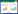扩展功能
文章信息
- 魏亚茹, 王怡静, 马巧丽, 李千雪, 李传虹, 李新, 夏晶晶, 芦燕, 于景丽, 希尼尼根
- WEI Ya-Ru, WANG Yi-Jing, MA Qiao-Li, LI Qian-Xue, LI Chuan-Hong, LI Xin, XIA Jing-Jing, LU Yan, YU Jing-Li, Xininigen
- 粉粒氨氮和水分影响浮霉菌门群落的空间分化
- Effects of silt particles, ammonia nitrogen and water content on structural and functional differentiation of Plantomycetes communities
- 微生物学通报, 2020, 47(9): 2732-2745
- Microbiology China, 2020, 47(9): 2732-2745
- DOI: 10.13344/j.microbiol.china.200338
-
文章历史
- 收稿日期: 2020-04-03
- 接受日期: 2020-07-24
- 网络首发日期: 2020-08-06
2. 内蒙古自治区环境污染控制与废物资源化重点实验室 内蒙古 呼和浩特 010021;
3. 内蒙古大学生态与环境学院 内蒙古 呼和浩特 010021;
4. 内蒙古农业大学兽医学院 内蒙古 呼和浩特 010018
2. Inner Mongolia Key Laboratory of Environmental Pollution Control & Waste Resource Reuse, Hohhot, Inner Mongolia 010021, China;
3. School of Ecology and Environment, Inner Mongolia University, Hohhot, Inner Mongolia 010021, China;
4. College of Veterinary Medicine, Inner Mongolia Agricultural University, Hohhot, Inner Mongolia 010018, China
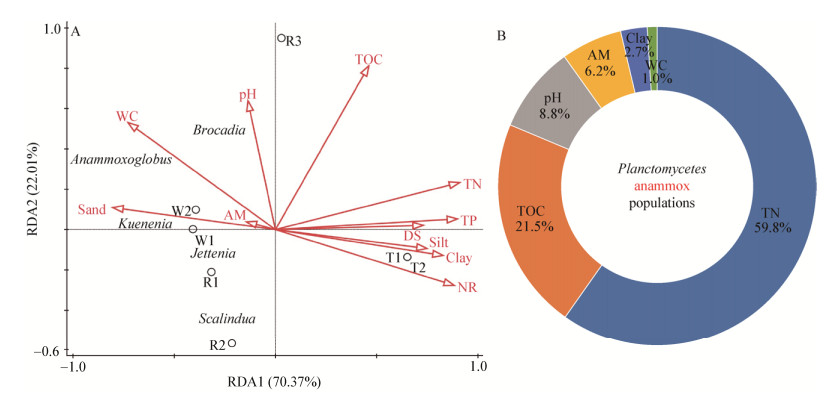
厌氧氨氧化(anaerobic ammonium oxidation,anammox)是氮素生物地球化学循环的重要组成部分[1]。迄今发现具有Anammox功能的细菌全部来自浮霉菌门(Planctomycetes)[2]。Planctomycetes广泛分布于咸水、海水、淡水、土壤等天然系统[1, 3-7]和废水等人工系统[8-12]。Anammox相关Planctomycetes能利用铵态氮(NH4+)和亚硝态氮(NO2-)生成氮气(N2)且不产生温室气体(N2O)[13],在高氨废水生物脱氮[8-12]及过量施用氮肥的农田土壤系统氮移除[3-4, 14]方面发挥重要作用,属生态友好型微生物。提升Anammox相关Planctomycetes反应活性和转化效率一直是废水生物脱氮和农田土壤过剩氮素安全移除的焦点问题[4, 14-15]。Kartal等[16]发现Planctomycetes不但具有Anammox功能,还可能在中间环节发挥反硝化(denitrification)功能(图 1)。He等[15]发现适合的有机碳源有利于Denitrification与Anammox相关Planctomycetes协同脱氮。大量研究证明,盐度[17]、碳源[15, 18]、NH4+/NO2-的比例[19]及粉粒[20]等是影响Anammox相关Planctomycetes种群分化的关键因素(图 1)。
|
| 图 1 影响Planctomycetes群落厌氧氨氧化或反硝化功能分化的相关环境因素(A)和采样点布局(B) Figure 1 Environmental factors affecting differentiation of Planctomycetes communities related to anammox or denitrification (A) and the design of sampling sites in this study (B) 注:T1、T2:粉黏粒丰富的全营养环境;W1、W2:粉黏粒缺乏的中营养环境;R2、R3:粉粒绝对优势的富营养环境;R1:砂粒绝对优势的寡营养环境. Note: T1 and T2: Copiotrophic environment with rich silt and clay particles; W1 and W2: Mesotrophic environment deficient in silt and clay particles; R2 and R3: Eutrophic environment dominated by rich silt particles; R1: Oligotrophic environment dominated by rich sand particles. |
|
|
为了探究我国温带半干旱区是否存在Planctomycetes群落结构和功能的空间分化以及驱动分化的关键环境要素,本研究运用16S rRNA基因高通量测序技术检测我国温带半干旱区水生环境下砂质沉积物与粉质沉积物、湿生环境下粉砂质土壤和旱生环境下粉质栗钙土Planctomycetes群落的种群组成、丰度及空间分布变化,同步运用生物统计学方法分析粉粒等理化因子对Planctomycetes群落结构空间分化的影响,结合文献资料探讨Planctomycetes群落可能具有的Anammox或反硝化功能,以期为以后准确跟踪Anammox相关Planctomycetes群落提供科学线索,最终为Anammox或反硝化相关Planctomycetes功能群在高氨废水和农田土壤过剩氮素的安全高效脱氮方面提供新的研究思路。
1 材料与方法 1.1 样品的采集及其理化特性检测选择我国华北温带半干旱区锡林河生态系统(43°39′-44°36′N、115°33′-117°28′E),沿着垂直河流主线的方向依次采集水生环境下河床中心砂质沉积物(R1)、河床边缘粉质沉积物(R2)、牛轭湖粉质沉积物(R3),湿生环境下近水侧河滨粗粉砂质土壤(W1)和远水侧河滨细粉砂质土壤(W2),旱生环境下二级阶地低坡处细粉质栗钙土(T1)和二级阶地高坡处粗粉质栗钙土(T2),沉积物/土壤的采集深度均为0-10 cm表层,采样时间为2017年7月上旬。其中,水生环境下河床中心砂质沉积物R1包括R11、R12、R13、R14、R15、R16等6个样品,水生环境下河床边缘粉质沉积物R2包括R21、R22、R23、R24等4个样品,水生环境下牛轭湖粉质沉积物R3包括R31、R32、R33、R34、R35等5个样品;湿生环境下近水侧河滨粗粉砂质土壤W1包括W11、W12、W13、W14等4个样品,湿生环境下远水侧河滨细粉砂质土壤W2包括W21、W22、W23等3个样品;旱生环境下细粉质栗钙土T1包括T11、T12、T13、T14、T15、T16等6个样品,旱生环境下粗粉质栗钙土T2包括T21、T22、T23、T24、T25、T26等6个样品。沉积物/土壤样品布点方案(图 1)及样品的检测方法参照文献[21],理化分析检测结果见表 1。
| 样本 Sample | 黏粒 Clay (%) | 粉粒 Silt (%) | 砂粒 Sand (%) | 水分 WC (%) | pH | 盐度 DS (mg/kg) | 氨氮 AM (mg/kg) | 硝态氮 NR (mg/kg) | 总有机碳 TOC (g/kg) | 总氮 TN (g/kg) | 总磷 TP (g/kg) |
| R11 | 0.00 | 0.03 | 99.97 | 17.95 | 8.03 | 0.31 | 1.04 | 5.17 | 2.57 | 0.44 | 0.08 |
| R12 | 0.00 | 0.06 | 99.94 | 17.85 | 8.05 | 0.29 | 1.12 | 5.83 | 2.53 | 0.42 | 0.11 |
