扩展功能
文章信息
- 邓木兰, 梁志成, 梁秀怡, 张智, 李芳红, 赵子建
- DENG Mu-Lan, LIANG Zhi-Cheng, LIANG Xiu-Yi, ZHANG Zhi, LI Fang-Hong, Allan Zijian ZHAO
- 基于URA3基因快速构建表达载体及其在里氏木霉的应用
- Rapid construction of an expression vector based on URA3 gene for application in Trichodermium reesei
- 微生物学通报, 2020, 47(2): 649-658
- Microbiology China, 2020, 47(2): 649-658
- DOI: 10.13344/j.microbiol.china.190475
-
文章历史
- 收稿日期: 2019-06-03
- 接受日期: 2019-09-19
- 网络首发日期: 2019-11-04
2. College of Pharmacy and Health Sciences, Saint John’s University, New York 11439, USA;
3. 深圳大学生命科学与海洋学院 广东 深圳 518060;
1. School of Biomedical and Pharmaceutical Sciences, Guangdong University of Technology, Guangzhou
2. Guangdong 510006, China;
3. College of Pharmacy and Health Sciences, Saint John's University, New York 11439, USA;
1. College of Life Science and Oceanography, Shenzhen University, Shenzhen, Guangdong 518060, China
基因工程(genetic engineering)突破了现有育种无法打破的物种之间的界限,可以使不同物种如原核与真核、动物与植物、人和其他生物等的遗传信息进行重组和转移[1-2]。基因工程必不可少的工具就是表达载体[3-5],虽然市面上已经有多种商品化的表达载体可供选购,例如Novagen Corporation用于大肠杆菌蛋白表达的pET系列载体;Invitrogen Corporation用于毕赤酵母蛋白表达的pPICZ系列载体;Clontech Laboratories用于哺乳类动物细胞的pDsRed1、pEYFP系列载体等,但是这些表达载体只针对某物种,一旦遇到里氏木霉等,则要面临没有相应的商品化表达载体可用的尴尬局面,严重影响研究进度。在构建复杂的多片段拼接载体时,传统的载体构建方法[6-7]需要恰当地挑选酶切位点,必要时需要选择多个转化载体,操作复杂,既费时又费力。因此,寻找一种快速、高效、便捷的载体构建方法对异源蛋白在里氏木霉中高效表达具有重要的意义。
URA3基因编码乳清苷5-磷酸脱羧酶(orotidine 5′-phosphate decarboxylase),该酶催化尿嘧啶核苷酸生物合成的最后一步反应,与乳清酸磷酸核糖基转移酶(Ura5p)结合,可以将5-氟-乳清酸(5-fluoro-orotic acid,5-FOA)转化成5-氟尿嘧啶(5-fluorouracil,5-FU),5-FU是一种嘧啶类似物,可以掺入核酸链中代替Uracil或Thymine,干扰RNA和DNA加工和功能,最终导致细胞死亡[8],广泛用于酵母的筛选标记[9-10]。Δura3菌株的生长需要额外提供外源尿嘧啶,并且可在含有5-FOA的培养基中生存。利用URA3基因作为基因转化选择标记的优点是含有URA3基因的菌株不能在5-FOA培养基中生长[11],可同时去除阴性转化子和野生型菌株。
基于URA3/5-FOA选择系统的机理,本文根据DNA重组酶和同源重组的原理提出了快速构建表达载体的方法,将线性的PCR片段定向重组,一步实现1−5个片段的高效无缝拼接构成完整的质粒。以URA3基因的启动子上游5′端序列和终止子下游3′端序列构建同源重组臂,利用强启动子和终止子,替换原来URA3基因的启动子和终止子序列,构建新的表达元件组合,并在强启动子和终止子之间添加多克隆位点(multiple cloning site,MCS),以方便外源基因的重组插入,将这些片段与大肠杆菌复制原点和氨苄抗性基因共同构建成新的载体,并以RFP mCherry为报告基因,分析和鉴定载体的功能。
里氏木霉(Trichoderma reesei) RUT-C30是无性型红褐肉座菌属嗜温丝状真菌[12],具有较好的蛋白合成和分泌能力,以及真核细胞的分泌机制[13],其N糖基化和高甘露糖型等可能和哺乳动物表达体系相似[14-16]。RUT-C30还获得美国食品药品监督管理局(Food and Drug Administration,FDA)的“一般认为安全(generally recognized as safe,GRAS)”认证,其在生产过程中无致病性、无真菌毒素和抗生素产生,这为外源蛋白在里氏木霉的高效表达及生产提供一条切实可行的方法[17-19]。虽然RUT-C30具有良好的工业化蛋白生产潜力,但目前科研界对其研究得仍不够深入[20-23],可供研究的商品化表达载体寥寥无几,使得研究受到很大局限。因此,本研究基于URA3基因快速构建表达载体,以里氏木霉作为表达菌株来展示具体的构建方法。
1 材料与方法 1.1 材料 1.1.1 菌株和载体里氏木霉(Trichoderma reesei) RUT-C30购自于中国科学院微生物研究所菌种保藏中心;大肠杆菌(JM109)由本实验室保存;mCherry基因来自由本实验室保存的pLVX-IRES-mCherry质粒,用JCAT在线工具进行密码子优化,并在mCherry的C端加入6×His-Tag蛋白标签,序列交给深圳华大基因科技公司合成DNA;本实验所用引物(表 1)均在深圳华大基因科技公司合成。
| 引物Primer | 引物序列Primers sequence (5′→3′) | 酶切位点Restriction enzyme site | 扩增的目的片段Amplified target fragment |
| pTRUC-F1 | CGGTACGCGCGGATCTTCCAGAGATCCGCGGCTCGCCTCTTCTTTGTGCTTTTCTCCTG | Sac Ⅱ | 5′URA3重组臂5′URA3 homologous reorganization arm |
