扩展功能
文章信息
- 许从峰, 张方政, 张伟, 申贵男, 袁媛, 晏磊, 王伟东
- XU Cong-Feng, ZHANG Fang-Zheng, ZHANG Wei, SHEN Gui-Nan, YUAN Yuan, YAN Lei, WANG Wei-Dong
- 合成微生物群落用于木质纤维素生物质转化的研究进展
- Synthetic microbial consortia for lignocellulosic biomass conversion
- 微生物学通报, 2020, 47(10): 3431-3441
- Microbiology China, 2020, 47(10): 3431-3441
- DOI: 10.13344/j.microbiol.china.200690
-
文章历史
- 收稿日期: 2020-07-06
- 接受日期: 2020-08-28
- 网络首发日期: 2020-09-09
2. 粮食副产物加工与利用教育部工程研究中心 黑龙江 大庆 163319
2. Engineering Research Center of Processing and Utilization of Grain By-products, Ministry of Education, Daqing, Heilongjiang 163319, China
目前,全球约80%的能源需求靠化石燃料满足,化石燃料的大量使用导致二氧化碳排放量持续升高,被认为是全球变暖的罪魁祸首[1-2]。生物燃料具备取代化石燃料的潜力,是近年来研究热点之一。世界各国都投入了大量的资金以加速生物燃料研究和商业化开发[3-4]。木质纤维素生物质是地球上最丰富的可再生能源,全球每年大约有1万亿t纤维素被合成和降解,被认为是生物燃料的潜在候选[5-6]。自然菌群中不同微生物之间相互协调,能产生复杂的协同酶系统,不同类型的酶共同催化能有效促进木质纤维素的转化。近年来,人们采用选择性培养的方法从自然环境中富集出高效降解木质纤维素的复合菌系,能有效强化木质纤维素的水解作用,使之转化成能源物质,如乙醇、甲烷、氢和丁醇等,为木质纤维素的资源化利用提供了基础[7-9]。
木质纤维素转化为生物燃料和化学用品的生物工艺通常包括4个步骤:生物质的预处理、糖化作用、可溶性糖的发酵和下游加工,复合菌系在前2个步骤中效果显著,但是其内部代谢通路复杂,很难对最终代谢产物进行控制。传统的联合生物加工方法试图将所有需要的生化功能整合到一个菌株中,这种想法很难实现,主要原因包括导入菌体的基因难以稳定遗传,工程菌株的酶活不高,菌株对代谢产物的耐受性比较低[10]。随着合成生物学的发展,人们尝试将所需的生化功能分配到不同的微生物,这些微生物所组成的菌群称为合成微生物群落,其能够一步将木质纤维素转化成所需的产物,这种策略的灵感来源于自然菌群[11-12]。利用合理的策略构建微生物菌群并赋予其新的功能,能有效减少生产成本,在生物质加工方面具有很大的优势。我们主要从合成微生物群落的设计、新的生物分析工具及其在生物燃料生产方面的应用与挑战方面系统总结近年来在木质纤维素联合生物加工领域的研究进展。
1 系统生物学为合成微生物群落的设计提供指导自然菌群在生物质能源生产方面具有绿色无污染和可持续发展等优点[13-14],然而环境中只有不足1%的微生物可以通过传统的培养技术获得,人们对于菌群内部协同关系的了解非常有限,使得预测与控制菌群中微生物行为的难度大大增加,阻碍了微生物资源的开发与利用[15]。合成微生物群落相较于自然菌群具有组成明确、功能稳定和代谢模型简单等优点,可以对微生物实现精准的控制与监测,通过对关键菌株的改造以及菌株之间协同关系的设计,能有效提高物种之间的合作,减少环境带来的生存压力,从而高效地转化木质纤维素,使合成微生物群落具有更多的应用。
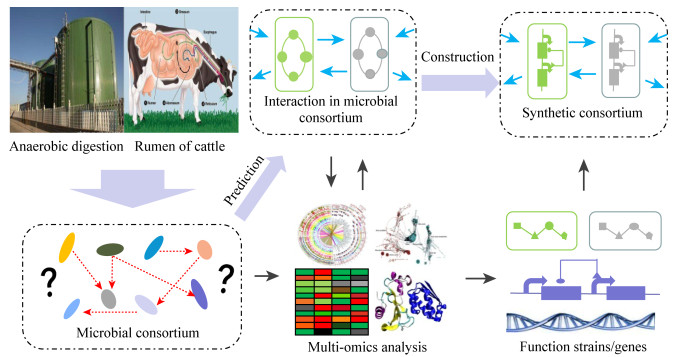
自然菌群是合成微生物群落的基础和来源,其内部尚未探明的复杂机制限制了合成微生物群落的研究进展,要想全面探究菌群的协同机制需要采取多种方法的联合。系统生物学试图用数学和计算方法在系统层面上分析、理解、建模和模拟复杂的生物系统,是全面解析微生物菌群及其发酵过程的重要研究方法[16],在解决合成微生物群落设计与优化的技术问题方面起主导作用,促进了合成微生物群落在联合生物加工领域的应用。近年来,由于高通量测序技术的迅速发展,组学技术(基因组、转录组、蛋白质组、代谢组和代谢流组等)已被广泛应用于菌群的研究,系统生物学方法能够借助组学数据对微生物群落中的功能多样性、基因表达水平、遗传和代谢通路进行全面描述[17-22],为合成微生物群落的设计奠定了基础,主要包括功能菌株的选择与代谢途径的设计、功能基因的筛选与表达以及菌株之间协同机制的探究与优化(图 1)。