| R13 | 0.00 | 0.04 | 99.96 | 17.92 | 8.04 | 0.32 | 1.06 | 5.25 | 2.55 | 0.43 | 0.13 |
| R14 | 0.00 | 0.02 | 99.98 | 17.84 | 7.91 | 0.22 | 1.00 | 3.77 | 2.34 | 0.32 | 0.08 |
| R15 | 0.00 | 0.03 | 99.97 | 17.92 | 8.04 | 0.31 | 1.05 | 5.17 | 2.57 | 0.43 | 0.09 |
| R16 | 0.00 | 0.02 | 99.98 | 17.94 | 8.02 | 0.32 | 1.03 | 5.16 | 2.58 | 0.45 | 0.07 |
| R21 | 1.61 | 4.40 | 93.99 | 20.23 | 8.36 | 1.27 | 38.42 | 14.53 | 7.17 | 0.99 | 0.22 |
| R22 | 1.65 | 4.60 | 93.75 | 20.18 | 8.31 | 1.18 | 26.48 | 5.89 | 7.19 | 1.06 | 0.27 |
| R23 | 1.61 | 4.40 | 93.99 | 20.22 | 8.35 | 1.29 | 39.47 | 14.58 | 7.17 | 0.98 | 0.23 |
| R24 | 1.61 | 4.30 | 94.09 | 20.23 | 8.37 | 1.26 | 36.38 | 14.49 | 7.15 | 0.99 | 0.22 |
| R31 | 1.66 | 4.50 | 93.84 | 23.17 | 8.75 | 1.24 | 23.74 | 7.63 | 24.54 | 1.97 | 0.36 |
| R32 | 1.64 | 4.70 | 93.66 | 24.05 | 8.67 | 1.21 | 26.89 | 11.15 | 24.27 | 1.92 | 0.33 |
| R33 | 1.63 | 4.60 | 93.77 | 24.14 | 8.65 | 1.18 | 20.74 | 5.91 | 24.35 | 1.88 | 0.31 |
| R34 | 1.67 | 4.50 | 93.83 | 23.19 | 8.74 | 1.23 | 23.91 | 8.28 | 24.60 | 1.98 | 0.36 |
| R35 | 1.65 | 4.40 | 93.95 | 22.16 | 8.73 | 1.22 | 23.86 | 8.22 | 24.57 | 1.96 | 0.37 |
| W11 | 0.00 | 1.53 | 98.47 | 22.24 | 8.62 | 1.08 | 7.73 | 4.51 | 8.88 | 1.14 | 0.26 |
| W12 | 0.02 | 1.59 | 98.39 | 21.19 | 8.72 | 1.13 | 10.95 | 6.31 | 9.13 | 1.23 | 0.31 |
| W13 | 0.00 | 1.51 | 98.49 | 22.21 | 8.59 | 1.07 | 8.18 | 5.01 | 8.89 | 1.12 | 0.27 |
| W14 | 0.00 | 1.56 | 98.44 | 22.25 | 8.66 | 1.09 | 8.17 | 5.07 | 8.87 | 1.16 | 0.28 |
| W21 | 2.86 | 16.19 | 80.95 | 20.88 | 8.64 | 1.32 | 7.92 | 9.42 | 10.69 | 1.26 | 0.33 |
| W22 | 2.17 | 16.62 | 81.21 | 20.69 | 8.67 | 1.29 | 6.41 | 10.68 | 10.91 | 1.29 | 0.32 |
| W23 | 3.89 | 19.96 | 76.15 | 19.28 | 8.49 | 1.39 | 6.86 | 8.08 | 11.05 | 1.37 | 0.38 |
| T11 | 13.22 | 24.84 | 61.94 | 15.78 | 8.35 | 1.81 | 6.73 | 17.69 | 14.54 | 2.83 | 0.61 |
| T12 | 15.18 | 22.14 | 62.68 | 15.19 | 8.23 | 1.84 | 6.50 | 17.94 | 15.02 | 2.88 | 0.66 |
| T13 | 14.74 | 23.29 | 61.97 | 15.26 | 8.19 | 1.76 | 5.73 | 19.28 | 14.50 | 2.92 | 0.63 |
| T14 | 16.02 | 24.01 | 59.97 | 15.37 | 8.28 | 1.79 | 5.38 | 16.31 | 15.05 | 2.95 | 0.64 |
| T15 | 13.21 | 24.91 | 61.88 | 15.76 | 8.31 | 1.82 | 6.76 | 17.73 | 14.52 | 2.79 | 0.63 |
| T16 | 13.17 | 24.68 | 62.15 | 15.79 | 8.37 | 1.81 | 6.74 | 17.68 | 14.57 | 2.86 | 0.57 |
| T21 | 6.15 | 22.04 | 71.81 | 14.37 | 8.41 | 1.75 | 6.91 | 16.17 | 13.97 | 2.59 | 0.69 |
| T22 | 6.88 | 22.19 | 70.93 | 14.96 | 8.39 | 1.83 | 5.44 | 18.86 | 13.99 | 2.64 | 0.72 |
| T23 | 5.79 | 21.67 | 72.54 | 14.55 | 8.42 | 1.74 | 9.17 | 21.24 | 13.45 | 2.55 | 0.68 |
| T24 | 6.18 | 22.03 | 71.79 | 14.36 | 8.43 | 1.75 | 6.88 | 16.64 | 13.98 | 2.56 | 0.71 |
| T25 | 6.16 | 22.06 | 71.78 | 14.38 | 8.42 | 1.76 | 6.97 | 16.67 | 13.95 | 2.58 | 0.68 |
| T26 | 5.18 | 20.15 | 74.67 | 14.42 | 8.49 | 1.81 | 6.38 | 16.33 | 13.35 | 2.43 | 0.65 |
| 注:表中数值代表 3个重复的平均值;黏粒:< 2 μm;粉粒:2-20 μm;砂粒:> 20 μm. Note: The values represented the average of three replicates. Clay: < 2 μm; Silt: 2-20 μm; Sand: > 20 μm. WC: Water content; DS: Dissolved salt; AM: Ammonia nitrogen; NR: Nitrate nitrogen; TOC: Total organic carbon; TN: Total nitrogen; TP: Total phosphorus. | |||||||||||
FastDNA® SPIN Kit for Soil,MP Biomedical公司;QIAquick PCR Purification Kit,Qiagen公司。QuantiFluorTM-ST蓝色荧光定量系统,Promega公司。