| pTRUC-F2 | GCCAAGGTAGGTACCTACCACTAGGCGCGCCGGAACCTGGATACATCCATCATCACGC | Asc Ⅰ | CBH I启动子 CBH I promoter (Pcbh1) |
| pTRUC-F3 | GTCAACCGCGGACTGCGCATCTTAAGATCGAT GCGGCCGCCCGGCATGAAGTCTGACCGGGTAG | Afl Ⅱ; Not Ⅰ | PDC终止子PDC terminator (Tpdc) |
| pTRUC-F4 | GAGGAAGACATCGAGGCGTCCAACTAGTGGC GGAGGCGAACTGCACATTGG | Spe Ⅰ | 3′URA3重组臂3′URA3 homologous reorganization arm |
| pTRUC-F5 | GTCCCCCCGGAACAGCGACCTGACCGCGGAA TCGTCGAACGGCAGGCGTGC | Sac Ⅱ | pUC复制起点和氨苄抗性基因pUC origin and ampicillin resistance gene |
| mCherry-F | GCGCATCTTAAGATGGTTTCTAAGGGTGAAG AAGAC | Afl Ⅱ | mCherry红色荧光蛋白Red fluorescent protein of mCherry |
| pTRUC-R1 | GCGTGATGATGGATGTATCCAGGTTCCGGCGCGCCTAGTGGTAGGTACCTACCTTGGC | Asc Ⅰ | 5′URA3重组臂5′URA3 homologous reorganization arm |
| pTRUC-R2 | CTACCCGGTCAGACTTCATGCCGGGCGGCCGC ATCGATCTTAAGATGCGCAGTCCGCGGTTGAC | Afl Ⅱ; Not Ⅰ | CBH I启动子CBH I promoter (Pcbh1) |
| pTRUC-R3 | CCAATGTGCAGTTCGCCTCCGCCACTAGTTGG ACGCCTCGATGTCTTCCTC | Spe Ⅰ | PDC终止子PDC terminator (Tpdc) |
| pTRUC-R4 | GCACGCCTGCCGTTCGACGATTCCGCGGTC AGGTCGCTGTTCCGGGGGGAC | Sac Ⅱ | 3′URA3重组臂3′URA3 homologous reorganization arm |
| pTRUC-R5 | CAGGAGAAAAGCACAAAGAAGAGGCGAGCCGCGGATCTCTGGAAGATCCGCGCGTACCG | Sac Ⅱ | pUC复制起点和氨苄抗性基因pUC origin and Ampicillin resistance gene |
| mCherry-R | CCGGCAGCGGCCGCTTAGTGATGGTGATGGT GATGCTTGTACAATTCGTCCATACCACC | Not Ⅰ | mCherry红色荧光蛋白Red fluorescent protein of mCherry |
| 注:下划线所标注序列为限制性酶切位点序列. Note: The underlined sequences are restriction enzyme site sequences. | |||
Afl Ⅱ和Not Ⅰ限制性内切酶、T4 DNA连接酶购自TaKaRa公司;pEASY-Uni Seamless Cloning and Assembly Kit购自北京全式金生物技术有限公司;真菌基因组提取试剂盒、胶回收试剂盒、质粒提取试剂盒均购自Omega公司;偶联辣根过氧化物酶的His-Tag (D3I1O) XP® Rabbit mAb (HRP Conjugate)抗体购自CST公司。超净工作台、恒温培养箱,上海龙跃仪器设备有限公司;空气浴摇床,上海天呈实验仪器制造有限公司;电泳转化膜系统,Bio-Rad公司。
1.1.3 主要培养基LB固体培养基(g/L):胰化蛋白胨10.0,酵母提取物5.0,氯化钠10.0,琼脂15.0,pH 7.0。PDA培养基(g/L):土豆浸出液200.0,葡萄糖10.0,琼脂15.0,用于里氏木霉的固体培养。里氏木霉液体基础培养基和原生质体再生培养基参照Penttilä等[14]的方法配制。5-FOA的PDA筛选培养基:含0.5% 5-FOA的PDA培养基。诱导产酶培养基(g/L):液体基础培养基加乳糖20.0 g/L,酵母提取物12.0 g/L。
1.2 方法pTRUC表达载体各部分元件来源如下:(1) CBHI启动子(Pcbh1)和PDC终止子序列(Tpdc)的获得,使用表达水平高的CBHI基因(glycoside hydrolase family 7)以及丙酮酸脱羧酶基因(pyruvate decarboxylase,PDC)的Pcbh1启动子和Tpdc终止子,5′与3′URA3重组臂扩增自RUT-C30基因组;(2) pMD-19T质粒提供pUC origin和氨苄抗性基因的来源,使目的载体能够在大肠杆菌中复制及保存。根据GenBank中里氏木霉基因组核苷酸序列设计引物,将各个载体片段PCR扩增出来,切胶纯化回收,然后通过重组酶将这5个片段连接起来,再转入大肠杆菌中扩增,pTRUC载体的结构示意图见图 1。
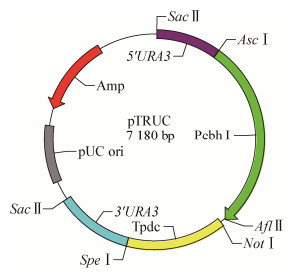
|
| 图 1 表达载体pTRUC的结构示意图 Figure 1 Structure diagram of expression carrier pTRUC |
|
|
pTRUC表达载体各部分元件PCR反应体系均为25 μL:ddH2O 18.2 μL,10×PCR缓冲液2.5 μL,dNTP Mix 2 μL,模板(20 μmol/L) 1 μL,正、反向引物(20 μmol/L)各0.5 μL,Taq酶(5 U/μL) 0.3 μL。
Pcbh1启动子PCR反应条件:95 ℃ 3 min;95 ℃ 30 s,55 ℃ 30 s,72 ℃ 2 min,35个循环;72 ℃ 20 min,4 ℃保存。
Tpdc终止子、5′URA3同源臂、3′URA3同源臂、pUC ori与Amp抗性基因PCR反应条件中72 ℃延伸时间分别为1 min、50 s、50 s、2 min 40 s,其他条件同Pcbh1启动子PCR反应条件。
1.2.1 RUT-C30的培养和基因组提取将里氏木霉接种于PDA平板上,于28 ℃恒温培养4−5 d后制备孢子悬液,接种于液体基础培养基中,于28 ℃、250 r/min条件下培养2 d,然后用试剂盒提取gDNA并用1%凝胶核酸电泳检测。