|
| 图 1 系统生物学为合成微生物群落的设计提供指导 Figure 1 Systems biology provides guidance in designing synthetic microbial consortia Note: The molecular interaction mechanisms between microorganisms in microbial consortia could be elucidated by multi-omics analysis (e.g., genomics, transcriptomics, proteomics, metabolomics, etc.), which could provide insights for the design and construction of synthetic consortia[11]. |
|
|
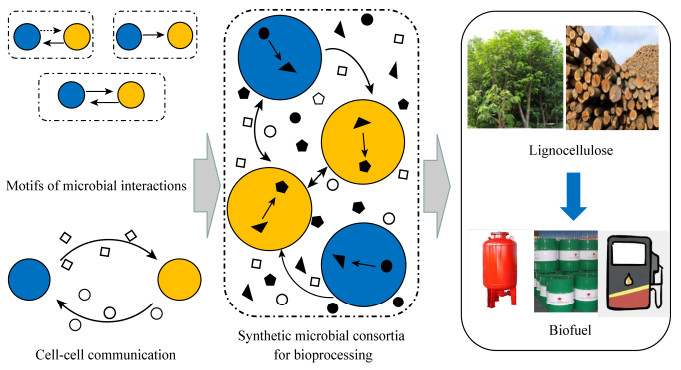
利用合成微生物群落进行联合生物加工的主要障碍是其需要同时控制个体微生物和整个菌群系统的稳定[23]。我们重点讨论由两种不同菌株构成的微生物群落,为了提高合成微生物群落的稳定性、高效性以及对代谢物的耐受性,我们将菌群的设计原则总结为细胞间的协同关系和细胞间信号交流(图 2)。
|
| 图 2 合成微生物群落的设计原则 Figure 2 The design principles of synthetic microbial consortia for consolidated bioprocessing 注:实线箭头和虚线箭头分别表示促进和抑制作用,空心的图形代表信号分子,实心的图形表示代谢物. Note: The solid arrow and the dashed arrow indicate promotion and inhibition, respectively. The hollow graphics represent signal molecules, and the solid graphics represent metabolites in consortia. Engineering of microbial interactions and cell-cell communication are combined to enable coordination and biofulet formation[24]. |
|
|
微生物群落中的许多相互作用模式(如共生、互利、竞争、中立、寄生或捕食)大部分是由微生物彼此之间的代谢物交换介导,其中基于互利关系的菌群更有可能实现群落的稳定和功能的强劲[25-28]。在底物的转化过程中,一个物种将底物代谢成废物,作为第二个物种的底物,如果第一个物种产生的废物对其自身是“有毒”的,那么菌株之间的关系是互利的,因为第一个物种为第二个物种提供基质,而第二个物种通过清除毒素促进两个物种的共存[29]。
Shou等[30]通过建立数学模型分析了两种酵母菌在菌群中共生的限制条件,结果表明两种菌株的初始生长率、存活率及代谢产物的生产是两者协同作用发生的结果,与任何一种纯培养菌株相比均有更高的稳定性和高效性。Bayer等[31]利用产纤维素酶的放线菌Actinotalea fermentans将纤维素转化成乙醇和乙酸盐,然后由工程菌株Saccharomyces cerevisiae转化成甲基卤化物,可以有效促进两者的共存(图 3A)。Minty等[32]组建了一个包含Escherichia coli和产纤维素酶的真菌Trichoderma reesei的合成微生物菌群,E. coli就像一个“骗子”,其不能产生纤维素酶但是却利用T. reesei的水解产物,合作-欺骗关系是该菌群保持稳定的关键,我们认为该方法有进一步被开发的潜力(图 3B)。
