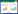扩展功能
文章信息
- 陈静, 陈英虎
- CHEN Jing, CHEN Ying-Hu
- CRISPR/Cas9在Epstein-Barr病毒感染免疫中的研究进展
- CRISPR-Cas9 in immunity of Epstein-Barr virus infection: a review
- 微生物学通报, 2020, 47(9): 3065-3074
- Microbiology China, 2020, 47(9): 3065-3074
- DOI: 10.13344/j.microbiolchina.200734
-
文章历史
- 收稿日期: 2020-07-17
- 接受日期: 2020-08-30
- 网络首发日期: 2020-09-02
EBV是1964年Epstein和Barr在伯基特淋巴瘤(Burkitt’s lymphoma,BL)中发现的肿瘤相关DNA病毒,属于人类疱疹病毒γ亚科[1-2]。人类是EBV唯一的天然宿主,患者和EBV潜伏感染者为传染源,经口密切接触传播,主要感染人类的口咽部上皮细胞和B淋巴细胞[3]。EBV在人群中的感染非常普遍,流行病学资料显示,成年人的EBV抗体阳性率高达90%以上[4]。临床上,EBV感染不同地区、不同年龄人群引起的疾病形式多样,预后差异较大;除少数急性EBV感染者呈自限性,预后较好外;大部分慢性潜伏感染者病程较长,迁延不愈;同时,EBV潜伏感染与多种恶性肿瘤密切相关,如伯基特淋巴瘤、鼻咽癌(nasopharyngeal carcinoma,NPC)、霍奇金淋巴瘤(Hodgkin lymphoma,HL)、弥漫性大B细胞淋巴瘤(diffuse large B cell lymphoma,DLBCL)、EBV相关胃癌(Epstein-Barr virus-associated gastric carcinoma,EBVaGC)和免疫抑制患者(如HIV感染或器官移植后)发生的B细胞淋巴瘤等[5-6]。目前,EBV的具体致病机制尚不明确,EBV慢性潜伏感染及相关恶性肿瘤仍缺乏有效的治疗药物和根治措施,总体预后不佳[4-6]。
CRISPR/Cas系统又称成簇的规律间隔短回文重复序列及相关核酸酶系统,源于细菌与古生菌中的适应性免疫系统,用于抵御外源噬菌体或质粒DNA入侵[7-8]。CRISPR/Cas9作为结构简化改造的Ⅱ型CRISPR/Cas系统,可针对目的基因序列按照碱基互补配对原则设计向导RNA (single guide RNA,sgRNA),在sgRNA引导下,由Cas9核酸酶进行靶向识别剪切,再通过同源重组(homologous recombination,HR)和/或非同源末端链接(non-homologous ending-joining,NHEJ)完成特定基因序列的靶向编辑[9-10]。作为第三代DNA核酸酶基因编辑技术,相较传统的DNA核酸酶基因编辑技术,如锌指核酸酶(Zinc-finger nucleases,ZFN)和类转录激活因子效应物核酸酶(transcription activator-like effector nucleases,TALENs)基因编辑技术等,CRISPR/Cas9系统具有简便高效、靶向位点选择灵活以及可同时编辑多个靶点等优势[10-11]。近年来,越来越多的学者致力于将CRISPR/Cas9技术应用于EBV感染免疫相关研究中并取得了一系列突破性进展。
1 运用CRISPR/Cas9技术筛选EBV致病基因亚型和宿主依赖基因EBV感染不同人群、不同宿主细胞后引发的疾病类型和临床预后存在较大差异,除宿主遗传背景和地理环境因素之外,不同的EBV致病基因亚型和宿主依赖基因是其关键因素[12-15]。以往根据基因多态性将EBV基因组粗略分为2个亚型,1型最常见,主要作用于B细胞,使其成为B淋巴细胞永生化细胞系,2型常见于非洲部分地区交叉感染病例;在体外细胞培养条件下,1型转化B淋巴细胞、使其发生癌变的能力强于2型[14, 16]。后有研究发现,EBV潜伏感染时可表达9种潜伏期抗原,包括6种核抗原(EBV nuclear antigens,EBNA1、2、3a、3b、3c和LP)和3种潜伏膜蛋白(latent membrane proteins,LMP1、2a和2b);根据病毒表达的潜伏期抗原种类,可将EBV基因组分为3型:Ⅰ型仅EBNA1表达,常见于伯基特淋巴瘤;Ⅱ型表达EBNA1和LMPs,见于鼻咽癌、EBV相关胃癌、EBV阳性霍奇金淋巴瘤、NK/T细胞淋巴瘤和弥漫性大B细胞淋巴瘤等;Ⅲ型表达全部EBNAs、LMPs和非编码RNA等,见于淋巴母细胞系(lymphoblastoid cell lines,LCLs)、EBV阳性移植后淋巴增殖病和HIV相关淋巴增殖病等[16-17]。最新研究发现,EBV基因亚型众多,远超上述分类,即使临床特征相近的同一EBV感染疾病类型,其致病基因亚型之间也存在不同程度的基因变异[12-13]。随着基因测序技术的快速发展和推广应用,针对EBV致病基因序列和宿主依赖基因的研究逐渐开展起来。但人类病毒基因组序列通常较长,EBV双链DNA更是长达175 kb,第一代基因测序技术即传统桑格法难以对其进行完整复制分析;第二代测序技术虽然不受序列长度限制,但因其探针测序较短,难以识别区分EBV基因组中的重要重复片段,如ir1–4和fr等;因此,针对EBV致病基因序列特征的测序技术备受期待[18-19]。
EBV感染宿主B淋巴细胞后,可使其恶变转化为伯基特淋巴瘤和淋巴母细胞系等,在其恶变转化过程中,EBV致病基因亚型和宿主依赖基因如何发挥作用目前仍不明确。Ma等[19]利用CRISPR/Cas9技术,对EBV感染阳性、B细胞发生恶变转化的BL和LCLs基因组进行基因亚型筛选,分别检测出与肿瘤细胞生长和存活至关重要的57个BL基因和87个LCLs基因;进一步通过基因功能缺失对比研究证实,EBV潜伏膜蛋白LMP-1诱导的cflip是LCLs抵御TNF-α介导的程序性细胞死亡的关键因子;除此之外,EBV诱导激活的batf/irf4是LCLs中抑癌基因bim抑制和癌基因myc激活的决定因素。在另一项研究中,Guo等[20]利用全基因组CRISPR/Cas9筛选技术在BL细胞中筛选出一个以宿主基因myc为中心的关键基因网络,其包括fact、staga和mediator等基因,它们相互协作,抑制关键基因bzlf1启动子。bzlf1是EBV溶细胞性裂解反应的主要调控基因,其编码的ZEBRA蛋白是激活EBV溶细胞性裂解级联反应的重要转录因子[21]。该基因表达下调可使B淋巴细胞免于溶细胞性裂解死亡,持续处于潜伏感染状态,最终介导宿主B淋巴细胞恶变转化[20-21]。上述研究表明,CRISPR/Cas9全基因筛选技术的应用有助于发现及分析EBV相关BL和LCLs的关键致病基因亚型和宿主依赖基因,从而加深对EBV致病机理的认识。