1.2.2 PCR引物设计及序列扩增载体pTRUC各个元件基因,包括Pcbh1启动子(约1 971 bp)、Tpdc终止子(约1 030 bp)、5′URA3同源臂(约658 bp)、3′URA3同源臂(约784 bp)、pUC ori与Amp抗性基因(约2 692 bp),通过重组酶将上述片段连接获得目的表达载体pTRUC,长度约为7 180 bp。
构建表达载体过程中需要用到的引物见表 1,具体扩增过程如下:(1)以RUT-C30基因组为模板,用引物对pTRUC-F1和pTRUC-R1、pTRUC-F2和pTRUC-R2、pTRUC-F3和pTRUC-R3、pTRUC-F4和pTRUC-R4,分别扩增出5′URA3重组臂DNA序列、CBH1启动子(Pcbh1) DNA序列、PDC终止子序列(Tpdc) DNA序列、3′URA3重组臂DNA序列;(2)以pMD-19T质粒DNA为模板,用引物对pTRUC-F5和pTRUC-R5,扩增出pUC origin和Ampicillin resistance gene DNA序列;(3)以密码子优化的人工合成的mCherry DNA为模板,用引物mCherry-R和mCherry-F扩增出mCherry红色荧光蛋白DNA序列。
PCR扩增完成后,取5 μl扩增产物进行1%凝胶核酸电泳鉴定,纯化回收扩增产物。
1.2.3 表达载体pTRUC的构建将纯化好的5个载体元件DNA片段配成等摩尔量的混合液,每个DNA片段的摩尔量均为0.1 pmol/μL,将此混合液与2×Assembly Mix反应液进行连接反应,然后将大肠杆菌JM109感受态细胞与反应液混合,铺于含50 μg/mL氨苄抗生素的LB平板,37 ℃恒温培养箱倒置培养过夜,挑选单菌落放大培养,提取质粒,然后进行电泳和测序鉴定。
1.2.4 表达载体pTRUC-mCherry的构建pTRUC质粒DNA和mCherry PCR产物DNA分别进行Afl Ⅱ和Not Ⅰ双酶切反应,用连接酶连接双酶切的pTRUC质粒DNA和mCherry DNA产物,重组载体命名为pTRUC-mCherry,将连接好的质粒转入大肠杆菌JM109感受态细胞中,涂氨苄抗性平板37 ℃倒置培养过夜。将平板上的单菌落接种于氨苄抗性的LB液体培养基中培养,按照DNA提取试剂盒说明书步骤提取pTRUC-mCherry质粒DNA进行Afl Ⅱ和Not Ⅰ酶切鉴定,反应完成后取5 μL扩增产物进行1%凝胶核酸电泳鉴定,部分质粒DNA送去深圳华大基因科技公司测序。
1.2.5 RUT-C30原生质体的制备及质粒pTRUC- mCherry的转化取108 RUT-C30孢子接种于30 mL液体基础培养基中,里氏木霉原生质体的制备和转化主要按照Penttilä等[14]的方法进行,将质粒pTRUC- mCherry转化至里氏木霉原生质体内,然后将原生质体悬浮液涂布于5个5-FOA PDA筛选培养基平板上,28 ℃培养3−5 d后观察阳性转化子生长情况。
1.2.6 RUT-C30 pTRUC-mCherry阳性转化子的鉴定(1) 显微镜观察红色荧光蛋白的表达。将5-FOA PDA筛选平板上长出的阳性转化子分别转接到PDA平板上进行扩大培养。一周后首先将各转化子分别接种于诱导产酶培养基中,28 ℃、220 r/min培养48 h后,将培养物置于激光共聚焦显微镜观察RFP在RUT-C30中的表达情况。
(2) PCR鉴定mCherry基因。选取一株表达红色荧光蛋白的阳性转化子接种于诱导产酶培养基中,28 ℃、220 r/min扩大培养48 h后去上清,将菌丝体置于研钵,用液氮研磨成粉,转至2.0 mL EP管中,用真菌基因组DNA快速抽提试剂盒提取其基因组DNA,另取RUT-C30亲本菌株和pTRUC空载体转化菌株作为对照,重复此操作,得到相应的基因组DNA。
将提取的基因组DNA作为模板,mCherry-R和mCherry-F分别进行PCR扩增,扩增产物进行1%凝胶核酸电泳鉴定。
(3) Western blotting鉴定RFP表达。将菌液抽滤,称取0.1 g菌丝体用200 μL RIPA裂解液充分振荡混匀菌丝体,将菌液抽滤,称取0.1 g菌丝体用200 μL RIPA裂解液充分振荡混匀菌丝体,加入40 μL 6×loading buffer,振荡混匀,95 ℃金属浴加热10 min,制备成蛋白样品,进行12% PAGE分离胶凝胶电泳,然后将蛋白印迹转移到PVDF膜上,5%的脱脂牛奶封闭1 h,用His-Tag一抗4 ℃孵育过夜,用1×TBST洗膜5 min,洗5次,然后ECL显色。
2 结果与分析 2.1 里氏木霉基因组DNA的提取用基因组DNA提取试剂盒提取RUT-C30基因组DNA,电泳检验,结果如图 2所示,表明成功提取到RUT-C30基因组DNA。
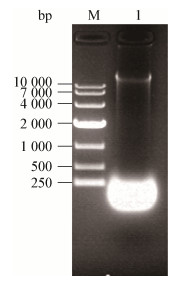
|
| 图 2 RUT-C30基因组DNA提取结果 Figure 2 Results of RUT-C30 genome DNA extraction 注:M:1 kb marker;1:RUT-C30基因组. Note: M: 1 kb marker; 1: RUT-C30 genome. |
|
|
通过扩增得到载体pTRUC各个元件基因,Pcbh1启动子、Tpdc终止子、5′URA3同源臂、3′URA3同源臂、pUC ori与氨苄抗性基因。通过重组酶将上述片段连接获得目的载体pTRUC,大小约为7 180 bp,核酸凝胶电泳结果如图 3所示,电泳及测序结果均表明各序列均已得到正确扩增,载体pTRUC成功构建。
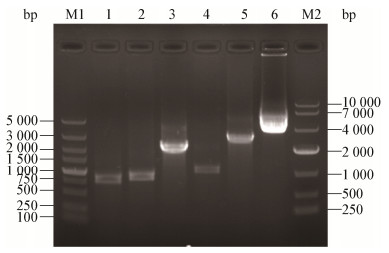
|
| 图 3 pTRUC载体及5个DNA片段扩增结果 Figure 3 Results of pTRUC vector and amplification five DNA fragments 注:M1:5 000 bp Marker;1:5′URA3同源臂基因DNA片段;2:3′URA3同源臂基因DNA片段;3:Pcbh1启动子DNA片段;4:Tpdc终止子DNA片段;5:pMD-19T DNA片段;6:pTRUC质粒;M2:1 kb Marker. Note: M1: 5 000 bp Marker; 1: DNA fragments of 5′URA3 homologous arm; 2: DNA fragments of 3′URA3 homologous arm; 3: DNA fragments of promoter Pcbh1; 4: DNA fragments of terminator Tpdc; 5: DNA fragments of pMD-19T; 6: Plasmid pTRUC; M2: 1 kb Marker. |