1.3 沉积物/土壤DNA的提取、PCR、高通量测序和测序数据分析沉积物/土壤样品总DNA提取按照FastDNA® SPIN Kit for Soil说明进行。利用16S rRNA基因V3- V4区通用引物338F (5′-ACTCCTACGGGAGGCAGCA-3′)和806R (5′-GGACTACHVGGGTWTCTAAT-3′)进行PCR扩增,每一个样品加8 bp的标签序列以示区分,待后续大规模样品平行测序,PCR反应体系和反应条件参照文献[21]。所有样品的PCR产物经纯化和定量后送往北京百迈客生物科技有限公司利用Illumina MiSeq PE300测序平台进行高通量测序。使用Trimmomatic和FLASH软件优化拼接获取高质量序列,使用UPARSE软件在97%的相似度水平上进行操作分类单元(operational taxonomic unit,OTU)聚类分析同步剔除嵌合体。利用RDP classifier对每条序列进行分类学注释,相似性达到97%的OTU归为同一种细菌,再将每条序列与SILVA数据库(SSU128)进行序列比对(比对阈值70%),获得具体的分类学名称,分析Planctomycetes群落全部种群在总细菌群落组成中的相对丰度,进行后续的统计分析。高通量详细分析参见文献[21-22]。
1.4 数据的统计分析用Excel 2010进行折线图、累计柱状图和线性回归分析等基本图形绘制。采用SPSS 20.0进行Pearson相关性分析。用百迈客生物云计算平台(BMKCloud)内置R语言进行Heatmap热图分析。用CANOCO 5.0对Planctomycetes群落与砂粒、粉粒、黏粒、水分、pH、溶解性盐、有机碳、全氮、全磷、氨氮、硝态氮环境因子进行冗余分析(redundancy analysis,RDA),以及单个环境因子影响Planctomycetes群落结构空间变化的变异权重分析(variation partitioning analysis,VPA)。
2 结果与分析 2.1 高通量测序结果分析34个沉积物/土壤样品共获得3 115 787条优质序列(459 bp),在97%相似性水平上划分为43 830个OTU和791种细菌,隶属于37个门类。物种组成数排序前5位的门类包括Proteobacteria 357种、Bacteroidetes 88种、Actinobacteria 87种、Acidobacteria 59种、Firmicutes 45种、Planctomycetes 9种和Tenericutes等11个门类均含有1个物种。37个门类中相对丰度最高的物种来自拟杆菌门(Bacteroidetes) (R16,18.79%),丰度第二的物种来自酸杆菌门(Acidobacteria) (T21,11.03%),浮霉菌门(Planctomycetes)中最优势的物种相对丰度排序为第191位,来自浮霉菌门(Planctomycetes)的9种细菌隶属于OM190、Phycisphaerae、Pla4_lineage、Planctomycetaci等4个纲和8个属,详见表 2。
| 门 Phylum | 纲 Class | 目 Order | 科 Family | 属 Genus | 种 Species | 编号 Codes |
| Planctomycetes | OM190 | Uncultured_bacterium | Other | Other | Other | OM190_ub_o_o. o1 |
| Other | Other | Other | Other | OM190_o_o_o. o2 | ||
| Phycisphaerae | Phycisphaerales | Phycisphaeraceae | AKYG587 | Uncultured_bacterium | AKYG587. ub3 | |
| Phycisphaera | Uncultured_bacterium | Phycisphaera ub4 | ||||
| SM1A02 | Uncultured_bacterium | SM1A02. ub5 | ||||
| Pla4_lineage | Other | Other | Other | Other | Pla4_lineage_o_o_o. o6 | |
| Planctomycetacia | Planctomycetales | Planctomycetaceae | Singulisphaera | Other | Singulisphaera o7 | |
| Uncultured_bacterium | Singulisphaera ub8 | |||||
| Uncultured | Uncultured_bacterium | Planctomycetaceae_u. ub9 | ||||
| Note: u: Uncultured; o: Other; ub: Uncultured_bacterium; The number represented the serial number of each species. | ||||||
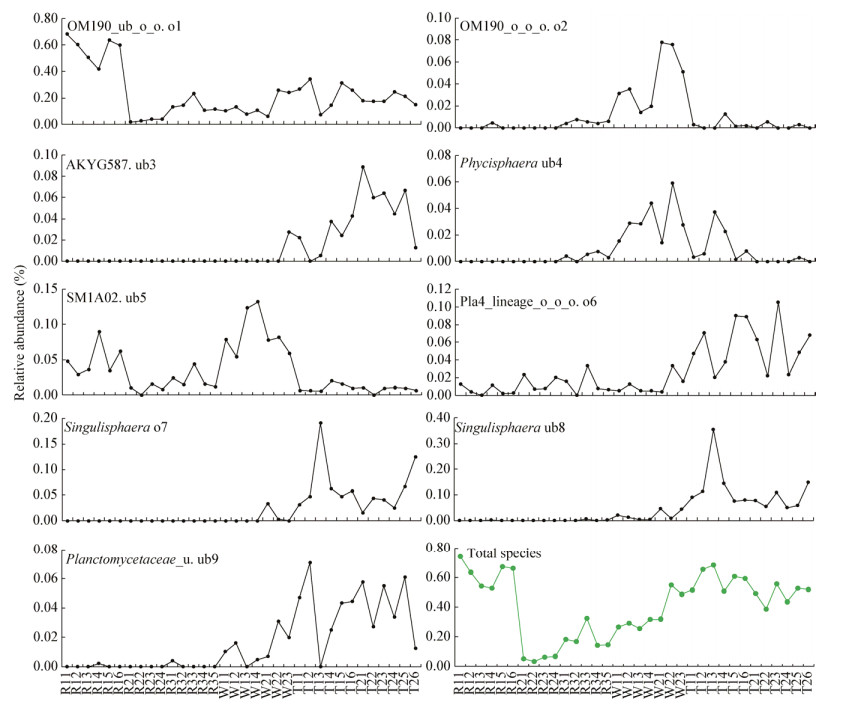
由图 2可知,9个种群的累积相对丰度依次为0.53%-0.74% (R1)、0.035%-0.069% (R2)、0.14%- 0.33% (R3)、0.26%-0.32% (W1)、0.32%-0.55% (W2)、0.51%-0.69% (T1)、0.39%-0.56% (T2)。沿着R1、R2、R3、W1、W2、T1、T2梯度,OM190_o_o_o. o2、AKYG587. ub3、Phycisphaera ub4、Singulisphaera o7、Singulisphaera ub8、Planctomycetaceae_u. ub9呈现出先增后降的分布趋势,在水生环境河床中心砂质沉积物R1和水生环境下河床边缘粉质沉积物R2中整体分布趋近于零,在旱生环境下细粉质栗钙土T1中分布最高;OM190_ub_o_o. o1、SM1A02. ub5、Pla4_lineage_ o_o_o. o6呈现出先降后升的分布趋势。
|
| 图 2 温带半干旱区河流系统Planctomycetes群落单个种群及其全部种群的空间分化 Figure 2 Spatial differentiation of single and total populations from Planctomycetes communities in river ecosystem, temperate semi-arid region |
|
|
由图 2可知,Planctomycetes群落中9个种群组成与丰度各不相同,9种菌的最高相对丰度顺序依次为OM190_ub_o_o. o1 (0.68%)、OM190_o_o_o. o2(0.08%)、AKYG587. ub3 (0.09%)、Phycisphaera ub4 (0.06%)、SM1A02. ub5 (0.13%)、Pla4_lineage_ o_o_o. o6 (0.11%)、Singulisphaera o7 (0.19%)、Singulisphaera ub8 (0.35%)、Planctomycetaceae_u. ub9 (0.07%)。依据单个种群在全部样品中的最高相对丰度划分OM190_ub_o_o. o1为最优势菌,相对丰度介于0.50%-1.00%;Singulisphaera ub8为次优势菌,相对丰度介于0.20%-0.50%;有3种菌属于中等优势菌,相对丰度介于0.10%-0.20%;剩余4种菌相对罕见,相对丰度低于0.10%。
具体而言,OM190_ub_o_o. o1、SM1A02. ub5、Pla4_lineage_o_o_o. o6最高相对丰度依次出现在R11 (0.68%)、W14 (0.13%)、T23 (0.11%)。OM190_o_o_o. o2、AKYG587. ub3、Phycisphaera ub4、Singulisphaera o7、Singulisphaera ub8、Planctomycetaceae_u. ub9主要分布在湿生环境下远水侧河滨细粉砂质土壤W2、旱生环境下细粉质栗钙土T1、旱生环境下粗粉质栗钙土T2,最高相对丰度依次出现在W21 (0.078%)、T21 (0.089%)、W22 (0.059%)、T13 (0.19%)、T13 (0.35%)、T12 (0.071%)。另外,Singulisphaera o7、Singulisphaera ub8均在T13出现最高峰。综上所述,OM190_ub_o_o. o1最适合于水生环境河床中心砂质沉积物R1;SM1A02. ub5最适合于湿生环境下近水侧河滨粗粉砂质土壤W1;OM190_o_o_o. o2、Phycisphaera ub4最适合于湿生环境下远水侧河滨细粉砂质土壤W2;Singulisphaera o7、Singulisphaera ub8、Planctomycetaceae_u. ub9最适合于旱生环境下细粉质栗钙土T1,AKYG587. ub3和Pla4_lineage_o_o_o. o6最适合于旱生环境下粗粉质栗钙土T2。