EBV除诱导B淋巴细胞恶变转化外,也可引发上皮细胞恶变[5, 12]。研究显示全球约10%胃癌患者存在EBV感染,EBVaGC每年新发病例超过8万人[22]。相较其他EBV非相关胃癌(Epstein-Barr virus non-associated gastric carcinoma,EBVnGC),EBVaGC基因特征包括bart miRNA高表达、ebna-1低表达、lmp1缺失等[23-24]。bart miRNA为EBV关键非编码基因,可通过靶向调控DNA双链断裂修复基因atm、抑癌基因tp53和胞核输入受体ipo7基因等机制,抑制细胞凋亡和逃避免疫杀伤,促进肿瘤演变进展[25-27]。Kanda等[28]将从胃癌患者分离培养的EBV阳性的胃癌细胞株SNU719和YCCEL1作为研究对象,利用CRISPR/Cas9技术,以bart miRNA病毒开放阅读框(bart miRNA viral open reading frames,bvrf1)作为靶点,用以往验证有效的pX330-sgEBV作为引导RNA,精准识别病毒基因组中的bart miRNA致病基因序列,对其进行靶向剪切和同源修复,再结合第三代测序技术PacBio单分子测序对包含重复片段的基因组进行完整读取,并进一步利用Western blotting蛋白免疫印迹等实验进行验证[29]。此方法克服了传统二代测序技术的缺陷,加深了对胃癌相关EBV致病基因亚型的认识;提示CRISPR/Cas9技术可用于不同疾病相应的EBV基因组异质性和表型变异的鉴别分析,进而阐明不同EBV致病基因亚型的生物学意义[30]。该研究也存在一些待改进的地方,首先用bvrf1作为CRISPR/Cas9技术的识别靶点,聚焦在BART miRNA病毒开放阅读框,而不是完整的EBV全基因组[30]。另外,缺少正常的健康组作为对照,而且相应的胃癌细胞类型只来源于亚洲病例(韩国),未涵盖其他地区和人群标本,无法综合且全面分析胃癌相关EBV基因变异特征;因此,只有CRISPR/Cas9技术和其他技术相互补充,才能全面深入地分析EBV相关疾病的致病基因亚型和宿主依赖基因[30-31]。
2 运用CRISPR/Cas9技术探索EBV关键致病机制EBV通过唾液传播进入体内后,主要感染人B淋巴细胞,也可感染上皮细胞、T淋巴细胞及NK细胞等[3-4, 32]。根据宿主细胞的转归,EBV感染可分为两类[33-37]:(1)溶细胞性感染(lytic infection,Lysis):主要在B淋巴细胞中,病毒DNA在宿主细胞核中复制、转录、成熟,编码大量病毒蛋白,促进宿主细胞“爆炸式增殖”,同时病毒编码蛋白可被体内抗原提呈细胞(antigen-presenting cells,APCs)识别,最终在细胞毒性T淋巴细胞(cytotoxic T lymphocytes,CTLs)作用下引起宿主细胞溶解、死亡,并释放大量新生病毒颗粒,常见于急性感染如儿童传染性单核细胞增多症(infectious mononucleosis,IM);(2)潜伏感染(latent infection,Latency),可见于B淋巴细胞、T淋巴细胞和NK细胞等,病毒DNA双链在宿主细胞核中完成克隆复制,但病毒蛋白编码基因大部分不表达,仅少量免疫原性低的抗原蛋白表达,可在宿主细胞中长期存活、持续存在,最终导致宿主细胞恶变转化,常见于慢性活动性EBV感染、EBV相关肿瘤如伯基特淋巴瘤、鼻咽癌等。EBV感染宿主细胞后的转归类型并非静止不变,呈潜伏感染状态的EBV在B细胞受体交联作用(BCR cross-linking)或化学诱导物激活后,可转变为溶细胞性感染,即溶细胞性重激活(lytic reactivation) (图 1)[36-37]。EBV感染机体后,病毒与宿主细胞之间通过多种关键基因及信号通路交互作用,最终引起不同类型的感染和疾病转归[3, 17, 38-39]。利用CRISPR/Cas9技术对其中的关键基因进行功能缺失(loss of function)和调控干预,再通过对比研究和综合分析,有助于阐明EBV感染的具体致病机理[40]。

|
| 图 1 EBV感染过程及其类型 Figure 1 The process of EBV infection and its types |
|
|
在终生感染EBV的记忆B细胞和相关肿瘤细胞中,如何使EBV长期维持在潜伏期,其相关机制仍不明确。以往研究发现,高免疫原性的EBNA2和LMPs在EBV新近感染的B淋巴细胞中显著高表达,而在既往感染过的记忆B细胞和伯基特淋巴瘤中明显下调[36]。为了了解高免疫原性病毒蛋白在潜伏感染期的调控机制,研究者将合并EBV感染的伯基特淋巴瘤细胞作为研究对象,利用CRISPR-Cas9技术靶向敲除调控组蛋白泛素化的prc1基因,使H2AK119Ub1泛素化受抑制,继而稳定lmps启动子,使lmps基因免于基因沉默,正常发挥作用[41]。同理,敲除DNA甲基化调控基因uhrf1和dnmt1可引起LMP1、EBNA2和bzlf1编码蛋白含量显著上调[41]。LMP1、LMPs和EBNA2均为高免疫原性病毒蛋白,其上调可触发机体免疫细胞对宿主细胞的识别杀伤;EBV裂解关键基因bzlf1的编码蛋白可激活宿主细胞溶细胞性裂解反应;这一系列级联效应最终可引起伯基特淋巴瘤细胞表型发生转变,使其无法维持在持续潜伏感染状态[21, 36]。有研究证实,在合并EBV感染的伯基特淋巴瘤中,DNA甲基转移酶1 (DNA methyltransferase,DNMT1)、泛素样含PHD和环指域1蛋白(ubiquitin-like ringfinger domains,UHRF1)和多梳抑制蛋白复合物1 (polycomb repressive complex 1,PRC1)可抑制高免疫原性病毒蛋白表达和bzlf1编码蛋白介导的EBV溶细胞性级联反应,从而有利于宿主细胞逃避免疫识别和免疫应答,长期维持潜伏感染状态,为其恶变转化提供前提条件[41]。
干扰素调节因子8 (interferon regulatory factor,IRF8),又名干扰素共有序列结合蛋白,作为IRF家族的重要转录因子,在正常B细胞分化中发挥关键调控作用[42-43]。Lv等[33]将EBV感染的B淋巴细胞系Akata作为研究对象,利用CRISPR/Cas9技术对其中的irf8基因进行靶向敲除,发现宿主细胞中的凋亡蛋白酶Caspase-1、Caspase-3和Caspase-8均显著下降,继而使KAP1、PAX5和DNMT3A等蛋白稳定性增加,免于裂解清除。KAP1、PAX5和DNMT3A可通过与TRIM28、bzlf1编码蛋白等共同作用,抑制宿主细胞溶细胞性裂解和重激活过程[44-46]。以上研究[33, 44-46]表明,在EBV感染的Akata细胞中,IRF8通过上调凋亡蛋白酶表达,诱导KAP1、PAX5和DNMT3A等蛋白裂解,此类蛋白的降解可使宿主细胞由潜伏感染向溶细胞感染转变,最终导致EBV的溶细胞性重激活。
EBV可以单独感染宿主细胞,也可与其他病原体或病毒共同感染于同一宿主细胞,它们之间相互作用,共同促进疾病的演变进展[5, 47]。伯基特淋巴瘤常伴有EBV和疟原虫的同时感染[35, 48]。研究发现疟原虫消化降解血液产物疟原虫色素可通过激活Toll样受体-9 (TLR-9)抑制伯基特淋巴瘤细胞中的EBV溶细胞性裂解,而其具体机制仍不明确[48]。研究者利用CRISPR/Cas9技术分别靶向编辑Akata细胞中的tlr9、myd88、irak4基因;经过对比分析发现,TLR9-MyD88-IRAK4信号通路激活后,通过翻译后修饰方式使bzlf1编码蛋白含量显著下降,从而抑制EBV溶细胞性裂解,使其维持潜伏感染状态;研究表明在EBV与疟原虫共同感染宿主细胞过程中,疟原虫色素通过激活宿主细胞的TLR9-MyD88-IRAK4信号通路,下调溶细胞裂解基因bzlf1编码的蛋白含量,使EBV处于持续潜伏于宿主细胞体内免于破坏,继而促进EBV和疟原虫共同感染后的病变进展[35]。
