microbiological Methods, 2011, 87(2): 176-183. DOI:10.1016/j.mimet.2011.08.008 |
| [26] |
Zhao DL, Yu ZC, Li PY, Wu ZY, Chen XL, Shi M, Yu Y, Chen B, Zhou BC, Zhang YZ. Characterization of a cryptic plasmid pSM429 and its application for heterologous expression in psychrophilic Pseudoalteromonas[J]. Microbial Cell Factories, 2011, 10(1): 30. DOI:10.1186/1475-2859-10-30 |
| [27] |
Piekarski T, Buchholz I, Drepper T, Schobert M, Wagner-Doebler I, Tielen P, Jahn D. Genetic tools for the investigation of Roseobacter clade bacteria[J]. BMC Microbiology, 2009, 9(1): 265. DOI:10.1186/1471-2180-9-265 |
| [28] |
Chen HX, Lin HZ, Jiang P, Li FC, Qin S. Genetic transformation of marine cyanobacterium Synechococcus sp. CC9311 (Cyanophyceae) by electroporation[J]. Chinese Journal of Oceanology and Limnology, 2013, 31(2): 416-420. DOI:10.1007/s00343-013-2164-5 |
| [29] |
Klevanskaa K, Bier N, Stingl K, Strauch E, Hertwig S. PVv3, a new shuttle vector for gene expression in Vibrio vulnificus[J]. Applied and Environmental Microbiology, 2014, 80(4): 1477-1481. DOI:10.1128/AEM.03720-13 |
| [30] |
Hamashima H, Nakano T, Tamura S, Arai T. Genetic transformation of Vibrio parahaemolyticus, Vibrio alginolyticus, and Vibrio cholerae non O-1 with plasmid DNA by electroporation[J]. Microbiology and Immunology, 1990, 34(8): 703-708. DOI:10.1111/j.1348-0421.1990.tb01047.x |
| [31] |
Smillie C, Garcillán-Barcia MP, Francia MV, Rocha EPC, De La Cruz F. Mobility of plasmids[J]. Microbiology and Molecular Biology Reviews, 2010, 74(3): 434-452. DOI:10.1128/MMBR.00020-10 |
| [32] |
Thomas CM, Nielsen KM. Mechanisms of, and barriers to, horizontal gene transfer between bacteria[J]. Nature Reviews Microbiology, 2005, 3(9): 711-721. DOI:10.1038/nrmicro1234 |
| [33] |
Wang PX, Yu ZC, Li BY, Cai XS, Zeng ZS, Chen XL, Wang XX. Development of an efficient conjugation-based genetic manipulation system for Pseudoalteromonas[J]. Microbial Cell Factories, 2015, 14(1): 11. DOI:10.1186/s12934-015-0194-8 |
| [34] |
Tolonen AC, Liszt GB, Hess WR. Genetic manipulation of Prochlorococcus strain MIT9313: green fluorescent protein expression from an RSF1010 plasmid and Tn5 transposition[J]. Applied and Environmental Microbiology, 2006, 72(12): 7607-7613. DOI:10.1128/AEM.02034-06 |
| [35] |
Yu ZC, Zhao DL, Ran LY, Mi ZH, Wu ZY, Pang XH, Zhang XY, Su HN, Shi M, Song XY, et al. Development of a genetic system for the deep-sea psychrophilic bacterium Pseudoalteromonas sp. SM9913[J]. Microbial Cell Factories, 2014, 13(1): 13. DOI:10.1186/1475-2859-13-13 |
| [36] |
Le Roux F, Binesse J, Saulnier D, Mazel D. Construction of a Vibrio splendidus mutant lacking the metalloprotease gene vsm by use of a novel counterselectable suicide vector[J]. Applied and Environmental Microbiology, 2007, 73(3): 777-784. DOI:10.1128/AEM.02147-06 |
| [37] |
Hoffart E, Grenz S, Lange J, Nitschel R, Müller F, Schwentner A, Feith A, Lenfers-Lücker M, Takors R, Blombach B. High substrate uptake rates empower Vibrio natriegens as production host for industrial biotechnology[J]. Applied and Environmental Microbiology, 2017, 83(22): e01614-17. |
| [38] |
Luo P, He XY, Liu QT, Hu CQ. Developing universal genetic tools for rapid and efficient deletion mutation in Vibrio species based on suicide T-vectors carrying a novel counterselectable marker, vmi480[J]. PLoS One, 2015, 10(12): e0144465. DOI:10.1371/journal.pone.0144465 |
| [39] |