2.3 温带半干旱区河流系统样品间Planctomycetes群落不同种群的相似性分析Planctomycetes群落9个种群的Heatmap (图 3)显示,来自水生环境的R1、R2、R3和湿生环境的W1、W2种群组成丰富且相似度较高,聚为第一大类;来自旱生环境的T1、T2种群组成较为丰富且相似度较高,聚为第二大类。在第一大类中,W1、W2 (W23除外)、R1 (R13、R14)和R3可划分为第1簇,其种群组成较为丰富且相似度较高;R1 (R13、R14除外)和R2可划分为第2簇,与第1簇相比其种群组成较为单一,相似度较高。在第二大类中,T1中T12、T13和T14划分为第3簇,T1 (T12、T13、T14除外)和T2划分为第4簇,第4簇较第3簇种群组成更为丰富。
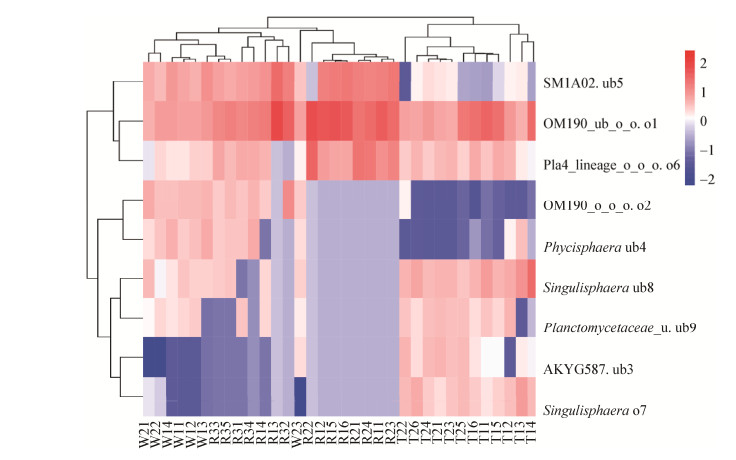
|
| 图 3 基于热图分析样品间Planctomycetes不同种群的相似性 Figure 3 Similarity analysis among samples based on heatmap of different populations from Planctomycetes |
|
|
从Planctomycetes群落9个种群的空间分布特征来看,OM190_ub_o_o. o1、SM1A02. ub5和Pla4_lineage_o_o_o. o6在各种环境均有分布,而且主要分布在R1水生环境中聚成第一大类。其中,OM190_ub_o_o. o1和Pla4_lineage_o_o_o. o6生存环境极为相似聚成一小簇。其他6个种群聚成第二大类,其中OM190_o_o_o. o2和Phycisphaera ub4主要分布在水生和湿生环境而非旱生环境聚成第1簇;AKYG587. ub3、Singulisphaera o7主要分布在旱生环境而非湿生和水生环境聚成第2簇;Singulisphaera ub8和Planctomycetaceae_u. ub9主要分布在旱生环境与湿生环境而非水生环境聚为第3簇。简言之,Planctomycetes群落能敏感指示生存环境的变化,是生存环境变化的敏感指示菌。
2.4 温带半干旱区河流系统Planctomycetes群落不同种群的空间分化及成因分析由Pearson相关性分析(表 3)可知,AKYG587. ub3、Pla4_lineage_o_o_o. o6、Singulisphaera o7、Singulisphaera ub8、Planctomycetaceae_u. ub9与黏粒(clay)、粉粒(silt)、盐度(dissolved salt)、全氮(total nitrogen)、全磷(total phosphorus)和硝态氮(nitrate nitrogen)都呈现极显著正相关关系(P < 0.01),与总有机碳(total organic carbon)呈不显著的正相关关系,与砂粒(sand)和水分(water content)都呈现极显著负相关关系(P < 0.01),与pH (Planctomycetaceae_u. ub9除外)和氨氮(ammonia nitrogen)呈负相关关系但未达到显著水平(P > 0.05)。OM190_ub_o_o. o1仅与砂粒呈正相关关系,与其他环境因子都呈负相关关系。
| 种群 Populations | 黏粒 Clay | 粉粒 Silt | 砂粒 Sand | 含水量 WC | pH | 溶解性盐 DS | 总有机碳 TOC | 全氮 TN | 全磷 TP | 氨氮 AM | 硝态氮 NR |
| OM190_ub_o_o. o1 | -0.122 | -0.213 | 0.188 | -0.286 | -0.697** | -0.621** | -0.475** | -0.380* | -0.363* | -0.623** | -0.314 |
| OM190_o_o_o. o2 | -0.168 | 0.070 | 0.011 | 0.330 | 0.450** | 0.023 | -0.032 | -0.165 | -0.113 | -0.150 | -0.235 |
| AKYG587. ub3 | 0.457** | 0.701** | -.0639** | -0.675** | -0.019 | 0.579** | 0.179 | 0.614** | 0.737** | -0.269 | 0.638** |
| Phycisphaera ub4 | 0.050 | 0.089 | -0.078 | 0.242 | 0.378* | 0.114 | 0.008 | 0.010 | 0.008 | -0.202 | -0.135 |
| SM1A02. ub5 | -0.488** | -0.446** | 0.476** | 0.455** | 0.144 | -0.472** | -0.349* | -0.539** | -0.507** | -0.306 | -0.663** |
| Pla4_lineage_o_o_o. o6 | 0.634** | 0.715** | -0.710** | -0.597** | -0.006 | 0.639** | 0.244 | 0.669** | 0.692** | -0.170 | 0.709** |
| Singulisphaera o7 | 0.702** | 0.649** | -0.689** | -0.616** | -0.152 | 0.560** | 0.187 | 0.645** | 0.651** | -0.269 | 0.651** |
| Singulisphaera ub8 | 0.769** | 0.695** | -0.744** | -0.598** | -0.144 | 0.583** | 0.194 | 0.665** | 0.665** | -0.297 | 0.654** |
| Planctomycetaceae_u. ub9 | 0.664** | 0.787** | -0.770** | -0.658** | 0.007 | 0.661** | 0.203 | 0.691** | 0.768** | -0.331 | 0.683** |
| 注:*:在0.05水平(双侧)上显著相关;**:在0.01水平(双侧)上显著相关. Note: *: Correlation was significant at the 0.05 level (2-tailed); **: Correlation was significant at the 0.01 level (2-tailed). WC: Water content; DS: Dissolved salt; TOC: Total organic carbon; TN: Total nitrogen; TP: Total phosphorus; AM: Ammonia nitrogen; NR: Nitrate nitrogen. | |||||||||||
OM190_o_o_o. o2与粉粒、砂粒、水分、盐度和pH均呈现正相关关系,其中pH为显著正相关关系(P < 0.01);与黏粒、总有机碳、全氮、全磷、氨氮和硝态氮含量均为负相关关系。Phycisphaera ub4与黏粒、粉粒、水分、pH、盐度、总有机碳、全氮和全磷呈正相关关系,与砂粒、氨氮和硝态氮呈负相关关系。SM1A02. ub5与砂粒和水分呈极显著正相关关系(P < 0.01),与pH呈正相关关系,与黏粒、粉粒、盐度、总有机碳、全氮、全磷和硝态氮呈极显著负相关关系(P < 0.01),与氨氮呈负相关关系。
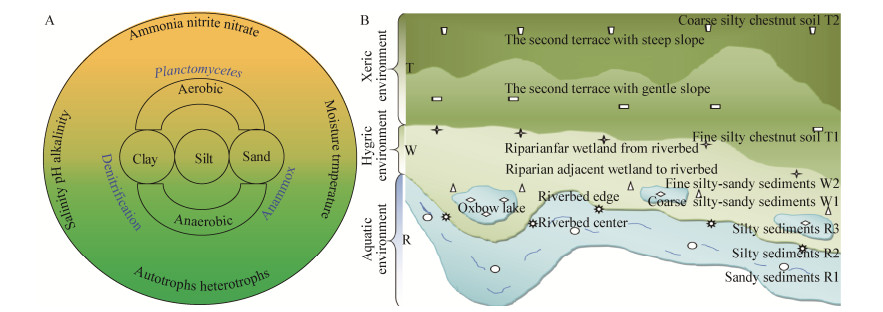
