扩展功能
文章信息
- PENG Zijia, YU An, LUO Zeyu, LIU Xiaoyong, CHEN Wanfu, YU Zhongdong
- 彭子嘉, 余安, 骆泽煜, 刘小勇, 陈万福, 余仲东
- Punctularia atropurpurascens (Punctulariaceae, Basidiomycota): a new record to China
- 担子菌门总革菌科中国新记录种紫黑点壳菌
- Microbiology China, 2021, 48(11): 4232-4239
- 微生物学通报, 2021, 48(11): 4232-4239
- DOI: 10.13344/j.microbiol.china.210155
-
文章历史
- Received: February 07, 2021
- Accepted: May 12, 2021
- Published online: June 15, 2021
2. College of Forestry, Central South University of Forestry and Technology, Changsha, Hunan 410004, China;
3. Institute of Microbiology, Chinese Academy of Sciences, Beijing 100101, China
2. 中南林业科技大学林学院 湖南 长沙 410004 ;
3. 中国科学院微生物研究所 北京 100101
The genus Punctularia consists of merely three accepted species, P. atropurpurascens (Berk. & Broome) Petch, P. bambusicola C.L. Zhao, and P. strigosozonata (Schwein.) P.H.B. Talbot[1-3]. In China, Punctularia bambusicola is reported from Yunnan province[2], P. strigosozonata is found in Shanxi province and Jilin province[4-5], but P. atropurpurascens has not been reported[6-7] or listed in the data-base Checklist of Fungi in China (http://fungalinfo.im.ac.cn/) till now. Its known distribution involved few countries in Africa, Asia, Europe, North America and South America[8-11] (https://www.gbif.org), and India[12] is the only Asia country documenting this species.
Studies on P. atropurpurascens involved morphological features[9], growth cycles[10], degradation function[13-14], antifungal and medicinal components, which have garnered huge attention in recent years due to its ecological, economical and medical values. For example, Anke et al.[15] extracted phlebiakauranol aldehyde from cultures of P. atropurpurascens, which exhibited strong antifungal, antibacterial and cytotoxic activities against several phytopathogens; Alborés et al.[16] detected and purified a novel lectin with specific compatibility towards N-acetyl-glucosamine from mycelia; latterly, Alborés et al.[17] continued extracting a laccase from the extracellular extract of P. atropurpurascens, which was able to degrade Remazol Brilliant Blue R and Acid Blue 25 dyes. In this study, we firstly reported this high-valuable P. atropurpurascens in China, and described its eye-catching morphologies, rDNA ITS phylogenetic relationship with the homogenesis species, and its growth cycles and hosts in details.
2 Materials and Methods 2.1 MaterialsWugong county, located in western Guanzhong plain in Shaanxi province, belongs to the semi-humid monsoon belt of warm temperate zone. Its average annual precipitation and sunshine duration are 552.6–663.3 mm and 2 094.9 h, respectively. From south to north, with an elevation from 415 m to 600 m, there are four parallel landforms, Weihe beach, terraced plain, loess tableland, and front and depression of piedmont pluvial fan. Weishui River, commonly known as back river in Wugong county, is a tributary of Weihe river, which traced back to the Yellow river[18].
The studied specimens were collected beside Weishui river. Voucher specimens were deposited in the Mycological Herbarium of Forestry College, Northwest A & F University (HMNWAFU-CF). Tiny sections were cut from the fruitbody and incubated on the potato-dextrose agar (PDA) medium at 25 ℃ in the dark for fungal isolation. About 5 days later, emerging fungal mycelia were transferred into fresh PDA plates to develop purified living cultures[19].