Van De Water JAJM, Voolstra CR, Rottier C, Cocito S, Peirano A, Allemand D, Ferrier-Pagès C. Seasonal stability in the microbiomes of temperate gorgonians and the red coral Corallium rubrum across the Mediterranean Sea[J]. Microbial Ecology, 2018, 75(1): 274-288. DOI:10.1007/s00248-017-1006-y |
| [40] |
Klindworth A, Mann AJ, Huang SX, Wichels A, Quast C, Waldmann J, Teeling H, Glöckner FO. Diversity and activity of marine bacterioplankton during a diatom bloom in the North Sea assessed by total RNA and pyrotag sequencing[J]. Marine Genomics, 2014, 18: 185-192. DOI:10.1016/j.margen.2014.08.007 |
| [41] |
Pollock FJ, Krediet CJ, Garren M, Stocker R, Winn K, Wilson B, Huete-Stauffer C, Willis BL, Bourne DG. Visualization of coral host-pathogen interactions using a stable GFP-labeled Vibrio coralliilyticus strain[J]. Coral Reefs, 2015, 34(2): 655-662. DOI:10.1007/s00338-015-1273-3 |
| [42] |
Bourne DG, Morrow KM, Webster NS. Insights into the coral microbiome: underpinning the health and resilience of reef ecosystems[J]. Annual Review of Microbiology, 2016, 70(1): 317-340. DOI:10.1146/annurev-micro-102215-095440 |
| [43] |
Dehio C, Meyer M. Maintenance of broad-host-range incompatibility group P and group Q plasmids and transposition of Tn5 in Bartonella henselae following conjugal plasmid transfer from Escherichia coli[J]. Journal of Bacteriology, 1997, 179(2): 538-540. DOI:10.1128/JB.179.2.538-540.1997 |
| [44] |
Guillard RRL, Ryther JH. Studies of marine planktonic diatoms: I. Cyclotella nana Hustedt, and Detonula confervacea (Cleve) Gran[J]. Canadian Journal of Microbiology, 1962, 8(2): 229-239. DOI:10.1139/m62-029 |
| [45] |
Wang F, Qiu RG, Tang L. Establishment and optimization for conjugal transfer system of tautomycetin-producing strain S. griseochromogenes[J]. Journal of Dalian University of Technology, 2012, 52(3): 338-342. (in Chinese) 王芬, 邱荣国, 唐莉. 变构菌素产生菌S. griseochromogenes接合转移体系建立与优化[J]. 大连理工大学学报, 2012, 52(3): 338-342. |
| [46] |
Ohtani N, Sato M, Tomita M, Itaya M. Restriction on conjugational transfer of pLS20 in Bacillus subtilis 168[J]. Bioscience, Biotechnology and Biochemistry, 2008, 72(9): 2472-2475. DOI:10.1271/bbb.80315 |
| [47] |
Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop Ⅱ RM, Petersona KM. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes[J]. Gene, 1995, 166(1): 175-176. DOI:10.1016/0378-1119(95)00584-1 |
| [48] |
Kovach ME, Phillips RW, Elzer PH, Roop RM, Peterson KM. pBBR1MCS: a broad-host-range cloning vector[J]. Biotechniques, 1994, 16(5): 800-802. |
| [49] |
Antoine R, Locht C. Isolation and molecular characterization of a novel broad-host-range plasmid from Bordetella bronchiseptica with sequence similarities to plasmids from Gram-positive organisms[J]. Molecular Microbiology, 1992, 6(13): 1785-1799. DOI:10.1111/j.1365-2958.1992.tb01351.x |
| [50] |
Richhardt J, Larsen M, Meinhardt F. An improved transconjugation protocol for Bacillus megaterium facilitating a direct genetic knockout[J]. Applied Microbiology and Biotechnology, 2010, 86(6): 1959-1965. DOI:10.1007/s00253-010-2503-9 |
| [51] |
Goldberg J, Wilkinson C. Global threats to coral reefs: coral bleaching, global climate change, disease, predator plagues, and invasive species[A]//Wilkinson CR. Status of Coral Reefs of the World[M]. Townsville, Australia: GCRMN Australian Institute of Marine Science, 2004: 67-92
|
 2021, Vol. 48
2021, Vol. 48