冗余分析(图 4A)相关结果显示,第1排序轴与第2排序轴的解释度分别为57.05%和19.27%。OM190_ub_o_o. o1优先分布在水生的R1、R2和R3环境,与水分、砂粒与氨氮关联度较高。OM190_o_o_o. o2、Phycisphaera ub4和SM1A02. ub5优先分布在湿生的W1和W2环境中,与pH和水分关联度高;AKYG587. ub3、Pla4_lineage_o_o_o. o6、Singulisphaera o7、Singulisphaera ub8和Planctomycetaceae_u. ub9优先分布在旱生的T1和T2环境中,与粉粒、黏粒、盐度、总有机碳、全氮、全磷、硝态氮的关联度较高。总之,RDA分析得到了和Pearson相关性分析一致性的结果。

|
| 图 4 Planctomycetes群落不同种群与环境因子的冗余分析(A)及环境因子的变异权重分析(B) Figure 4 Redundancy analysis of different populations from Planctomycetes communities and environmental factors (A) and variation partitioning analysis of environmental factors (B) Note: Plan-1: OM190_ub_o_o. o1; Plan-2: OM190_o_o_o. o2; Plan-3: AKYG587. ub3; Plan-4: Phycisphaera ub4; Plan-5: SM1A02. ub5; Plan-6: Pla4_lineage_o_o_o. o6; Plan-7: Singulisphaera o7; Plan-8: Singulisphaera ub8; Plan-9: Planctomycetaceae_u. ub9. |
|
|
变异权重分析(图 4B)结果显示,已知环境因子对Planctomycetes群落空间变异贡献了85.1%的解释度,其中粉粒、氨氮、水分、盐度、总有机碳、黏粒的解释度分别为52.7%、10.4%、10.3%、3.5%、2.6%、2.0%,蒙特尔检验达到P < 0.01。其中,粉粒、氨氮、水分三者的解释度高达73.4%,占已知环境因子总解释度的86.3%,说明粉粒、氨氮、水分是驱动Planctomycetes群落结构空间分化的主要环境因子。
线性回归分析(图 5)结果进一步证实,粉粒、氨氮、水分与Planctomycetes群落单个种群的相对丰度变化高度相关。其中,粉粒的相关系数R2明显高于氨氮、水分因子的相关系数R2,而粉粒的P值远低于氨氮、水分因子的P值。总体而言,线性回归分析得到了和变异权重分析一致性的结果,粉粒、氨氮、水分是已知环境因子中驱动Planctomycetes群落结构空间分化的主要环境因子。
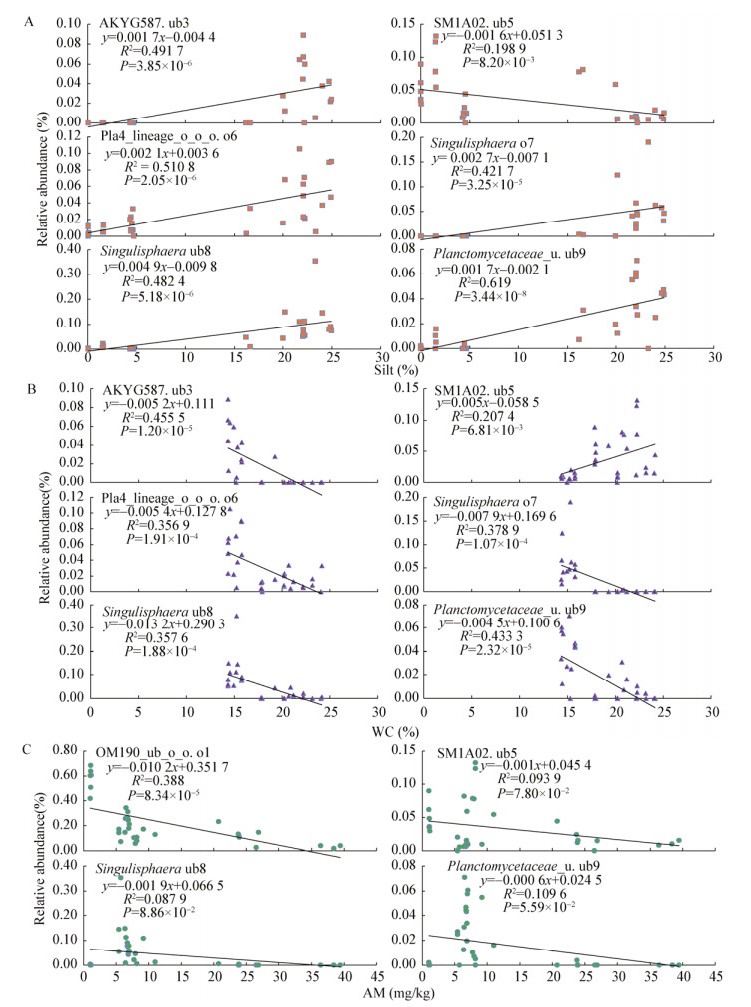
|
| 图 5 基于线性回归分析Planctomycetes群落不同种群与粉粒(A)、水分(B)、氨氮(C)的关系 Figure 5 The relationship between different populations from Planctomycetes communities and silt particles (A), water content (WC) (B) and ammonia nitrogen (AM) (C) based on linear regression analysis |
|
|
16S rRNA基因是研究Anammox相关Planctomycetes群落的重要分子钟[23-24],16S rRNA基因中有87%-99%的序列都来自于Planctomycetes。本研究基于16S rRNA基因V3-V4区引物338F (5′-ACTCCTACGGGAGGCAGCA-3′)和806R (5′-GGACTACHVGGGTWTCTAAT-3′)进行PCR扩增子高通量测序未检测到Anammox相关的Brocadia、Kuenenia、Anammoxoglobus、Jettenia和Scalindua成员,曾静[25]2016年利用Planctomycetales (pla46R/630R)及Anammox细菌特异性16S rRNA基因(Amx368f/Amx820)进行巢式PCR扩增子克隆测序在本研究区域水生表层砂质或粉质沉积物及湿生表层粉砂质土壤检测到了Brocadia、Anammoxoglobus、Jettenia、Kuenenia和Scalindua (图 6),说明Planctomycetales 16S rRNA基因特异性引物是研究Anammox菌群空间分化的重要保障,选择细菌16S rRNA基因高通量测序并非研究Anammox细菌在内全部细菌的万能钥匙。本研究发现主要分布在旱生粉质土壤环境中的AKYG587. ub3、Pla4_lineage_o_o_o. o6、Singulisphaera o7、Singulisphaera ub8和Planctomycetaceae_u. ub9虽不是Planctomycetales相关Anammox菌群,但可能为利用硝酸盐作为底物的反硝化菌群(由硝酸盐含量正向驱动),这和Kartal等[16]2007年、Flores等[26]2014年及He等[15] 2018年证明Planctomycetes群落具有反硝化功能的研究结果是一致的。Zhou等[22]2018年研究农田系统发现Planctomycetes群落增加,可能和Planctomycetes群落参与反硝化作用有关系,这为本研究发现Planctomycetes群落具有潜在的反硝化作用提供了间接证据。
Youssef等[27]2014年报道的浮霉菌门(Planctomycetes)包括Planctomycetacia和Phycisphaerae等2个纲,Phycosphaerales、Planctomycetales和Brocadiales等3个目,以及Anammox细菌相关和不相关的12个属。本研究基于16S rRNA基因高通量测序检测到Planctomycetes门共包括4个纲和5个目。纲水平上除了包括Phycisphaerae和Planctomycetacia外,还包括OM190纲和Pla4_lineage纲;目水平上除了包括Phycisphaerales和Planctomycetales外,还包括OM190纲中的Uncultured_bacterium目和Other目以及Pla4_lineage纲中的Other目。简言之,纲和目高级分类单元水平上,本研究发现Planctomycetes群落包括了更为丰富的类群。这说明随着高通量测序和生物信息学技术的不断发展,Planctomycetes群落中能被检测的类群日趋丰富。然而本研究发现Planctomycetes群落中只有8个属和9个种,低于2014年报道的类群数[28]。Planctomycetes群落中9个种群的最高相对丰度(0.06%-0.68%)较低,说明种属水平上Planctomycetes类群的空间扩散能力更容易受到环境条件限制,导致在本研究区域Planctomycetes群落中可检测到的种群并非优势类群。
本研究发现在属水平上有明确分类学名称的细菌包括隶属于Phycisphaeraceae的Phycisphaera和隶属于Planctomycetaceae的Singulisphaera。Planctomycetes群落属水平上Phycisphaera ub4主要分布在湿生环境,与盐度呈不显著的正相关关系,这和Fukunaga等[29]从海洋中分离到的Phycisphaera mikurensis有相似性,说明Phycisphaera可能是中等嗜盐菌。Kulichevskaya等[30]研究发现Singulisphaera rosea适合生存于酸性湿地环境,这为本研究检测到的Singulisphaera ub8适合于相对酸性土壤环境(与pH呈负相关关系)的结果提供了科学证据,说明Planctomycetes群落中的Singulisphaera适于酸性环境。Ivanova等[7]证明Planctomycetes在以地衣为优势的西伯利亚寒温带土壤中具有高度的生物多样性,这和本研究中国华北温带半干旱区湿地土壤Planctomycetes的分布特征具有相似性。本研究发现Planctomycetes群落在纲分类水平上的Planctomycetacia主要分布在旱生和水生环境中,由有机碳、总氮、总磷和pH等环境因子驱动,这和刘洋等[31]发现Planctomycetes受土壤碳、氮、磷等影响和Kulichevskaya等[30]发现Plactomycetes适于低pH北方湿地环境的研究结果高度吻合。