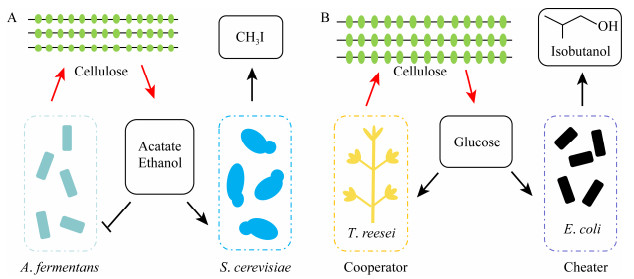
|
| 图 3 合成微生物群落的互利共生模型 Figure 3 The cooperation and commensalism model of synthetic microbial consortia 注:A:A. fermentans将纤维素转化成醋酸盐和乙醇,这些产物对A. fermentans有抑制作用,S. cerevisiae能利用这些产物为能源生产CH3I[31]. B:T. reesei产生纤维素酶将纤维素水解成葡萄糖,葡萄糖同时是T. reesei和E. coli的生长底物,E. coli可以将葡萄糖发酵成异丁醇[32]. Note: A: A. fermentans ferments cellulosic to acetate and ethanol, and that has inhibition to A. fermentans, but S. cerevisiae can converte that as energy sources to CH3I[31]. B: T. reesei produces cellulases that hydrolyze cellulose to glucose which as growth substrates for T. reesei and E. coli, and E. coli ferments glucose into isobutanol[32]. |
|
|
信息的交换在影响微生物群落的种群行为方面起中心作用[33],合成微生物群落的稳定性通常需要通过生物体之间的信号分子和细胞间接触来调节。微生物可以通过群体感应机制与其他微生物进行信息交流,部分自然信号分子已经被用于工程菌株的开发,使通过人工合成信号分子来控制菌群成为可能[34-35]。
合成微生物群落面临的一个重大挑战是如何通过细胞之间传递信号来控制整个微生物群体。Groß等[36]构建了一个由两株酵母菌共培养的菌群,利用一种天然酵母信息素(α因子)控制细胞间信号交流和目标蛋白的表达,通过基因模块化和细胞间信号交流的结合使每个菌株的功能独立优化成为可能。Weber等[37]以挥发性乙醛为信号,使细菌、酵母和哺乳动物细胞之间能进行信号交流,更多新的信号系统应该能够实现跨物种的通信,包括基于细胞间直接接触和不接触的信号。Kylilis等[38]利用一个自动选择正交通信的软件(ChemmComms- calculator-v1.0)识别出4个正交通道,并通过实验证明了3个通道同时存在于合成微生物群落。开发多个非干扰的细胞间通信通道是一个可行的步骤,有助于合成微生物群落中细胞间信号传递效率的提升。
人工合成细胞-细胞的信号分子是调整合成微生物群落的一个有价值的工具。但是,导入或者敲除的基因容易发生失活或修复,细胞间信号交流受阻的合成微生物群落往往很难维持物种的稳定,在微流控设备之外难以培养[25, 39-40]。
3 合成微生物群落的优化我们相信,深入理解合成微生物群落中的代谢相互作用,最终将使我们更好地了解和开发微生物菌群的代谢潜力。为了实现合成微生物群落的广泛优化,阐明群落中微生物的生理和代谢模型是很重要的[41]。例如,代谢组学和蛋白质组学最近被用来阐明Bacillus megaterium和Ketogulonicigenium vulgare之间的相互作用[42-43],结果表明,两者在微生物群落中的生理特征和通过代谢物进行的信息交流是决定种群动态和功能稳定的关键。Chignell等[44]利用定量蛋白质组学方法对由Pseudomonas putida KT2440和Bacillus atrophaeus ATCC 9372组成的人工菌群进行了研究,结果发现,与纯培养菌株相比,在包含两种菌株的群落中有100多种蛋白质出现上调或下调。另外,种群动态模型、空间模型和IBM模型(individual-based models)也被用来描述细胞间协同关系[45],与代谢工程的联合将有助于微生物群落针对目标功能进行合理的优化。