2.2 Morphological observationThe colorful photographs were captured by LUMIX G8M during each stage in the growth cycles, and fungal histological sections were observed at magnification up to 1 000× using an Olympus CX41 microscope. The microscopic observation followed Guan et al.[2] and Cui et al.[20]. Hand drawings were implemented with the aid of a digital pen tablet (GAOMON WH850) and the software Adobe Photoshop 2020. Slides of basidiocarps were stained by Cotton Blue and Melzer's reagent for microscopic observation, measurement and drawing. Conidia from the culture and basidiospores from basidiocarps were observed and measured randomly. To determine the size variation of spores, 5% of the data were excluded from each end of the measurement range and noted in parentheses. In the text, the following abbreviations are used: IKI=Melzer's reagent, IKI–=inamyloid and indextrinoid, KOH=5% potassium hydroxide, CB=Cotton Blue, CB–=acyanophilous reaction of CB, L=mean spore length (arithmetic average of all spores), W=mean spore width (arithmetic average of all spores), Q=variation in the L/W ratios, n=number of spores from the given numbers of specimens. The width of a basidium was measured at its thickest part, and the length was measured from the apex (sterigmata excluded) to the basal septum. Terminological description of color followed Petersen[21].
2.3 DNA extraction and PCR amplificationA total of 0.02 g thalli from the specimen and a pure colony were separately ground to extract the genomic DNA by CTAB method according to Doyle et al.[22] and Qi et al.[23]. The concentrations of DNA extract from specimen and colony are 535.960 ng/μL (OD260/280=1.95) and 462.237 ng/μL (OD260/280=1.90), respectively. Internal transcribed spacer (ITS) regions were amplified using the universal primers ITS1/ITS4[24]. The PCRs were carried out in a final volume of 30 μL, containing 15 μL 2×Taq PCR MasterMix (CoWin Biosciences, China), 1.5 μL of 10 μmol/L each primer (Shanghai Sangon Biotech Company Limited, China), 3 μL template DNA, and 9 μL ddH2O. Thermal cycling was implemented in a GeneAmp PCR TC-96 instrument (Bioer Technology Company Limited, Hangzhou, China) for pre-denaturation at 94 ℃ for 3 min, a subsequent PCR cycle lasting for 30 s at 94 ℃, 30 s at 55 ℃, and 30 s at 72 ℃. Such 35 cycles and a final extension at 72 ℃ for 10 min were carried out until the procedure was terminated at 4 ℃. The PCR products were then sequenced by Shanghai Sangon Biotech Company Limited, China, with primers ITS1 and ITS4 same as used in PCR reaction.
2.4 Sequence alignment and phylogenetic analysisSequences were assembled by BioEdit 7.2.5 (Bioedit Limited, UK), searched by BLAST, compared in the NCBI database and deposited in GenBank (Accession No. MW556264, MW556465). Other homologous sequences used for phylogenetic analysis were downloaded from GenBank (https://www.ncbi.nlm.nih.gov/genbank/), UNITE (https://unite.ut.ee) and Mushroom Observer (https://mushroomobserver.org/)[9-10]. All sequences were aligned using the MUSCLE algorithm in MEGA-X (https://www.megasoftware.net/home), and were manually adjusted to allow maximum alignment and minimum gaps, with all positions containing gaps or missing data eliminated completely. Two sequences of Vuilleminia alni Boidin, Lanq. & Gilles downloaded from GenBank were used as the outgroup. Aligned sequence matrix was exported as FASTA and MEGA format. Phylogenetic trees were constructed with maximum-likelihood (ML), maximum parsimony (MP) and bayesian inference (BI) methods.
Topologies were constructed based on ML analysis with MEGA formatted file[25]. Twenty-four different nucleotide substitution models were considered, and the model with the lowest bayesian information criterion (BIC) score was chosen as the best-fit evolutionary model. Initial trees for the heuristic search were obtained automatically by applying neighbor-joinning and BioNJ algorithms to calculate a matrix of pairwise distances, which was further estimated using the maximum composite likelihood (MCL) approach; and the topology with superior log-likelihood value was selected. The MP tree was obtained by using the subtree-pruning- regrafting (SPR) algorithm[26] with search level 1, in which the initial trees were obtained by the random addition of sequences (10 replicates). Both MP and ML bootstrap analysis were conducted 1 000 replicates by software MEGA-X[27].