Yang等[23]2006年利用厌氧活性污泥、好氧活性污泥和河流沉积物研究Anammox的特性,发现颗粒物微结构对Anammox能否形成附着型生物膜、解离型无机颗粒核、进行自凝聚作用至关重要。Khramenkov等[20]2013年发现河流粉质沉积物有利于获取Anammox相关Planctomycetes富集培养物。2014年Morrissey等[32]发现粘土中加入胞外DNA (extracellular DNA)会增加Planctomycetes的相对丰度,说明Planctomycetes能有效降解黏土中的胞外DNA。综上,黏粒、粉粒、砂粒可能对Anammox相关Planctomycetes群落有影响,为本研究粉粒主要能驱动Planctomycetes群落空间分化的结果提供了间接证据[20, 23, 32]。
本研究发现旱生环境粉质栗钙土中的优势菌AKYG587. ub3、Pla4_lineage_o_o_o. o6、Singulisphaera o7、Singulisphaera ub8、Planctomycetaceae_u. ub9全部由有机碳正向驱动,说明来自于Planctomycetes群落的这5个种群可能为异养菌[33-35];由硝态氮正向驱动,说明来自于Planctomycetes群落的这5个种群可能为反硝化菌[16, 36];偏好于黏粒和粉粒较为丰富且全氮和全磷及盐度相对较高的旱生环境粗粉质或细粉质栗钙土。Zhou等[22]2018年在研究农田系统氨氧化和反硝化微生物及细菌群落结构时发现麦田Planctomycetes群落增加,可能和Planctomycetes群落参与反硝化作用有关系,这为本研究发现Planctomycetes群落具有潜在的反硝化作用提供了间接证据。本研究发现Planctomycetes群落全部种群由氨氮负向驱动,说明温带半干旱河流系统的Planctomycetes群落可能类似于完全氨氧化菌(complete ammonia oxidizers,Comammox),对氨有高度的亲和能力,适合在低氨寡营养动态环境下缓慢生长,高氨和营养丰富的环境不利于生存,属于K策略菌群[21, 37]。该菌对氨氮的亲和力高,最大氨氮速率低,适合在低营养的动态环境中缓慢生长。Planctomycetaceae_u. ub9与黏粒、粉粒、盐度、全氮、全磷和硝态氮都呈现极显著正相关关系(P < 0.01),与总有机碳呈不显著的正相关关系(P > 0.05)。
4 结论温带半干旱区河流系统Planctomycetes群落不同种群存在明显的空间分化特征,9个种群对生存环境变化敏感,是生存环境变化的敏感指示菌,而且全部由氨氮负向驱动,并不具有Anammox的功能。OM190_ub_o_o. o1种群主要分布在寡营养的水生环境砂质沉积物中,与水分、砂粒与氨氮关联度较高,由砂粒正向驱动,可能为对氨氮有高度亲和力的游离型自养氨氧化菌;OM190_o_o_o. o2、Phycisphaera ub4和SM1A02. ub5种群主要分布在中营养的湿生环境粉砂质土壤中,与pH和水分关联度高;其中OM190_o_o_o. o2由盐度和粉粒正向驱动,Phycisphaera ub4种群表现出由中营养向全营养微生物过渡的趋势,由硝态氮负向驱动;AKYG587. ub3、Pla4_lineage_o_o_o. o6、Singulisphaera o7、Singulisphaera ub8、Planctomycetaceae_u. ub9种群主要分布在旱生全营养粉质土壤中,与粉粒、黏粒、盐度、总有机碳、全氮、全磷、硝态氮的关联度较高,由黏粒、粉粒、盐度、有机碳、全氮、全磷、硝态氮正向驱动,可能为附着型的异养反硝化菌。粉粒、氨氮、水分是驱动温带半干旱区河流生态系统Planctomycetes群落结构及功能空间分化的主要因子。
| [1] |
Kuypers MMM, Sliekers AO, Lavik G, et al. Anaerobic ammonium oxidation by anammox bacteria in the Black Sea[J]. Nature, 2003, 422(6932): 608-611. DOI:10.1038/nature01472 |
| [2] |
van Niftrik L, Geerts WJC, van Donselaar EG, et al. Cell division ring, a new cell division protein and vertical inheritance of a bacterial organelle in anammox planctomycetes[J]. Molecular Microbiology, 2009, 73(6): 1009-1019. DOI:10.1111/j.1365-2958.2009.06841.x |
| [3] |
Nie SA, Lei XM, Zhao LX, et al. Response of activity, abundance, and composition of anammox bacterial community to different fertilization in a paddy soil[J]. Biology and Fertility of Soils, 2018, 54(8): 977-984. DOI:10.1007/s00374-018-1320-7 |
| [4] |
Tsushima I, Ogasawara Y, Kindaichi T, et al. Development of high-rate anaerobic ammonium-oxidizing (anammox) biofilm reactors[J]. Water Research, 2007, 41(8): 1623-1634. DOI:10.1016/j.watres.2007.01.050 |
| [5] |
Huang PB, Jiao NZ, Feng J, et al. Research progress on Planctomycetes' diversity and ecological function in marine environments[J]. Microbiology China, 2014, 41(9): 1891-1902. (in Chinese) 黄佩蓓, 焦念志, 冯洁, 等. 海洋浮霉状菌多样性与生态学功能研究进展[J]. 微生物学通报, 2014, 41(9): 1891-1902. |
| [6] |
Spring S, Bunk B, Spröer C, et al. Genome biology of a novel lineage of Planctomycetes widespread in anoxic aquatic environments[J]. Environmental Microbiology, 2018, 20(7): 2438-2455. DOI:10.1111/1462-2920.14253 |
| [7] |
Ivanova AO, Dedysh SN. Abundance, diversity, and depth distribution of Planctomycetes in acidic northern wetlands[J]. Frontiers in Microbiology, 2012, 3: 5. |
| [8] |
Kuenen JG. Anammox bacteria: from discovery to application[J]. Nature Reviews Microbiology, 2008, 6(4): 320-326. DOI:10.1038/nrmicro1857 |