3 运用CRISPR/Cas9技术靶向编辑基因治疗EBV相关疾病临床资料显示,成人体内EBV感染以潜伏感染为主。目前,EBV持续潜伏感染的宿主细胞仍缺乏有效的预防和治疗手段,预后较差[5, 47]。传统预防性疫苗是以EBV的包膜糖蛋白GP350或其他潜伏蛋白作为靶蛋白,刺激机体产生中和抗体,从而阻止病毒感染,干预疾病进展;治疗性疫苗则主要针对病毒感染相关恶性肿瘤,以潜伏感染过程中产生的核抗原和潜伏膜蛋白作为靶点,刺激机体产生特异性细胞免疫应答,提高CTLs对肿瘤细胞的杀伤能力[49-50]。但EBV基因组序列较长,基因亚型种类多、差异大,编码蛋白种类繁杂;而且由于程序性死亡受体1 (programmed death factor-1,PD-1)/程序性死亡受体配体1 (programmed death factor ligand,PD-L1)等肿瘤免疫逃逸分子作用,导致预防性疫苗和治疗性疫苗研制存在诸多限制[49-50]。另外,传统抗病毒药物仅可缓解部分急性感染患者症状,却难以彻底清除体内病毒,对慢性感染和潜伏感染患者疗效欠佳[5, 47]。因此,开发新型EBV感染治疗新策略成为目前研究热点和难题。近年来,随着CRISPR/Cas9技术在人类疾病治疗方面研究的深入,一些临床疗效不佳、预后较差的疾病,如病毒相关性疾病、基因遗传性疾病和肿瘤相关疾病等有望在治疗领域取得新突破[9-10, 40, 51-54]。利用新型CRISPR/Cas9基因编辑技术,对EBV致病基因序列进行靶向编辑,可有效降低EBV基因含量,提高肿瘤细胞对化疗、放疗的敏感性,干预肿瘤进展[10]。
EBV感染是非洲儿童地方性伯基特淋巴瘤最主要的致病因素,并可通过多种方式促进肿瘤的恶变进展。van Diemen等[55]研究发现ebna1 (EBV核抗原位点)和orip (EBV复制起源位点)均可以控制EBV复制和维持游离病毒存活,其中orip包括了ebna1的结合位点和部分双重对称原件[56-57]。通过设计针对ebna1区域的CRISPR/Cas9系统,对致病基因序列进行靶向敲除,发现EB病毒感染的伯基特淋巴瘤Akata-Bx1细胞系中的病毒基因含量降低了40%−60%;进一步对比研究发现,设计联合针对ebna1区域和orip特定区域的靶向编辑系统,可使细胞内病毒基因含量下降超过95%以上,伴随病毒荷载量的显著下降,该淋巴瘤细胞增殖速度显著抑制,进而导致细胞凋亡;该研究表明通过联合设计针对不同识别位点基因区域的靶向RNA,CRISPR/Cas9技术可以最大程度地消除伯基特淋巴瘤中的潜伏感染病毒,从而抑制肿瘤细胞生长增殖[55]。
原发性渗出性淋巴瘤(primary effusion lymphoma,PEL)起源于B细胞,常伴有EBV和卡波西肉瘤相关疱疹病毒(Kaposi’s sarcoma-associated herpes virus,KSHV)的共同感染[5, 58-59]。Bigi等[59]研究发现,将外源性EBV作用于单独KSHV感染的PEL细胞中,KSHV基因组稳定性和KSHV潜伏相关核抗原(latency-associated nuclear antigen,LANA)含量均可显著上调;再通过构建针对EBV基因组的CRISPR/Cas9质粒,将其转染到KSHV和EBV双重感染的PEL细胞中,检测其中EBV含量显著下降的同时,经对比研究发现,该肿瘤细胞活力、KSHV基因组拷贝数及潜伏相关核抗原均显著下调,而导入EBV核抗原-1位点基因ebna1则可逆转上述表型变化。该研究表明在原发性渗出性淋巴瘤中,EBV和卡波西肉瘤相关疱疹病毒相互作用,共同促进疾病进展;利用靶向EBV基因组的CRISPR/Cas9技术,去除EBV含量的同时,可显著下调LANA含量和KSHV荷载量,降低肿瘤细胞活力,为探索临床治疗合并EBV感染的原发性渗出性淋巴瘤提供理论依据[59]。
在我国华南地区,鼻咽癌发病率与长期慢性EBV感染密切相关,其转移和复发率较高,预后欠佳[3, 5-6]。研究发现,bart miRNA含量在鼻咽癌、胃癌等EBV阳性上皮肿瘤组织中显著高表达。Yuen等[53, 60]设计出针对EBV基因组中bart启动子区域的引导RNA,利用CRISPR/Cas9技术对EBV阳性鼻咽癌细胞系C666-1进行靶向编辑,结果证实bart 编码的病毒miRNA含量显著下降,荧光素酶报告基因检测结果显示,bart基因表达活力也显著降低,进一步深度基因测序证实未发现脱靶效应。如前所述,作为EBV关键非编码基因,bart miRNA可通过抑制细胞凋亡和逃避免疫杀伤等机制,促进肿瘤恶变进展[25-27]。该研究通过靶向编辑降低bart含量,为探索干预鼻咽癌进展新策略提供理论支持[53, 60]。该研究团队另一项研究发现,设计针对EBV DNA基因组不同位点的靶向CRISPR/Cas9系统,对鼻咽癌细胞系C666-1进行靶向基因编辑,可使其中的EBV DNA含量下降50%,虽不能将感染的C666-1细胞系直接杀死,但可显著增强它们对化疗药物顺铂和5-氟尿嘧啶的化疗敏感性,表明CRISPR/Cas9技术虽不能直接阻止鼻咽癌进展,但可为其辅助治疗(如化疗)提供新的治疗窗[61]。
近来,免疫细胞检查点抑制剂PD-1和PD-L1的临床应用开启了肿瘤免疫治疗的新时代,也引起了人们对免疫疗法治疗各种癌症的广泛关注。EBV相关性胃癌相较于EBV非相关性胃癌,具有淋巴细胞浸润率高、PD-L1扩增率高等特点。Su等[62]将EBV-LMP2A诱导的CTLs作为研究对象,利用CRISPR-Cas9技术靶向破坏其中的PD-1,经体外实验证实,改造后的CTLs可显著增加对EBV-LMP2A抗原的免疫应答反应,从而增强对EBVaGC细胞的杀伤毒性;更为重要的是,当结合低剂量放疗时,这种靶向破坏PD-1的CTLs在EBVaGC移植瘤小鼠模型中发挥出显著的抗肿瘤作用;他们利用CRISPR-Cas9技术构建出新型细胞毒性T细胞CTLs,并证实其在EBVaGC动物模型中具有抗肿瘤作用,为进一步利用T细胞过继疗法治疗EBVaGC提供了新思路。
4 结语与展望综上所述,EBV感染疾病种类多,关键致病基因亚型差异大。病毒在宿主细胞中的感染状态并非单一固定,在疾病演变过程中可通过多种途径及机制逃避体内免疫监控,促进疾病进展[63-64];另外,EBV感染也可合并其他病原体、寄生虫共同感染,而其中的关键基因及信号通路也并非静止不变,利用CRISPR/Cas9基因编辑技术可以实时动态监测宿主细胞内的EBV致病基因亚型和宿主依赖基因,深入探讨致病基因的作用机制,联合设计针对不同致病基因序列的引导RNA,构建多靶点CRISPR/Cas9系统,为临床治疗EBV相关疾病,尤其是EBV相关恶性肿瘤提供新型临床干预靶点和策略。