代谢模型必须建立在准确获取微生物群落的生物量、底物和代谢物的基础之上。Klitgord等通过代谢流平衡分析(flux balance analysis,FBA)预测了微生物在不同环境条件下的相互作用[46],结果表明,该方法不仅能证明已知的相互作用,还能预测新的相互作用,有可能确定菌群的最佳培养条件。该结果有助于充分了解合成微生物群落的动态和进化,开发新的代谢途径并应用于代谢工程。近年来,基因工程(包括CRISPR/Cas系统,用于有效的基因删除、插入和转录控制)[47]和快速组装DNA片段[48]的方法取得飞跃式的进步,尤其是单细胞测序技术的普及[49-50],越来越多的跨学科研究策略使得通过合理构建代谢途径来控制微生物行为成为可能[51-52],因此,合成微生物群落的概念不仅在联合生物加工方面得到更多的应用,在污染物降解和生物医药等方面也有很大的开发空间[53-54]。
4 合成微生物群落在联合生物加工方面的应用与挑战在单个反应器中,利用合成微生物群落将木质纤维素转化为生物燃料的方法具有效率高、成本低的巨大优势,相关实例见表 1,但是大部分合成微生物群落的研究仅停留在实验室条件下,其工业化应用仍然处于起步阶段,要想充分发挥微生物的潜力还需将菌群动态和代谢模型与实际生产进一步结合。我们认为除了考虑合成微生物群落中细胞间协同关系、信号交流以及菌株耐受性和外来物种入侵等问题,在实际生产中还需要以灵活的方式对菌群的空间生态位提供必要的控制。
| 糖化菌株/发酵菌株 Saccharolytic strain/Fermentation strain |
基质 Substrate |
产物 Product |
产率 Yield (g/g) |
滴度 Titer (g/L) |
来源 Source |
| Clostridium phytofermentans/S. cerevisiae cdt-1 | α-cellulose | Ethanol | 0.31 | 22.0 | [55] |
| Acremonium cellulolyticus C-1/S. cerevisiae ATCC 4126 | Solka floc (cellulose) | Ethanol | 0.18 | 46.3 | [56] |
| Kluyveromyces fragilis LOCK 0027/Zymomonas mobilis 3881 | Jerusalem artichoke | Ethanol | 0.48 | 99.0 | [57] |
| Clostridium thermocellum ATCC 27405/Clostridium saccharoperbutylacetonicum N1-4 | Avicel | Butanol | 0.20 | 7.9 | [58] |
| Clostridium celevecrescens/C. acetobutylicum | Filter paper | Butanol | 0.09 | 2.7 | [59] |
| Clostridium cellulovorans 743B/Clostridium beijerinckii NCIMB 8052 | Corn cobs | Butanol | 0.12 | 8.3 | [60] |
| Thermoanaerobacterium sp. M5/Clostridium acetobutylicum | Hemicellulose | Butanol | 0.14 | 8.3 | [61] |
| Engineered consortium of C. cellulovorans/C. beijerinckii | Alkali extracted corn cobs | Butanol | 0.14 | 11.5 | [62] |
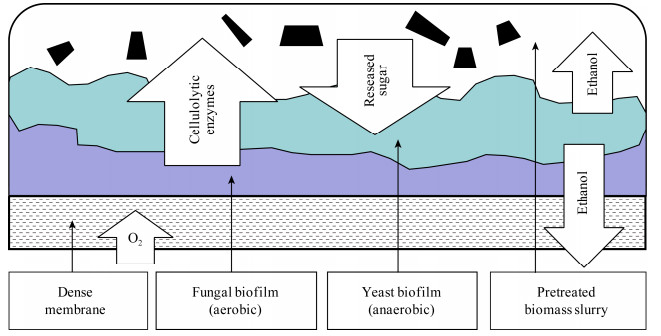
商业纤维素酶通常由真菌产生,而Trichoderma reesei因其产生纤维素酶的能力更强而被广泛研究。一般来说,真菌对木质纤维素的降解是在好氧情况下发生,而微生物的发酵作用是在厌氧的情况下发生,因此,好氧-厌氧菌群结构被认为是联合生物加工最具潜力的组合之一。为了同时满足两者的生存条件,可以设计一种多物种生物膜反应器,好氧菌在紧靠膜的位置,为上方的厌氧菌创造厌氧环境和提供相关底物(图 4)。例如,通过Trichoderma reesei、Saccharomyces cerevisiae和Scheffersomyces stipitis的联合作用,以经稀酸预处理后的小麦秸秆为底物,生产了9.8 g/L乙醇[63]。