|
|
将构建好的pTRUC载体质粒和红色荧光蛋白mCherry的PCR产物分别用Afl Ⅱ和Not Ⅰ双酶切后由连接酶连接,形成重组载体pTRUC-mCherry后转化至大肠杆菌JM109,提取质粒进行酶切鉴定,电泳结果见图 4,在约7 000 bp和750 bp处可见条带,符合载体pTRUC及mCherry基因的长度,测序结果表明重组载体构建成功。
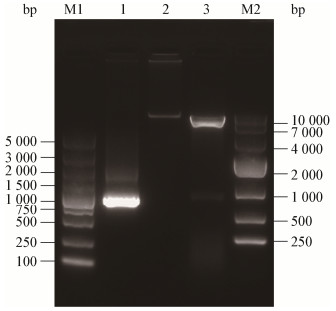
|
| 图 4 pTRUC-mCherry质粒双酶切结果 Figure 4 Result of pTRUC-mCherry plasmid digested 注:M1:5 000 bp Marker;1:mCherry DNA片段;2:pTRUC-mCherry质粒DNA;3:Afl Ⅱ和Not Ⅰ双酶切的pTRUC-mCherry质粒DNA;M2:1 kb Marker. Note: M1: 5 000 bp Marker; 1: DNA fragments of mCherry; 2: pTRUC-mCherry plasmid; 3: pTRUC-mCherry plasmid digested by Afl Ⅱ and Not Ⅰ; M2: 1 kb Marker. |
|
|
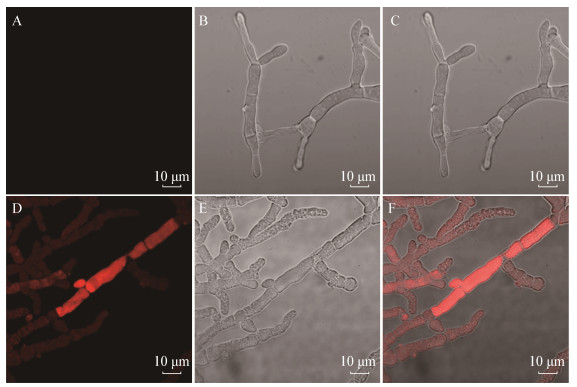
将重组载体pTRUC-mCherry转化至RUT-C30中进行表达鉴定,对载体pTRUC的表达功能进行鉴定,挑取菌丝体在激光共聚焦显微镜使用587 nm激发光条件下,可观察到发出红色荧光的菌丝体,而亲本菌株在相同条件下没有观察到红色荧光(图 5),表明载体pTRUC对插入的外源基因可进行有效的转录,并在RUT-C30中表达出红色荧光蛋白质,说明pTRUC表达载体能实现外源蛋白在RUT-C30中的表达。
|
| 图 5 阳性转化子红色荧光的观察 Figure 5 Observation of red fluorescence of positive transformants 注:A:RUT-C30在587 nm激发光光源下拍摄结果;B:RUT-C30在普通光源下拍摄结果;C:A和B合并图;D:mCherry重组菌在587 nm激发光光源下拍摄结果;E:mCherry重组菌在普通光源下拍摄结果;F:D和E合并图(放大倍数63×). Note: A: Shoot in 587 nm excitation light of RUT-C30; B: Shoot in ordinary light of RUT-C30; C: Merge A and B; D: Shoot in 587 nm excitation light of mCherry positive transformants; E: Shoot in ordinary light of mCherry positive transformants; F: Merge D and E (enlargement factor 63×). |
|
|
由于里氏木霉是多细胞的丝状体,其菌丝体任何一点都能发生分枝,在菌落发育的后期,菌丝之间相互接触,在菌丝接触点相近的壁局部降解而发生菌丝的结网现象,使菌落形成一个完整的网状结构,其意义在于将菌丝用放射状二维平面扩散到三维网状,使得营养物质等可以运送到菌落的任一地方[24]。重组mCherry里氏木霉菌株的菌丝体同样也有菌丝融合现象,当菌丝发生结网融合时,其表达的mCherry红色荧光蛋白也会互相分享,高表达mCherry红色荧光的转化子会与低表达的转化子或者是非转化子之间分享细胞质中的mCherry蛋白,从而表现出某一段菌丝表达量很高,其他菌丝表达量较低的情况,这也是丝状真菌表达较难筛选纯高表达菌株的原因之一。
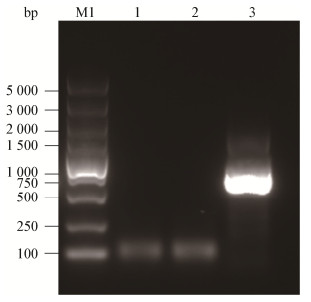
2.5 阳性转化子的PCR鉴定分别以RUT-C30亲本菌株、pTRUC空载体转化菌株、mCherry重组菌基因组DNA为模板,PCR扩增mCherry基因片段,PCR产物电泳检验结果见图 6,结果表明只有pTRUC-mCherry重组菌的基因组DNA含mCherry基因片段,片段大小约在750 bp,而RUT-C30亲本菌株、pTRUC空载体转化菌株均没有mCherry基因,说明pTRUC载体成功将mCherry基因重组并整合到RUT-C30基因组中。
|
| 图 6 mCherry基因组鉴定 Figure 6 Genome identification of mCherry 注:M:5 000 bp Marker;1:RUT-C30 gDNA为模板的PCR产物;2:pTRUC空载体转化菌株gDNA为模板的PCR产物;3:mCherry重组菌gDNA为模板的PCR产物. Note: M: 5 000 bp Marker; 1: PCR production of RUT-C30 gDNA; 2: PCR production of pTRUC gDNA; 3: PCR production of mCherry gDNA. |
|
|
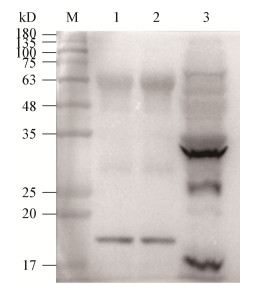
分别以RUT-C30亲本菌株、pTRUC空载体转化菌株、mCherry重组菌的菌体蛋白进行了Western blotting检测,结果表明只有mCherry重组菌的菌体蛋白有红色荧光蛋白mCherry的表达,在分子量约为30 kD处有特异性反应的条带,见图 7,该条带的大小与预测的目标蛋白分子量大小一致,说明红色荧光蛋白mCherry能在RUT-C30中表达,同时也说明pTRUC表达载体能将异源基因重组到RUT-C30中,并成功表达出相应蛋白。
|
| 图 7 蛋白印迹验证mCherry在RUT-C30中的表达情况 Figure 7 Western blotting analysis of expressing mCherry in RUT-C30 注:M:PR1930 Marker;1:RUT-C30空白对照;2:pTRUC空载体转化菌株;3:mCherry重组菌. Note: M: PR1930 Marker; 1: Control of RUT-C30; 2: Positive transformants of pTRUC; 3: Positive transformants of mCherry. |
|
|