CRISPR/Cas9技术应用于EBV相关研究领域仍存在以下几个问题:(1)如何选取安全合适的运输载体。目前,腺相关病毒(adeno-associated virus,AAV)、慢病毒(lentivirus,LV)和质粒作为相应CRISPR/Cas9系统运输载体存在诸多弊端,如整合基因组到靶细胞时具有潜在的诱导癌变风险,缺少靶细胞特异识别机制和代价昂贵等。最近研究发现,微囊泡、外泌体以及人工合成脂质体等载体系统有望成为有效、安全、经济可行的CRISPR/Cas9系统运输载体[65-67]。(2) CRISPR/Cas9技术的脱靶效应和副作用问题。目前研究认为,选择高效和特异的靶向引导RNA至关重要,研究者需要通过对靶基因序列与人类完整基因组进行比对,找寻尽量多的潜在靶基因识别序列,充分完备相应实验验证,争取找到最高效、毒副作用最小的靶向sgRNA序列[10, 68-69]。(3)生物安全性仍是目前CRISPR/Cas9技术用于临床转化前最值得慎重思考的问题[70-71]。近来有报道利用CRISPR/Cas9技术对人类胚胎直接进行编辑,虽然其初衷是使其编辑的胚胎后代免遭HIV病毒感染,但却严重违反了相关法律法规,更违反了医学伦理基本原则[72-73]。
CRISPR/Cas9系统因其技术优势,自问世以来便被寄予厚望,经过不断改造和深入探索,目前已在EBV致病机理与潜在治疗研究方面取得了一系列进展。但我们要理性地认识到这种从基础到临床的转化并非一蹴而就,需要充分考虑和验证其潜在副作用,尤其在生物安全性方面对其潜在影响进行综合分析,在充分证实安全有效的前提下,谨慎推行CRISPR/Cas9技术在人类疾病相关研究领域的实践与探索,促进人类健康事业可持续发展。
| [1] |
Vockerodt M, Yap LF, Shannon-Lowe C, et al. The Epstein-Barr virus and the pathogenesis of lymphoma[J]. The Journal of Pathology, 2015, 235(2): 312-322. DOI:10.1002/path.4459 |
| [2] |
Mui UN, Haley CT, Vangipuram R, et al. Human oncoviruses: Mucocutaneous manifestations, pathogenesis, therapeutics, and prevention: Hepatitis viruses, human T-cell leukemia viruses, herpesviruses, and Epstein-Barr virus[J]. Journal of the American Academy of Dermatology, 2019, 81(1): 23-41. DOI:10.1016/j.jaad.2018.10.072 |
| [3] |
Saha A, Robertson ES. Mechanisms of B-cell oncogenesis induced by Epstein-Barr virus[J]. Journal of Virology, 2019, 93(13): e00238-19. |
| [4] |
Dunmire SK, Verghese PS, Balfour Jr HJ. Primary Epstein-Barr virus infection[J]. Journal of Clinical Virology, 2018, 102: 84-92. DOI:10.1016/j.jcv.2018.03.001 |
| [5] |
Fugl A, Andersen CL. Epstein-Barr virus and its association with disease — a review of relevance to general practice[J]. BMC Family Practice, 2019, 20: 62. DOI:10.1186/s12875-019-0954-3 |
| [6] |
Chen YP, Chan A, Le QT, et al. Nasopharyngeal carcinoma[J]. The Lancet, 2019, 394(10192): 64-80. DOI:10.1016/S0140-6736(19)30956-0 |
| [7] |
Jiang WY, Bikard D, Cox D, et al. RNA-guided editing of bacterial genomes using CRISPR-Cas systems[J]. Nature Biotechnology, 2013, 31(3): 233-239. DOI:10.1038/nbt.2508 |
| [8] |
Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems[J]. Science, 2013, 339(6121): 819-823. DOI:10.1126/science.1231143 |
| [9] |
Platt RJ, Chen SD, Zhou Y, et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling[J]. Cell, 2014, 159(2): 440-455. DOI:10.1016/j.cell.2014.09.014 |
| [10] |
Gilani U, Shaukat M, Rasheed A, et al. The implication of CRISPR/Cas9 genome editing technology in combating human oncoviruses[J]. Journal of Medical Virology, 2019, 91(1): 1-13. |
| [11] |
Metje-Sprink J, Menz J, Modrzejewski D, et al. DNA-free genome editing: Past, present and future[J]. Frontiers in Plant Science, 2019, 9: 1957. DOI:10.3389/fpls.2018.01957 |
| [12] |
Kanda T, Yajima M, Ikuta K. Epstein-Barr virus strain variation and cancer[J]. Cancer Science, 2019, 110(4): 1132-1139. |
| [13] |
Correia S, Palser A, Elgueta KC, et al. Natural variation of Epstein-Barr virus genes, proteins, and primary MicroRNA[J]. Journal of Virology, 2017, 91(15): e00375-17. |
| [14] |