The FASTA formatted file was firstly transformed into NEXUS format using SequenceMatrix 1.8 and then was applied for calculating BI using MrBayes V3.1.2[28-29]. Four Markov chains were run from random starting trees for 1 000 000 generations till the average standard deviation of split frequencies was below 0.01, and trees were sampled every 100 generations. The first 25% of the sampled trees were discarded as burn-in and the remaining ones were used to reconstruct a majority-rule consensus and to calculate BI posterior probabilities (Bpps) of the clades. Tree reconstruction, visualization and editing were implemented with FIGTREE V1.4.0 (http://tree.bio.ed.ac.uk/software/figtree/) and MEGA-X. The Bpps were followed by bootstrap values of ML and MP at the nodes in the topology. Branches that received bootstrap support for ML (≥75%), MP (≥75%) and Bpp (≥0.95) were considered as significantly supported[20].
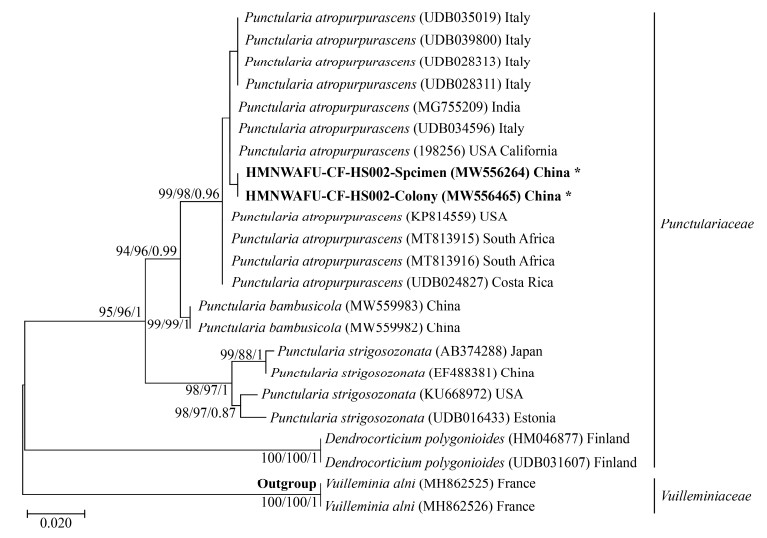
3 Results 3.1 Phylogenetic analysisTwenty-three sequences of rDNA ITS were employed to construct the phylogenetic tree, eight of which were downloaded from UNITE (prefix with UDB), twelve from GenBank, one from Mushroom Observer (without prefix) and two newly generated from our collections. Tamura 3-parameter model was estimated as the best-fit evolutionary model for the aligned dataset. ML analysis generated a tree topology that was almost identical to the MP and Bayesian tree. Phylogenetic analysis revealed that two sequences from the specimen (Accession No. MW556264) and the colony (Accession No. MW556465) of the HMNWAFU-CF-HS002 were finely fitted in the Punctularia atropurpurascens clade, which included ITS sequences from Italy, India, USA, South Africa and Costa Rica, and formed an independent sub-clade (Figure 1).