里氏木霉因其具有分泌大量水解酶的能力而成为同源和异源蛋白表达有吸引力的宿主[13, 25-26],但里氏木霉表达重组蛋白的质粒完全是由自己实验室构建或别人赠送,缺点是费时费力、质粒质量参差不齐、其他研究者难以获得,从而导致实验进展缓慢,结果不理想。目前构建里氏木霉表达体系的质粒主要是用Pcbh1启动子,它是目前公认的表达量较高且使用最多的诱导型强启动子,它由几种糖类如纤维素、槐糖、乳糖等诱导,并受分解代谢抑制的调节[27-28],当Pcbh1启动子用于蛋白质表达时,必须加入诱导物(或诱导剂)以触发靶基因的表达[15-16, 29]。外源蛋白在里氏木霉中的表达量达到50–150 mg/L[30-31],已被用于产生各种同源或异源蛋白,因此本文选择Pcbh1启动子构建载体。
抗药基因是发展最早、应用最广的一类筛选标记,这类筛选标记应用方便,选择效率高,功能稳定,在生物遗传转化中已经被广泛应用于植物、动物、微生物等遗传转化[32-34]。随着生物技术的发展和应用,生物安全性及抗药基因类筛选标记可能带来的不良影响已经受到人们的广泛关注,抗药基因产生的安全性问题体现在基因横向转移使病原微生物获得抗药性,对人类及其生存环境产生潜在的危害;植物遗传转化中使用的除草剂筛选标记基因也有可能改变生物的遗传多样性,造成生态危害[35]。但是可以预见的是,分子生物技术将被广泛地应用于发酵、制药、食品/饲料工程、废水处理、环境修复、生物防治等领域的菌株构建或基因改良,但这些通过基因转化改良的菌株是不能带有抗药基因的[36],为了避免这一安全问题,本研究构建的pTRUC载体主要目的是实现人源药用蛋白在里氏木霉中的高效表达,从而选择营养缺陷型筛选基因。营养缺陷型筛选基因的优点主要是:(1)安全性问题的考虑;(2)因为真核生物的抗生素不仅价格比较昂贵,而且对动植物的毒性也比较强,所以选择营养缺陷型筛选基因;(3)酵母、构巢曲霉等作为模式菌株已经成功构建营养缺陷型菌株。营养缺陷型筛选标记在细菌重组改造中的使用也已有研究。de Vos等[37]将乳酸乳球菌糖类代谢途径中的关键基因lacF在宿主菌染色体上失活,再将该基因克隆到游离表达质粒中,当质粒导入lacF缺陷型宿主菌后,克隆在质粒上的基因表达使宿主菌的乳糖缺陷得到互补,从而得到一种食品级的选择标记。因此,本研究采用Pcbh1启动子和URA3基因缺陷构建表达载体,以期实现药用蛋白在里氏木霉中安全、高效表达。
重组酶对DNA片段连接的高效性和位点特异性使其在基因工程中受到关注[38-41],构建载体的相邻片段之间有一定的重叠片段,利用重组酶实现了一步多个片段的高效无缝拼接构建成目的载体。整个构建过程快速高效,只需要进行PCR扩增和重组酶连接两步即可生成目标载体,比起传统的载体构建方法更加高效、便捷。
mCherry是一种红色荧光蛋白,将mCherry基因重组到目标表达载体上进行表达鉴定能对该载体的蛋白表达效果进行直观的评价[42-44]。载体插入红色荧光蛋白mCherry基因,结果显示转化后的木霉菌丝体在587 nm激发光下可观察到明亮的红色荧光,基因组PCR和Western blotting结果分析可知成功建立了能够表达外源基因的表达载体,利用URA3基因的负筛选原理,同源重组整合到RUT-C30的基因组上,在含有尿嘧啶的培养基中正常生长并稳定表达外源蛋白。
本研究使RUT-C30更容易成为表达异源基因的表达宿主,为促进更多药用和食用蛋白在里氏木霉中的表达应用奠定了基础,相信本文提供的方法会使更多有经济价值的物种成为基因工程蛋白表达产品产业化、商品化进程中的有力工具。
| [1] |
Alexander DR. Uses and abuses of genetic engineering[J]. Postgraduate Medical Journal, 2003, 79(931): 249-251. |
| [2] |
González-Fraguela ME. Bioethical considerations of genetic engineering[J]. Revista de Neurología, 2002, 35(8): 701-704. DOI:10.33588/rn.3508.2001231 |
| [3] |
Ashwini M, Murugan SB, Balamurugan S, et al. Advances in molecular cloning[J]. Molecular Biology, 2016, 50(1): 1-6. |
| [4] |
Sharma K, Mishra AK, Mehraj V, et al. Advances and applications of molecular cloning in clinical microbiology[J]. Biotechnology and Genetic Engineering Reviews, 2014, 30(1/2): 65-78. |
| [5] |
Xie JK, Kong XL, Bao JS, et al. Recent advances in molecular mapping and cloning of useful genes from wild rice and their application in breeding[J]. Hereditas (Beijing), 2004, 26(1): 115-121. (in Chinese) 谢建坤, 孔祥礼, 包劲松, 等. 水稻野生种质优异基因分子标记定位和利用的研究进展[J]. 遗传, 2004, 26(1): 115-121. DOI:10.3321/j.issn:0253-9772.2004.01.022 |
| [6] |
Zhang YP, Yang SS. Methods for construction of transgenic plant expression vector:a review[J]. Chinese Journal of Biotechnology, 2015, 31(3): 311-327. (in Chinese) 张阳璞, 杨淑慎. 几种新型植物基因表达载体的构建方法[J]. 生物工程学报, 2015, 31(3): 311-327. |
| [7] |
Nakayama H, Shimamoto N. Modern and simple construction of plasmid:saving time and cost[J]. Journal of Microbiology, 2014, 52(11): 891-897. DOI:10.1007/s12275-014-4501-6 |
| [8] |