Neves M, Marinho-Dias J, Ribeiro J, et al. Epstein-Barr virus strains and variations: Geographic or disease-specific variants?[J]. Journal of Medical Virology, 2017, 89(3): 373-387. |
| [15] |
Farrell PJ. Epstein-Barr virus strain variation[A]//Münz A. Epstein Barr Virus Volume 1: One Herpes Virus: Many Diseases[M]. Cham: Springer, 2015: 45-69
|
| [16] |
Palser AL, Grayson NE, White RE, et al. Genome diversity of Epstein-Barr virus from multiple tumor types and normal infection[J]. Journal of Virology, 2015, 89(10): 5222-5237. DOI:10.1128/JVI.03614-14 |
| [17] |
Tsao SW, Tsang CM, To KF, et al. The role of Epstein-Barr virus in epithelial malignancies[J]. The Journal of Pathology, 2015, 235(2): 323-333. |
| [18] |
Kwok H, Chiang AKS. From conventional to next generation sequencing of Epstein-Barr virus genomes[J]. Viruses, 2016, 8(3): 60. |
| [19] |
Ma YJ, Walsh MJ, Bernhardt K, et al. CRISPR/Cas9 screens reveal Epstein-Barr virus-transformed B cell host dependency factors[J]. Cell Host & Microbe, 2017, 21(5): 580-591.e7. |
| [20] |
Guo R, Jiang C, Zhang YC, et al. MYC controls the Epstein-Barr virus lytic switch[J]. Molecular Cell, 2020, 78(4): 653-669.e8. DOI:10.1016/j.molcel.2020.03.025 |
| [21] |
Countryman J, Jenson H, Seibl R, et al. Polymorphic proteins encoded within BZLF1 of defective and standard Epstein-Barr viruses disrupt latency[J]. Journal of Virology, 1987, 61(12): 3672-3679. DOI:10.1128/JVI.61.12.3672-3679.1987 |
| [22] |
Chen JN, He D, Tang F, et al. Epstein-Barr virus-associated gastric carcinoma: a newly defined entity[J]. Journal of Clinical Gastroenterology, 2012, 46(4): 262-271. DOI:10.1097/MCG.0b013e318249c4b8 |
| [23] |
Morales-Sanchez A, Fuentes-Panana EM. Epstein-Barr virus-associated gastric cancer and potential mechanisms of oncogenesis[J]. Current Cancer Drug Targets, 2017, 17(6): 534-554. |
| [24] |
Shinozaki-Ushiku A, Kunita A, Fukayama M. Update on Epstein-Barr virus and gastric cancer (review)[J]. International Journal of Oncology, 2015, 46(4): 1421-1434. DOI:10.3892/ijo.2015.2856 |
| [25] |
Lung RWM, Hau PM, Yu KHO, et al. EBV-encoded miRNAs target ATM-mediated response in nasopharyngeal carcinoma[J]. The Journal of Pathology, 2018, 244(4): 394-407. DOI:10.1002/path.5018 |
| [26] |
Zheng X, Wang J, Wei LY, et al. Epstein-Barr virus microRNA miR-BART5-3p inhibits p53 expression[J]. Journal of Virology, 2018, 92(23): e01022-18. |
| [27] |
Dölken L, Malterer G, Erhard F, et al. Systematic analysis of viral and cellular microRNA targets in cells latently infected with human γ-herpesviruses by RISC immunoprecipitation assay[J]. Cell Host & Microbe, 2010, 7(4): 324-334. DOI:10.1016/j.chom.2010.03.008 |
| [28] |
Kanda T, Furuse Y, Oshitani H, et al. Highly efficient CRISPR/Cas9-mediated cloning and functional characterization of gastric cancer-derived Epstein-Barr virus strains[J]. Journal of Virology, 2016, 90(9): 4383-4393. DOI:10.1128/JVI.00060-16 |
| [29] |
Mashiko D, Fujihara Y, Satouh Y, et al. Generation of mutant mice by pronuclear injection of circular plasmid expressing Cas9 and single guided RNA[J]. Scientific Reports, 2013, 3(1): 3355. DOI:10.1038/srep03355 |