| [9] |
Nishimura F, Hidaka T, Nakagawa A, et al. Removal of high concentration ammonia from wastewater by a combination of partial nitrification and anammox treatment[J]. Environmental Technology, 2012, 33(13/15): 1485-1489. |
| [10] |
Jenni S, Vlaeminck SE, Morgenroth E, et al. Successful application of nitritation/anammox to wastewater with elevated organic carbon to ammonia ratios[J]. Water Research, 2014, 49: 316-326. DOI:10.1016/j.watres.2013.10.073 |
| [11] |
Strous M, van Gerven E, Zheng P, et al. Ammonium removal from concentrated waste streams with the anaerobic ammonium oxidation (Anammox) process in different reactor configurations[J]. Water Research, 1997, 31(8): 1955-1962. DOI:10.1016/S0043-1354(97)00055-9 |
| [12] |
Su SH, Daverey A, Lin JG, et al. Application of simultaneous partial nitrification and anammox process for treatment of high strength nitrogen containing optoelectronic wastewater[J]. Proceedings of the Water Environment Federation, 2013, 2013(16): 1888-1906. DOI:10.2175/193864713813674009 |
| [13] |
van Teeseling MCF, Mesman RJ, Kuru E, et al. Anammox Planctomycetes have a peptidoglycan cell wall[J]. Nature Communications, 2015, 6: 6878. DOI:10.1038/ncomms7878 |
| [14] |
de Cocker P, Bessiere Y, Hernandez-Raquet G, et al. Enrichment and adaptation yield high anammox conversion rates under low temperatures[J]. Bioresource Technology, 2018, 250: 505-512. DOI:10.1016/j.biortech.2017.11.079 |
| [15] |
He SL, Yang W, Qin M, et al. Performance and microbial community of anammox in presence of micro-molecule carbon source[J]. Chemosphere, 2018, 205: 545-552. DOI:10.1016/j.chemosphere.2018.04.136 |
| [16] |
Kartal B, Kuypers MMM, Lavik G, et al. Anammox bacteria disguised as denitrifiers: nitrate reduction to dinitrogen gas via nitrite and ammonium[J]. Environmental Microbiology, 2007, 9(3): 635-642. DOI:10.1111/j.1462-2920.2006.01183.x |
| [17] |
Wu ZY, Meng H, Huang XW, et al. Salinity-driven heterogeneity toward anammox distribution and growth kinetics[J]. Applied Microbiology and Biotechnology, 2019, 103(4): 1953-1960. DOI:10.1007/s00253-018-9521-4 |
| [18] |
Ge CH, Sun N, Kang Q, et al. Bacterial community evolutions driven by organic matter and powder activated carbon in simultaneous anammox and denitrification (SAD) process[J]. Bioresource Technology, 2018, 251: 13-21. DOI:10.1016/j.biortech.2017.12.017 |
| [19] |
Gasa NP, Nnadozie CF, Kosgey K, et al. Effect of ammonium to nitrite ratio on reactor performance and microbial population structure in anammox reactors[J]. Environmental Technology, 2019. DOI:10.1080/09593330.2019.1610076 |
| [20] |
Khramenkov SV, Kozlov MN, Kevbrina MV, et al. A novel bacterium carrying out anaerobic ammonium oxidation in a reactor for biological treatment of the filtrate of wastewater fermented sludge[J]. Microbiology, 2013, 82(5): 628-636. DOI:10.1134/S002626171305007X |
| [21] |
Yu JL, Xia JJ, Li CH, et al. Niche differentiation of Nitrospira and associated environmental driving forces in Xilin river basin[J]. Microbiology China, 2020, 47(5): 1418-1429. (in Chinese) 于景丽, 夏晶晶, 李传虹, 等. 锡林河流域Nitrospira的生态位分化及环境驱动力[J]. 微生物学通报, 2020, 47(5): 1418-1429. |
| [22] |
Zhou XG, Wang ZL, Jia HT, et al. Continuously monocropped Jerusalem artichoke changed soil bacterial community composition and ammonia-oxidizing and denitrifying bacteria abundances[J]. Frontiers in Microbiology, 2018, 9: 705. DOI:10.3389/fmicb.2018.00705 |
| [23] |
Yang Y, Zuo JE, Quan ZX, et al. Study on performance of granular anammox process and characterization of the microbial community in sludge[J]. Water Science & Technology, 2006, 54(8): 197-207. |