Ying SH, Feng MG, Keyhani NO. Use of uridine auxotrophy (URA3) for markerless transformation of the mycoinsecticide Beauveria bassiana[J]. Applied Microbiology and Biotechnology, 2013, 97(7): 3017-3025. DOI:10.1007/s00253-012-4426-0 |
| [9] |
Park EH, Seo JH, Kim MD. Cloning and characterization of the orotidine-5′-phosphate decarboxylase gene (URA3) from the osmotolerant yeast Candida magnoliae[J]. Journal of Microbiology and Biotechnology, 2012, 22(5): 642-648. DOI:10.4014/jmb.1111.11071 |
| [10] |
Ko N, Nishihama R, Pringle JR. Control of 5-FOA and 5-FU resistance by Saccharomyces cerevisiae YJL055W[J]. Yeast, 2008, 25(2): 155-160. DOI:10.1002/yea.1554 |
| [11] |
Poon BPK, Mekhail K. Effects of perinuclear chromosome tethers in the telomeric URA3/5FOA system reflect changes to gene silencing and not nucleotide metabolism[J]. Frontiers in Genetics, 2012, 3: 144. DOI:10.3389/fgene.2012.00144 |
| [12] |
Li CC, Lin FM, Zhou L, et al. Cellulase hyper-production by Trichoderma reesei mutant SEU-7 on lactose[J]. Biotechnology for Biofuels, 2017, 10: 228. DOI:10.1186/s13068-017-0915-9 |
| [13] |
de Paula RG, Antonieto ACC, Ribeiro LFC, et al. New genomic approaches to enhance biomass degradation by the industrial fungus Trichoderma reesei[J]. International Journal of Genomics, 2018, 2018: 1974151. |
| [14] |
Penttilä M, Nevalainen H, Rättö M, et al. A versatile transformation system for the cellulolytic filamentous fungus Trichoderma reesei[J]. Gene, 1987, 61(2): 155-164. DOI:10.1016/0378-1119(87)90110-7 |
| [15] |
Li JX, Wang J, Wang SW, et al. Achieving efficient protein expression in Trichoderma reesei by using strong constitutive promoters[J]. Microbial Cell Factories, 2012, 11: 84. DOI:10.1186/1475-2859-11-84 |
| [16] |
Du J, Zhang X, Li XZ, et al. The cellulose binding region in Trichoderma reesei cellobiohydrolase I has a higher capacity in improving crystalline cellulose degradation than that of Penicillium oxalicum[J]. Bioresource Technology, 2018, 266: 19-25. DOI:10.1016/j.biortech.2018.06.050 |
| [17] |
Ramoni J, Marchetti-Deschmann M, Seidl-Seiboth V, et al. Trichoderma reesei xylanase 5 is defective in the reference strain QM6a but functional alleles are present in other wild-type strains[J]. Applied Microbiology and Biotechnology, 2017, 101(10): 4139-4149. DOI:10.1007/s00253-017-8161-4 |
| [18] |
Li CC, Lin FM, Sun W, et al. Constitutive hyperproduction of sorbicillinoids in Trichoderma reesei ZC121[J]. Biotechnology for Biofuels, 2018, 11: 291. DOI:10.1186/s13068-018-1296-4 |
| [19] |
Wang MY, Zhang ML, Li L, et al. Role of Trichoderma reesei mitogen-activated protein kinases (MAPKs) in cellulase formation[J]. Biotechnology for Biofuelsvolume, 2017, 10: 99. DOI:10.1186/s13068-017-0789-x |
| [20] |