| [30] |
Kanda T, Miyata M, Kano M, et al. Clustered microRNAs of the Epstein-Barr virus cooperatively downregulate an epithelial cell-specific metastasis suppressor[J]. Journal of Virology, 2015, 89(5): 2684-2697. DOI:10.1128/JVI.03189-14 |
| [31] |
Borozan I, Zapatka M, Frappier L, et al. Analysis of Epstein-Barr virus genomes and expression profiles in gastric adenocarcinoma[J]. Journal of Virology, 2018, 92(2): e01239-17. |
| [32] |
Hu BF, Zhang L, Shi D, et al. Clinical analysis of 18 children with chronic active Epstein-Barr virus infection[J]. Chinese Journal of Evidence-Based Pediatrics, 2019, 14(6): 434-437. (in Chinese) 胡波飞, 张丽, 施丹, 等. 儿童慢性活动性EB病毒感染18例病例系列报告[J]. 中国循证儿科杂志, 2019, 14(6): 434-437. |
| [33] |
Lv DW, Zhang K, Li RF. Interferon regulatory factor 8 regulates caspase-1 expression to facilitate Epstein-Barr virus reactivation in response to B cell receptor stimulation and chemical induction[J]. PLoS Pathogens, 2018, 14(1): e1006868. |
| [34] |
Al Masud HMA, Watanabe T, Yoshida M, et al. Epstein-Barr virus BKRF4 gene product is required for efficient progeny production[J]. Journal of Virology, 2017, 91(23): e00975-17. |
| [35] |
Jordi M, Marty J, Mordasini V, et al. IRAK4 is essential for TLR9-induced suppression of Epstein-Barr virus BZLF1 transcription in Akata Burkitt's lymphoma cells[J]. PLoS One, 2017, 12(10): e0186614. DOI:10.1371/journal.pone.0186614 |
| [36] |
Odumade OA, Hogquist KA, Balfour Jr HJ. Progress and problems in understanding and managing primary Epstein-Barr virus infections[J]. Clinical Microbiology Reviews, 2011, 24(1): 193-209. DOI:10.1128/CMR.00044-10 |
| [37] |
Münz C. Latency and lytic replication in Epstein-Barr virus-associated oncogenesis[J]. Nature Reviews Microbiology, 2019, 17(11): 691-700. DOI:10.1038/s41579-019-0249-7 |
| [38] |
Zhou HF, Schmidt SCS, Jiang SZ, et al. Epstein-Barr virus oncoprotein super-enhancers control B cell growth[J]. Cell Host & Microbe, 2015, 17(2): 205-216. |
| [39] |
Jha HC, Pei YG, Robertson ES. Epstein-Barr Virus: diseases linked to infection and transformation[J]. Frontiers in Microbiology, 2016, 7: 1602. DOI:10.3389/fmicb.2016.01602 |
| [40] |
Chen SL, Yu X, Guo DY. CRISPR-Cas targeting of host genes as an antiviral strategy[J]. Viruses, 2018, 10(1): 40. |
| [41] |
Guo R, Zhang YC, Teng MX, et al. DNA methylation enzymes and PRC1 restrict B-cell Epstein-Barr virus oncoprotein expression[J]. Nature Microbiology, 2020, 5(8): 1051-1063. DOI:10.1038/s41564-020-0724-y |
| [42] |
Tamura T, Yanai H, Savitsky D, et al. The IRF family transcription factors in immunity and oncogenesis[J]. Annual Review of Immunology, 2008, 26: 535-584. DOI:10.1146/annurev.immunol.26.021607.090400 |
| [43] |
Xu HP, Chaudhri VK, Wu ZG, et al. Regulation of bifurcating B cell trajectories by mutual antagonism between transcription factors IRF4 and IRF8[J]. Nature Immunology, 2015, 16(12): 1274-1281. DOI:10.1038/ni.3287 |
| [44] |
Li XF, Burton EM, Bhaduri-Mcintosh S. Chloroquine triggers Epstein-Barr virus replication through phosphorylation of KAP1/TRIM28 in Burkitt lymphoma cells[J]. PLoS Pathogens, 2017, 13(3): e1006249. DOI:10.1371/journal.ppat.1006249 |