| [24] |
Fuerst JA, Sagulenko E. Beyond the bacterium: Planctomycetes challenge our concepts of microbial structure and function[J]. Nature Reviews Microbiology, 2011, 9(6): 403-413. DOI:10.1038/nrmicro2578 |
| [25] |
Zeng J. Studies on anammox bacteria diversity, abundance and influence of environmental factors in Xilin River wetland[D]. Hohhot: Master's Thesis of Inner Mongolia University, 2016 (in Chinese) 曾静.锡林河湿地厌氧氨氧化菌群多样性、丰度及空间特征研究[D].呼和浩特: 内蒙古大学硕士学位论文, 2016 http://cdmd.cnki.com.cn/Article/CDMD-10126-1016133831.htm |
| [26] |
Flores C, Catita JAM, Lage OM. Assessment of planctomycetes cell viability after pollutants exposure[J]. Antonie van Leeuwenhoek, 2014, 106(2): 399-411. DOI:10.1007/s10482-014-0206-4 |
| [27] |
Youssef NH, Elshahed MS. The phylum Planctomycetes[A]//Rosenberg E, de Long EF, Lory S, et al. The Prokaryotes[M]. Berlin, Heidelberg: Springer, 2014: 759-810
|
| [28] |
Gu C, Zhou HF, Zhang QC, et al. Effects of various fertilization regimes on abundance and activity of anaerobic ammonium oxidation bacteria in rice-wheat cropping systems in China[J]. Science of the Total Environment, 2017, 599-600: 1064-1072. DOI:10.1016/j.scitotenv.2017.04.240 |
| [29] |
Fukunaga Y, Kurahashi M, Sakiyama Y, et al. Phycisphaera mikurensis gen. nov., sp. nov., isolated from a marine alga, and proposal of Phycisphaeraceae fam. nov., Phycisphaerales ord. nov. and Phycisphaerae classis nov. in the phylum Planctomycetes[J]. The Journal of General and Applied Microbiology, 2009, 55(4): 267-275. DOI:10.2323/jgam.55.267 |
| [30] |
Kulichevskaya IS, Detkova EN, Bodelier PLE, et al. Singulisphaera rosea sp. nov., a planctomycete from acidic Sphagnum peat, and emended description of the genus Singulisphaera[J]. International Journal of Systematic and Evolutionary Microbiology, 2012, 62(1): 118-123. DOI:10.1099/ijs.0.025924-0 |
| [31] |
Liu Y, Huang YM, Zeng QC. Soil bacterial communities under different vegetation types in the Loess Plateau[J]. Environmental Science, 2016, 37(10): 3931-3938. (in Chinese) 刘洋, 黄懿梅, 曾全超. 黄土高原不同植被类型下土壤细菌群落特征研究[J]. 环境科学, 2016, 37(10): 3931-3938. |
| [32] |
Morrissey EM, McHugh TA, Schwartz E, et al. Microbial decomposition of extracellular DNA in clay soils[A]//AGU Fall Meeting Abstracts[M]. Washington: AGU, 2014
|
| [33] |
Naumoff DG, Ivanova AA, Dedysh SN. Phylogeny of β-хylanases from Planctomycetes[J]. Molecular Biology, 2014, 48(3): 439-447. DOI:10.1134/S0026893314030145 |
| [34] |
Bondoso J, Albuquerque L, Nobre MF, et al. Roseimaritima ulvae gen. nov., sp. nov. and Rubripirellula obstinata gen. nov., sp. nov. two novel Planctomycetes isolated from the epiphytic community of macroalgae[J]. Systematic and Applied Microbiology, 2015, 38(1): 8-15. DOI:10.1016/j.syapm.2014.10.004 |
| [35] |
Delmont TO, Quince C, Shaiber A, et al. Nitrogen-fixing populations of Planctomycetes and Proteobacteria are abundant in surface ocean metagenomes[J]. Nature Microbiology, 2018, 3(7): 804-813. DOI:10.1038/s41564-018-0176-9 |
| [36] |
Hong X, Hong YW, Chen ZW, et al. Phylogenetic diversity of nirS-type denitrifying bacteria in Jiulong River estuary[J]. Microbiology China, 2015, 42(9): 1639-1650. (in Chinese) 洪璇, 洪有为, 陈仲巍, 等. 九龙江河口区nirS型反硝化细菌多样性及系统发育学分析[J]. 微生物学通报, 2015, 42(9): 1639-1650. |
| [37] |
Kits KD, Sedlacek CJ, Lebedeva EV, et al. Kinetic analysis of a complete nitrifier reveals an oligotrophic lifestyle[J]. Nature, 2017, 549(7671): 269-272. DOI:10.1038/nature23679 |
 2020, Vol. 47
2020, Vol. 47