|
| Figure 1 Phylogenetic tree constructed by the maximum-likelihood (ML) method based on sequences of the ITS region of nuclear ribosomal DNA 图 1 基于rDNA ITS区序列构建的最大似然法系统发育树 Note: The phylogenetic tree is drawn to scale, with branch lengths measured in the number of substitutions per site[27]. The Bayesian inference posterior probabilities (Bpps) are followed by bootstrap values of ML and maximum parsimony (MP) at the nodes in the topology. Sequence data are shown with species name, accession number in GenBank or UNITE (prefix with UDB) or Mushroom Observer (without prefix), and location of the voucher specimen 注:发育树按比例绘制,用每个位点的替换次数来衡量树枝的长度[27]。在拓扑结构的节点处标出了最大似然法(ML)、最大简约法(MP)的自展值,以及贝叶斯后验概率(Bpps)。序列信息有:物种名称、GenBank或UNITE (前缀为UDB)或Mushroom Observer (无前缀)登录号,以及凭证标本所在地 |
|
|
Punctularia atropurpurascens (Berk. & Broome) Petch, Ann. R. bot. Gdns Peradeniya 6: 160, 1916. Figure 2
|
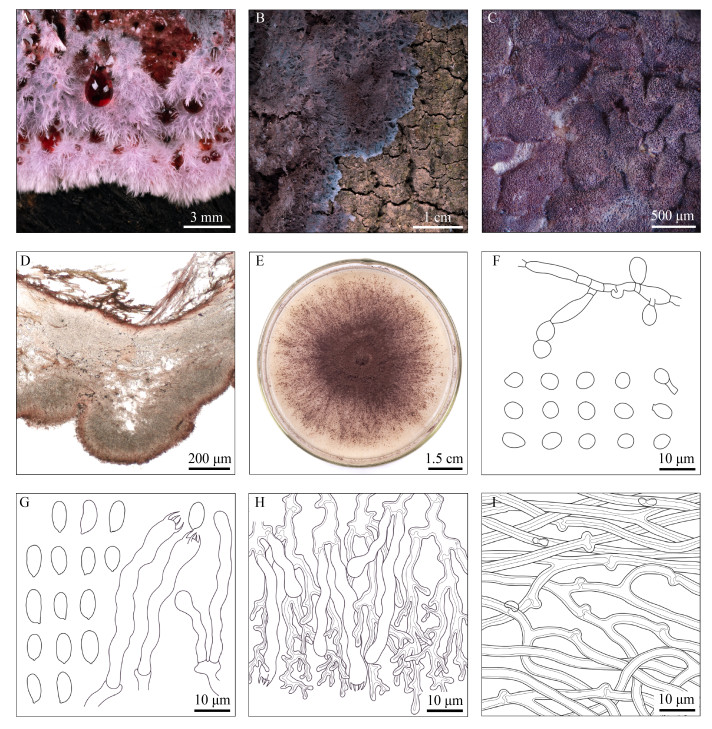
| Figure 2 Features of Punctularia atropurpurascens (drawn from HMNWAFU-CF-HS002) 图 2 紫黑点壳菌形态特征(绘自HMNWAFU-CF-HS002) Note: A: Morphological characteristics in autumn with red droplets secreted; B: Morphological characteristics in winter; C: Hymenium; D: Vertical section through the basidiocarp; E: Colony; F: Conidia and segments of supporting hyphae; G: Basidiospores, basidia and basidioles; H: Subhymenial generative hyphae, dendrohyphidia, basidia and basidioles; I: Subicular generative hyphae and abhymenial generative hyphae 注:A:秋季形态特征,伴有红色液滴渗出;B:冬季形态特征;C:子实层;D:担子果切片;E:菌落;F:分生孢子及支撑菌丝片段;G:担孢子、担子及幼担子;H:子实基层生殖菌丝、树状子实层端菌丝、担子及幼担子;I:菌丝层生殖菌丝及远子实层生殖菌丝 |
|
|
=Thelephora subhepatica Berk., London J. Bot. 5: 3, 1846.
≡Thelephora atropurpurascens Berk. & Broome, J. Linn. Soc., Bot. 14: 64, 1873.
=Corticium tuberculosum Pat., in Patouillard & Lagerheim, Bull. Soc. mycol. Fr. 8: 118, 1892.
=Punctularia tuberculosa (Pat.) Pat. & Lagerh., Bull. Herb. Boissier 3: 57, 1895.