| [45] |
Raver RM, Panfil AR, Hagemeier SR, et al. The B-cell-specific transcription factor and master regulator Pax5 promotes Epstein-Barr virus latency by negatively regulating the viral immediate early protein BZLF1[J]. Journal of Virology, 2013, 87(14): 8053-8063. DOI:10.1128/JVI.00546-13 |
| [46] |
Gray KS, Forrest JC, Speck SH. The de novo methyltransferases DNMT3a and DNMT3b target the murine gammaherpesvirus immediate-early gene 50 promoter during establishment of latency[J]. Journal of Virology, 2010, 84(10): 4946-4959. DOI:10.1128/JVI.00060-10 |
| [47] |
Nowalk A, Green M. Epstein-Barr virus[J]. Microbiology Spectrum, 2016, 4(3). DOI:10.1128/microbiolspec.DMIH2-0011-2015 |
| [48] |
Zauner L, Melroe GT, Sigrist JA, et al. TLR9 triggering in Burkitt's lymphoma cell lines suppresses the EBV BZLF1 transcription via histone modification[J]. Oncogene, 2010, 29(32): 4588-4598. DOI:10.1038/onc.2010.203 |
| [49] |
van Zyl DG, Mautner J, Delecluse HJ. Progress in EBV vaccines[J]. Frontiers in Oncology, 2019, 9: 104. DOI:10.3389/fonc.2019.00104 |
| [50] |
Cohen JI. Vaccine development for Epstein-Barr virus[A]//Kawaguchi Y, Mori Y, Kimura H. Human Herpesviruses[M]. Singapore: Springer, 2018: 477-493
|
| [51] |
Johansen AK, Molenaar B, Versteeg D, et al. Postnatal cardiac gene editing using CRISPR/Cas9 with AAV9-mediated delivery of short guide RNAs results in mosaic gene disruption[J]. Circulation Research, 2017, 121(10): 1168-1181. DOI:10.1161/CIRCRESAHA.116.310370 |
| [52] |
Savić N, Schwank G. Advances in therapeutic CRISPR/Cas9 genome editing[J]. Translational Research, 2016, 168: 15-21. DOI:10.1016/j.trsl.2015.09.008 |
| [53] |
Yuen KS, Chan CP, Wong NHM, et al. CRISPR/Cas9-mediated genome editing of Epstein-Barr virus in human cells[J]. The Journal of General Virology, 2015, 96(Pt 3): 626-636. DOI:10.1099/jgv.0.000012 |
| [54] |
Xue W, Chen SD, Yin H, et al. CRISPR-mediated direct mutation of cancer genes in the mouse liver[J]. Nature, 2014, 514(7522): 380-384. DOI:10.1038/nature13589 |
| [55] |
van Diemen FR, Kruse EM, Hooykaas MJG, et al. CRISPR/Cas9-mediated genome editing of herpesviruses limits productive and latent infections[J]. PLoS Pathogens, 2016, 12(6): e1005701. |
| [56] |
Reisman D, Yates J, Sugden B. A putative origin of replication of plasmids derived from Epstein-Barr virus is composed of two cis-acting components[J]. Molecular and Cellular Biology, 1985, 5(8): 1822-1832. DOI:10.1128/MCB.5.8.1822 |
| [57] |
Rawlins DR, Milman G, Hayward SD, et al. Sequence-specific DNA binding of the Epstein-Barr virus nuclear antigen (EBNA-1) to clustered sites in the plasmid maintenance region[J]. Cell, 1985, 42(3): 859-868. DOI:10.1016/0092-8674(85)90282-X |
| [58] |
Manzano M, Patil A, Waldrop A, et al. Gene essentiality landscape and druggable oncogenic dependencies in herpesviral primary effusion lymphoma[J]. Nature Communications, 2018, 9(1): 3263. DOI:10.1038/s41467-018-05506-9 |
| [59] |