Ivanova C, Ramoni J, Aouam T, et al. Genome sequencing and transcriptome analysis of Trichoderma reesei QM9978 strain reveals a distal chromosome translocation to be responsible for loss of vib1 expression and loss of cellulase induction[J]. Biotechnology for Biofuelsvolume, 2017, 10: 209. DOI:10.1186/s13068-017-0897-7 |
| [21] |
Liu R, Chen L, Jiang YP, et al. A novel transcription factor specifically regulates GH11 xylanase genes in Trichoderma reesei[J]. Biotechnology for Biofuelsvolume, 2017, 10: 194. DOI:10.1186/s13068-017-0878-x |
| [22] |
Xiong LL, Kameshwar AKS, Chen X, et al. The ACEII recombinant Trichoderma reesei QM9414 strains with enhanced xylanase production and its applications in production of xylitol from tree barks[J]. Microbial Cell Factories, 2016, 15(1): 215. DOI:10.1186/s12934-016-0614-4 |
| [23] |
Zhang X, Xia LM. Expression of Talaromyces thermophilus lipase gene in Trichoderma reesei by homologous recombination at the cbh1 locus[J]. Journal of Industrial Microbiology & Biotechnology, 2017, 44(3): 377-385. |
| [24] |
Xing LJ, Li MC, Wei DS. Common Mycology[M]. Beijing: Higher Education Press, 2010: 11-15. (in Chinese) 邢来君, 李明春, 魏东盛. 普通真菌学[M]. 北京: 高等教育出版社, 2010: 11-15. |
| [25] |
Gupta VK, Steindorff AS, de Paula RG, et al. The post-genomic era of Trichoderma reesei:what's next?[J]. Trends in Biotechnology, 2016, 34(12): 970-982. DOI:10.1016/j.tibtech.2016.06.003 |
| [26] |
Gao F, Hao ZZ, Sun XH, et al. A versatile system for fast screening and isolation of Trichoderma reesei cellulase hyperproducers based on DsRed and fluorescence-assisted cell sorting[J]. Biotechnology for Biofuels, 2018, 11: 261. DOI:10.1186/s13068-018-1264-z |
| [27] |
Liu T, Wang TH, Li X, et al. Improved heterologous gene expression in Trichoderma reesei by cellobiohydrolase I gene (cbh1) promoter optimization[J]. Acta Biochimica et Biophysica Sinica, 2008, 40(2): 158-165. DOI:10.1111/j.1745-7270.2008.00388.x |
| [28] |
Margolles-Clark E, Harman GE, Penttila M. Enhanced expression of endochitinase in Trichoderma harzianum with the cbh1 promoter of Trichoderma reesei[J]. Applied and Environmental Microbiology, 1996, 62(6): 2152-2155. DOI:10.1128/AEM.62.6.2152-2155.1996 |
| [29] |
Kiesenhofer DP, Mach RL, Mach-Aigner AR. Influence of cis element arrangement on promoter strength in Trichoderma reesei[J]. Applied and Environmental Microbiology, 2018, 84(1): e01742-17. DOI:10.1128/AEM.01742-17 |
| [30] |
Du FY, Wolger E, Wallace L, et al. Determination of product inhibition of CBH1, CBH2, and EG1 using a novel cellulase activity assay[J]. Applied Biochemistry and Biotechnology, 2010, 161(1/8): 313-317. DOI:10.1007/s12010-009-8796-4 |
| [31] |
Fitz E, Wanka F, Seiboth B. The promoter toolbox for recombinant gene expression in Trichoderma reesei[J]. Frontiers in Bioengineering and Biotechnology, 2018, 6: 135. DOI:10.3389/fbioe.2018.00135 |
| [32] |
Li H, Wang BC, Xu WJ, et al. Identification and network of outer membrane proteins regulating streptomysin resistance in Escherichia coli[J]. Journal of Proteome Research, 2008, 7(9): 4040-4049. DOI:10.1021/pr800310y |