=Punctularia subhepatica (Berk.) Hjortstam, Mycotaxon 54: 191, 1995.
Fruitbody—Basidiocarps effused to effuse- reflexed, adherent on the substrate, in patches about a few centimeters across, up to 1 mm thick. Subiculum thin layered, 17–84 (–105) µm thick, fibrose, coffee brown to dark brown. Hymenium surface papillate or shallowly tuberculate, becoming crustose to corneous when dry, reddish brown to dark purplish brown. Superior surface cottony, fluffy, membranous, light pink to rose, with reddish aqueous droplets secreted from pores in autumn; compact, woolly, partly powdered, pinkish purple to purplish brown in winter.
Hyphal structure—Hyphal system monomitic, generative hyphae with clamp connections. Subicular generative hyphae 2.3–3.9 µm wide, with thick solid walls, yellowish brown to coffee brown, sometimes with multiple clamp connections on the single septum. Abhymenial generative hyphae 1.8–3.7 µm wide, thick-walled, branched distantly from septa, hyaline. Subhymenial generative hyphae 1.9–4.3 µm wide, thin-walled to slightly thick-walled, subhyaline or very pale brown, frequently branched.
Hymenium—Dendrohyphidia numerous, luxuriant, dendritic or coralloid, up to 75 µm long, 3.0–6.6 µm wide, with thick walls and very narrow or absent lumens at tips, ochraceous to coffee brown, darker from bases to extremities. Basidia tubular or clavate, flexuose, infrequent, (39.1–66.3) µm×(5.1–6.9) µm; 4 sterigmata up to 5 µm long. Basidioles similar to basidia in shape, but smaller in size, without obvious sterigmata. Cystidia and cystidioles absent.
Spores—Basidiospores infrequent, ellipsoid to narrowly ellipsoid, (7.0–9.2 (–9.9)) µm×(3.5–4.6 (–4.9)) µm in size, L=8.0, W=4.0, Q=2 (n=30/1), smooth, thin-walled, hyaline. Conidia prevalent, abundant, in branched chains, subglobose to ellipsoid, sometimes irregular, (4.3–6.9 (–7.8)) µm×(3.6–4.6 (–4.8)) µm, L=5.5, W=4.1, Q=1.3 (n=30/1), with thick walls, pale brownish to pale purplish brown, with lipid bodies inside, breaking off from each one and showing the remains of the linking walls at both ends, sometimes shed with segments of supporting hyphae.
Colonies—Colonies tufted, pulverulent, violet at first, becoming dark reddish-brown later, with white margin, forming a thick mass of conidia.
Chemical reaction—CB–; IKI–; tissues darkening in KOH.
Specimen examined—China. Shaanxi province, Xianyang city, Wugong county, Weishui river, on rotten stumps of Paulownia tomentosa Steud., 4 October 2020 & 3 January 2021, Peng ZJ, Yu ZD & Luo ZY (HMNWAFU-CF-HS002).