本实验室利用从木质纤维素降解复合菌系WSC-6中分离的厌氧菌(Clostridium sp.)和好氧菌(Bacillu sp.和Clostridiaceae sp.)共培养,以滤纸为底物,通过乙醇和甲酸等代谢物探索三者之间的协同关系,结果表明,好氧菌可以通过消耗共培养体系中氧气和小分子有机酸来促进厌氧菌的生长,进而提高木质纤维素降解率,促进乙醇的累积[28, 64-67]。除了生物燃料的生产,这种概念的验证设计可以用于菌群从木质纤维素中生产其他生物产品(例如乳酸)[68],证明了基于合成微生物群落的联合生物加工技术利用木质纤维素生产高价值化学物质的可能性。
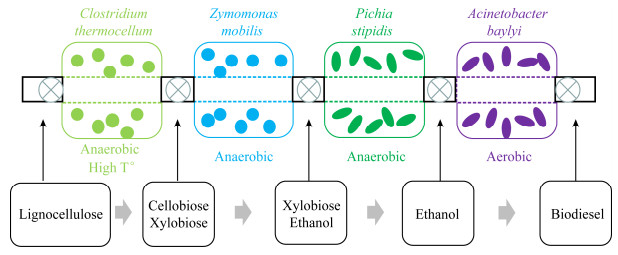
当不同物种在同一发酵装置培养时,各菌种除了对氧气需求量不同之外,对营养物质、温度和pH等需求的差异化也将不同程度地影响整体的生产效率[69]。空间连接微生物群落(spatially linked microbial consortium,SLMC)根据目标功能的差异来选择不同的菌株和培养环境,不需要为每个物种提供一致的生长条件[70]。SLMC允许在空间分布上对微生物进行控制,可以为每个纯培养微生物提供最佳的生存环境以强化单个物种的代谢效率,同时微生物之间可以通过代谢物和信号分子进行交流,有效提升整体的发酵效率(图 5)[71]。由于其模块化设计,SLMC将允许在乙醇生产之后附加额外的生物处理流程以转化乙醇,例如成为生物柴油[72-73]。SLMC旨在提供一个多样化的生存环境,以促进菌群中每个成员的生长和代谢功能,结合空间生态位,甚至可以将原本不可共存的物种联系起来以满足生产特殊产品的需求,在联合生物加工方面具有巨大的开发潜力。
在过去的几年里,合成微生物群落在木质纤维素联合生物加工方面的研究进展快速,证明其在开发生物燃料生产方面的巨大潜力。然而,在合成微生物群落用于大规模联合生物加工之前有以下几个问题需要注意,主要包括:(1)继续筛选降解木质纤维素纯培养菌株,不同菌株可能存在竞争或者生存条件差异较大,可以尽量采用从同一生存环境中获取的菌株进行菌群构建;(2)强化不同菌株之间协同机制的探究,分别从微生物个体、菌群和基因层面建立数学模型,为构建功能稳定的合成微生物菌群提供理论基础;(3)设计多物种合成微生物群落以强化菌群的功能与稳定性,重点在于开发满足多种微生物生存的工艺和培养基,例如空间连接的微生物群落(SLME)。随着系统生物学和合成生物学的快速发展,尤其是组学技术,在挖掘新的物种以及探究菌株之间协同机制方面取得巨大进步(图 1),同时对微生物进行基因编辑和基因精细调控的新工具正在出现[74-75],为未来设计稳定高效的多物种合成微生物群落奠定基础。
利用合成微生物群落干扰自然形成的生态系统可以调节其行为和功能,甚至创建出原本不存在的协同关系,从而开发新的应用。例如,在原厌氧发酵系统中外源性加入合成微生物群落,将是提高木质纤维素降解率和提升甲烷产率的有效方法,同时对新形成的整个体系进行系统生物学分析将有利于合成微生物群落的再设计和优化。最后,合成微生物群落不仅在联合生物加工领域应用广泛,而且在生物修复、生物制药等领域均有广阔的开发前景,这一策略很可能是利用木质纤维素生物质生产生物燃料和化学用品最具成本效益的途径。
| [1] |
Edrisi SA, Abhilash PC. Exploring marginal and degraded lands for biomass and bioenergy production:an indian scenario[J]. Renewable and Sustainable Energy Reviews, 2016, 54: 1537-1551. DOI:10.1016/j.rser.2015.10.050 |
| [2] |
Letcher TM.Introduction with a focus on atmospheric carbon dioxide and climate change[A]//Letcher TM.Future Energy[M].2nd ed.Amsterdam: Elsevier, 2014: 3-16
|
| [3] |