| [33] |
Peng XX, Xu CX, Ren HX, et al. Proteomic analysis of the sarcosine-insoluble outer membrane fraction of Pseudomonas aeruginosa responding to ampicilin, kanamycin, and tetracycline resistance[J]. Journal of Proteome Research, 2005, 4(6): 2257-2265. DOI:10.1021/pr050159g |
| [34] |
Xu CX, Lin XH, Ren HX, et al. Analysis of outer membrane proteome of Escherichia coli related to resistance to ampicillin and tetracycline[J]. Proteomics, 2006, 6(2): 462-473. DOI:10.1002/pmic.200500219 |
| [35] |
Zhang X, Li XQ, Xia LM. Heterologous expression of an alkali and thermotolerant lipase from Talaromyces thermophilus in Trichoderma reesei[J]. Applied Biochemistry and Biotechnology, 2015, 176(6): 1722-1735. DOI:10.1007/s12010-015-1673-4 |
| [36] |
Ramessar K, Peremarti A, Gómez-Galera S, et al. Biosafety and risk assessment framework for selectable marker genes in transgenic crop plants:a case of the science not supporting the politics[J]. Transgenic Research, 2007, 16(3): 261-280. DOI:10.1007/s11248-007-9083-1 |
| [37] |
de Vos WM. Safe and sustainable systems for food-grade fermentations by genetically modified lactic acid bacteria[J]. International Dairy Journal, 1999, 9(1): 3-10. DOI:10.1016/S0958-6946(99)00038-2 |
| [38] |
Ow DW. The long road to recombinase-mediated plant transformation[J]. Plant Biotechnology Journal, 2016, 14(2): 441-447. DOI:10.1111/pbi.12472 |
| [39] |
Wang YJ, Yau YY, Perkins-Balding D, et al. Recombinase technology:applications and possibilities[J]. Plant Cell Reports, 2011, 30(3): 267-285. DOI:10.1007/s00299-010-0938-1 |
| [40] |
Nandy S, Zhao S, Pathak BP, et al. Gene stacking in plant cell using recombinases for gene integration and nucleases for marker gene deletion[J]. BMC Biotechnology, 2015, 15: 93. DOI:10.1186/s12896-015-0212-2 |
| [41] |
Srivastava V, Thomson J. Gene stacking by recombinases[J]. Plant Biotechnology Journal, 2016, 14(2): 471-482. DOI:10.1111/pbi.12459 |
| [42] |
Karimi S, Ahl D, Vågesjö E, et al. In vivo and In vitro detection of luminescent and fluorescent Lactobacillus reuteri and application of red fluorescent mCherry for assessing plasmid persistence[J]. PLoS One, 2016, 11(3): e0151969. DOI:10.1371/journal.pone.0151969 |
| [43] |
van Zyl WF, Deane SM, Dicks LMT. Use of the mCherry fluorescent protein to study intestinal colonization by Enterococcus mundtii ST4SA and Lactobacillus plantarum 423 in mice[J]. Applied and Environmental Microbiology, 2015, 81(17): 5993-6002. DOI:10.1128/AEM.01247-15 |
| [44] |
Vacas A, Sugden C, Velasco-Rodriguez O, et al. Construction of two mCherry plasmids (pXG-mCherry) for transgenic Leishmania:valuable tools for future molecular analysis[J]. Journal of Parasitology Research, 2017, 2017: 1964531. DOI:10.1155/2017/1964531 |
 2020, Vol. 47
2020, Vol. 47