4 DisscussionFor the first time, the species P. atropurpurascens was reported in China based on morphological and phylogenetic evidences in this study. Phylogenetically, P. atropurpurascens is more closely related to P. bambusicola than P. strigosozonata based on rDNA ITS sequence analysis (Figure 1), which is in accordance with that of Knijn et al.[9] and Guan et al.[2]. Morphologically, the genus Punctularia lacks cystidia and cystidioles, while usually has obvious sterile dendrohyphidia, however, three species in this genus displayed diverse morphologies in basidiocarps thickness, color of hymenial surface and margin, basidia size, and dendrohyphidia length. P. atropurpurascens appeared to be thicker basidiocarps (up to 1 mm), larger basidia ((39.1–66.3) µm×(5.1–6.9) μm) and longer dendrohyphidia (up to 75 µm) than P. bambusicola, of which is 100–300 µm thick, (17.5–21.5) µm×(4.0–5.5) μm size and up to 42 μm long[2], respectively. The hymenial surface in the former is reddish brown to dark purplish brown or bluish[10], but from pink to rose in the latter. And both of them have basidiocarps with more or less whitish fibrillose margin[2, 9-10]. Punctularia strigosozonata differs from the above two species in its resupinate to effuse-reflexed basidiocarps, with orangish brown to maroon or dark violaceous hymenial surface and brown velutinous margin[2, 30-31]. Among them, only P. atropurpurascens developed the distinct structure of subiculum. Interestingly, P. atropurpurascens displays morphological changes during the annual growth cycle[10]. Two cycles were found under the different habitat humidity, a long one in wet conditions and a short one under drought stress[10]. The superior surface of P. atropurpurascens showed pink feathery morphology and exuded reddish aqueous droplets from pores in autumn (Figure 2A), but compact, woolly, partly powdered, pinkish purple to purplish brown without exudates when given an enough humidity in winter (Figure 2B), which were as same as Knijn et al.[9] reported. In addition, Knijn et al.[10] reported that these droplets were composed of phlebiarubrone derivatives and lipid molecules involved in various cellular signal transduction pathways in fungi and plants[32-35].
All three species of Punctularia displayed excellent abilities of degradation and were considered as white rot saprobes according to the previous studies[2, 9-10, 14]. Punctularia strigosozonata was found on branch of Pistacia, Populus and Quercus[3, 31], P. bambusicola on a dead bamboo, P. atropurpurascens on Acacia[1-2], Celtis australis L.[10], Juglans regia L.[4], Mangifera[1-2], P. radiata D. Don[14], Platanus acerifolia (Aiton) Willd., Quercus ilex L., Q. cerris L., and Q. suber L.[10]. Additionally, we found P. atropurpurascens can colonize and gravely decompose the stumps of P. tomentosa, an indigenous tree in Guanzhong Plain. This is the first report of the novel host P. tomentosa in China.
| [1] |
Petch T. Revisions of ceylon fungi (Part IV)[J]. Annals of the Royal Botanic Gardens Peradeniya, 1916, 6: 153-183. |
| [2] |
Guan QX, Zhao W, Zhao CL. A new species of Punctularia (Punctulariaceae, Basidiomycota) from southwest China[J]. Phytotaxa, 2021, 489(3): 285-292. DOI:10.11646/phytotaxa.489.3.5 |
| [3] |
Talbot PHB. Studies of some South African resupinate hymenomycetes[J]. Bothalia, 1958, 7(1): 131-187. DOI:10.4102/abc.v7i1.1652 |
| [4] |
Li ZY. An investigation on fungi of, Jiangxian County of Shanxi Province (I)[J]. Journal of Shanxi University: Natural Science Edition, 1985, 8(4): 83-89. 李宗英. 山西绛县真菌调查报告(Ⅰ)[J]. 山西大学学报: 自然科学版, 1985, 8(4): 83-89. |
| [5] |