Bigi R, Landis JT, An H, et al. Epstein-Barr virus enhances genome maintenance of Kaposi sarcoma-associated herpesvirus[J]. Proceedings of the National Academy of Sciences of the United States of America, 2018, 115(48): E11379-E11387. |
| [60] |
Yuen KS, Chan CP, Kok KH, et al. Mutagenesis and genome engineering of Epstein-Barr virus in cultured human cells by CRISPR/Cas9[A]//Reeves A. In Vitro Mutagenesis: Methods and Protocols[M]. New York: Humana Press, 2017, 1498: 23-31
|
| [61] |
Yuen KS, Wang ZM, Wong NM, et al. Suppression of Epstein-Barr virus DNA load in latently infected nasopharyngeal carcinoma cells by CRISPR/Cas9[J]. Virus Research, 2018, 244: 296-303. DOI:10.1016/j.virusres.2017.04.019 |
| [62] |
Su S, Zou ZY, Chen FJ, et al. CRISPR-Cas9-mediated disruption of PD-1 on human T cells for adoptive cellular therapies of EBV positive gastric cancer[J]. OncoImmunology, 2017, 6(1): e1249558. |
| [63] |
Ressing ME, van Gent M, Gram AM, et al. Immune evasion by Epstein-Barr virus[A]//Münz C. Epstein Barr Virus Volume 2: One Herpes Virus: Many Diseases[M]. Cham: Springer, 2015, 391: 355-381
|
| [64] |
Gram AM, Oosenbrug T, Lindenbergh MFF, et al. The Epstein-Barr virus glycoprotein gp150 forms an immune-evasive glycan shield at the surface of infected cells[J]. PLoS Pathogens, 2016, 12(4): e1005550. DOI:10.1371/journal.ppat.1005550 |
| [65] |
Lin Y, Wu JH, Gu WH, et al. Exosome-liposome hybrid nanoparticles deliver CRISPR/Cas9 system in MSCs[J]. Advanced Science, 2018, 5(4): 1700611. DOI:10.1002/advs.201700611 |
| [66] |
Givens BE, Naguib YW, Geary SM, et al. Nanoparticle-based delivery of CRISPR/Cas9 genome-editing therapeutics[J]. The AAPS Journal, 2018, 20(6): 108. DOI:10.1208/s12248-018-0267-9 |
| [67] |
Kim SM, Yang Y, Oh SJ, et al. Cancer-derived exosomes as a delivery platform of CRISPR/Cas9 confer cancer cell tropism-dependent targeting[J]. Journal of Controlled Release, 2017, 266: 8-16. DOI:10.1016/j.jconrel.2017.09.013 |
| [68] |
Koujah L, Shukla D, Naqvi AR. CRISPR-Cas based targeting of host and viral genes as an antiviral strategy[J]. Seminars in Cell & Developmental Biology, 2019, 96: 53-64. DOI:10.1016/j.semcdb.2019.04.004 |
| [69] |
Salsman J, Dellaire G. Precision genome editing in the CRISPR era[J]. Biochemistry and Cell Biology, 2017, 95(2): 187-201. |
| [70] |
Wang HF, La Russa M, Qi LS. CRISPR/Cas9 in genome editing and beyond[J]. Annual Review of Biochemistry, 2016, 85: 227-264. DOI:10.1146/annurev-biochem-060815-014607 |
| [71] |
Taguchi I, Yamada T, Akaishi R, et al. Attitudes of clinical geneticists and certified genetic counselors to genome editing and its clinical applications: A nation-wide questionnaire survey in Japan[J]. Journal of Human Genetics, 2019, 64(9): 945-954. DOI:10.1038/s10038-019-0635-z |
| [72] |
Lovell-Badge R. CRISPR babies: a view from the centre of the storm[J]. Development, 2019, 146(3): dev175778. DOI:10.1242/dev.175778 |
| [73] |
Getz LJ, Dellaire G. Angels and devils: dilemmas in dual-use biotechnology[J]. Trends in Biotechnology, 2018, 36(12): 1202-1205. DOI:10.1016/j.tibtech.2018.07.016 |
 2020, Vol. 47
2020, Vol. 47