Bau T, Liu WZ, Fan YG, Kang GP. A checklist of macrofungi collected from different forests in Changbai mountains (Ⅴ): deciduous forest[J]. Journal of Fungal Research, 2011, 9(2): 77-87, 99. 图力古尔, 刘文钊, 范宇光, 康国平. 长白山大型真菌物种多样性调查名录Ⅴ阔叶林带[J]. 菌物研究, 2011, 9(2): 77-87, 99. DOI:10.3969/j.issn.1672-3538.2011.02.004 |
| [6] |
Dai YC. A revised checklist of corticioid and hydnoid fungi in China for 2010[J]. Mycoscience, 2011, 52(1): 69-79. DOI:10.1007/S10267-010-0068-1 |
| [7] |
Li Y, Li TY, Yang ZL, Bau T, Dai YC. Atlas of Chinese Macrofungal Resources[M]. Zhengzhou: Central China Farmer's Publishing House, 2015. 李玉, 李泰辉, 杨祝良, 图力古尔, 戴玉成. 中国大型菌物资源图鉴[M]. 郑州: 中原农民出版社, 2015. |
| [8] |
Berkeley MJ. Decades of fungii. Decade XI[J]. Hooker's London Journal of Botany, 1846, 5: 1-6. |
| [9] |
Knijn A, Ferretti A. Punctularia atropurpurascens in the Villa Ada urban park in Rome, Italy[EB/OL]. 2018
|
| [10] |
Knijn A, Saar I, Ferretti A. Biological characteristics of Punctularia atropurpurascens through morphological and molecular analyses during development[EB/OL]. 2019
|
| [11] |
Spies CFJ, Mostert L, Carlucci A, Moyo P, Van Jaarsveld WJ, Du Plessis IL, Van Dyk M, Halleen F. Dieback and decline pathogens of olive trees in South Africa[J]. Persoonia - Molecular Phylogeny and Evolution of Fungi, 2020, 45(1): 196-220. DOI:10.3767/persoonia.2020.45.08 |
| [12] |
Vu D, Groenewald M, De Vries M, Gehrmann T, Stielow B, Eberhardt U, Al-Hatmi A, Groenewald JZ, Cardinali G, Houbraken J, et al. Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation[J]. Studies in Mycology, 2019, 92: 135-154. DOI:10.1016/j.simyco.2018.05.001 |
| [13] |
Ferraz A, Rodríguez J, Freer J, Baeza J. Formic acid/acetone-organosolv pulping of white-rotted Pinus radiata softwood[J]. Journal of Chemical Technology & Biotechnology, 2000, 75(12): 1190-1196. |
| [14] |
Ferraz A, Rodríguez J, Freer J, Baeza J. Biodegradation of Pinus radiata softwood by white- and brown-rot fungi[J]. World Journal of Microbiology and Biotechnology, 2001, 17(1): 31-34. DOI:10.1023/A:1016646802812 |
| [15] |
Anke H, Casser I, Steglich W, Pommer EH. Antibiotics from basidiomycetes. 26 Phlebiakauranol aldehyde an antifungal and cytotoxic metabolite from Punctularia atropurpurascens[J]. The Journal of Antibiotics, 1987, 40(4): 443-449. DOI:10.7164/antibiotics.40.443 |
| [16] |
Alborés S, Mora P, Cerdeiras MP, Fraguas LF. Screening for lectins from basidiomycetes and isolation of Punctularia atropurpurascens lectin[J]. Journal of Basic Microbiology, 2014, 54(2): 89-96. DOI:10.1002/jobm.201200229 |
| [17] |
Alborés S, Bustamante MJ, Fraguas LF, Cerdeiras MP. Proteins from Punctularia atropurpurascens with biotechnological applications[J]. Natural Resources, 2014, 5(15): 915-925. DOI:10.4236/nr.2014.515078 |
| [18] |
Zhao YT, Chang QR, Chen XX, Ma TG. Study on the spatial pattern of available potassium in county farmland: Wugong County as an example[J]. Journal of Northwest A & F University: Natural Science Edition, 2011, 39(3): 157-162, 167. 赵业婷, 常庆瑞, 陈学兄, 马廷刚. 县域耕地土壤速效磷空间格局研究: 以武功县为例[J]. 西北农林科技大学学报(自然科学版), 2011, 39(3): 157-162, 167. |
| [19] |
Cai MZ, Idrees M, Zhou Y, Zhang CL, Xu JZ. First report of green mold disease caused by Trichoderma hengshanicum on Ganoderma lingzhi[J]. Mycobiology, 2020, 48(5): 427-430. DOI:10.1080/12298093.2020.1794230 |
| [20] |
Cui BK, Li HJ, Ji X, Zhou JL, Song J, Si J, Yang ZL, Dai YC. Species diversity, taxonomy and phylogeny of Polyporaceae (Basidiomycota) in China[J]. Fungal Diversity, 2019, 97(1): 137-392. DOI:10.1007/s13225-019-00427-4 |
| [21] |
Petersen JH. Farvekort. The Danish Mycological Society's Colour-Chart[M]. Greve: Foreningen til Svampekundskabens Fremme, 1996.
|
| [22] |
Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissues[J]. Phytochemical Bulletin, 1987, 19: 11-15. |
| [23] |
Qi M, Xie CX, Chen QW, Yu ZD. Pestalotiopsis trachicarpicola, a novel pathogen causes twig blight of Pinus bungeana (Pinaceae: Pinoideae) in China[J]. Antonie Van Leeuwenhoek, 2021, 114(1): 1-9. DOI:10.1007/s10482-020-01500-8 |
| [24] |
White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics[A]//PCR Protocols[M]. Amsterdam: Elsevier, 1990: 315-322
|
| [25] |
Yadav S, Verma SK, Singh R, Singh VK, Kushwaha P. A new species of Podosphaera sect. Sphaerotheca subsect. Magnicellulatae from India and a key to all species reported on Fabaceae[J]. Phytotaxa, 2020, 453(2): 108-120. DOI:10.11646/phytotaxa.453.2.2 |
| [26] |
Nei M, Kumar S. Molecular Evolution and Phylogenetics[M]. New York: Oxford University Press, 2000.
|
| [27] |
Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms[J]. Molecular Biology and Evolution, 2018, 35(6): 1547-1549. DOI:10.1093/molbev/msy096 |
| [28] |
Huelsenbeck JP, Ronquist F. MRBAYES: bayesian inference of phylogenetic trees[J]. Bioinformatics, 2001, 17(8): 754-755. DOI:10.1093/bioinformatics/17.8.754 |
| [29] |
Ji JX, Li Z, Li Y, Kakishima M. Two new species of Pucciniastrum producing dimorphic sori and spores from northeast of China[J]. Mycological Progress, 2019, 18(4): 529-540. DOI:10.1007/s11557-018-1460-z |
| [30] |
Bondartseva MA, Lositskaya VM, Zmitrovich IV. Punctularia strigosozonata (Punctulariaceae) in Europe[J]. Karstenia, 2000, 40(1/2): 9-10. |
| [31] |
Bernicchia A, Gorjón SP. Fungi Europaei 12: Corticiaceae s.l[M]. Lomazzo: Edizioni Candusso, 2010.
|
| [32] |
Rodrigues ML, Travassos LR, Miranda KR, Franzen AJ, Rozental S, De Souza W, Alviano CS, Barreto-Bergter E. Human antibodies against a purified glucosylceramide from Cryptococcus neoformans inhibit cell budding and fungal growth[J]. Infection and Immunity, 2000, 68(12): 7049-7060. DOI:10.1128/IAI.68.12.7049-7060.2000 |
| [33] |
Gutierrez AL, Farage L, Melo MN, Mohana-Borges RS, Guerardel Y, Coddeville B, Wieruszeski JM, Mendonça-Previato L, Previato JO. Characterization of glycoinositolphosphoryl ceramide structure mutant strains of Cryptococcus neoformans[J]. Glycobiology, 2007, 17(6): 1C. DOI:10.1093/glycob/cwm030 |
| [34] |
Alvarez-Vasquez F, Riezman H, Hannun YA, Voit EO. Mathematical modeling and validation of the ergosterol pathway in Saccharomyces cerevisiae[J]. PLoS One, 2011, 6(12): e28344. DOI:10.1371/journal.pone.0028344 |
| [35] |
Gronnier J, Germain V, Gouguet P, Cacas JL, Mongrand S. GIPC: glycosyl inositol phospho ceramides, the major sphingolipids on earth[J]. Plant Signaling & Behavior, 2016, 11(4): e1152438. |
 2021, Vol. 48
2021, Vol. 48