Mehmood MA, Ibrahim M, Rashid U, et al. Biomass production for bioenergy using marginal lands[J]. Sustainable Production and Consumption, 2017, 9: 3-21. DOI:10.1016/j.spc.2016.08.003 |
| [4] |
Bryngemark E. Second generation biofuels and the competition for forest raw materials:a partial equilibrium analysis of Sweden[J]. Forest Policy and Economics, 2019, 109: 102022. DOI:10.1016/j.forpol.2019.102022 |
| [5] |
Kucharska K, Słupek E, Cieśliński H, et al. Advantageous conditions of saccharification of lignocellulosic biomass for biofuels generation via fermentation processes[J]. Chemical Papers, 2020, 74(4): 1199-1209. DOI:10.1007/s11696-019-00960-1 |
| [6] |
Mahmood H, Moniruzzaman M, Iqbal T, et al. Recent advances in the pretreatment of lignocellulosic biomass for biofuels and value-added products[J]. Current Opinion in Green and Sustainable Chemistry, 2019, 20: 18-24. DOI:10.1016/j.cogsc.2019.08.001 |
| [7] |
Sharma HK, Xu CB, Qin WS. Biological pretreatment of lignocellulosic biomass for biofuels and bioproducts:an overview[J]. Waste and Biomass Valorization, 2019, 10(2): 235-251. DOI:10.1007/s12649-017-0059-y |
| [8] |
Wang YX, Liu Q, Yan L, et al. A novel lignin degradation bacterial consortium for efficient pulping[J]. Bioresource Technology, 2013, 139: 113-119. DOI:10.1016/j.biortech.2013.04.033 |
| [9] |
Wang HP, Lim TT, Duong C, et al. Long-term mesophilic anaerobic co-digestion of swine manure with corn stover and microbial community analysis[J]. Microorganisms, 2020, 8(2): 188. DOI:10.3390/microorganisms8020188 |
| [10] |
Olson DG, McBride JE, Shaw AJ, et al. Recent progress in consolidated bioprocessing[J]. Current Opinion in Biotechnology, 2012, 23(3): 396-405. DOI:10.1016/j.copbio.2011.11.026 |
| [11] |
Song H, Ding MZ, Jia XQ, et al. Synthetic microbial consortia:from systematic analysis to construction and applications[J]. Chemical Society Reviews, 2014, 43(20): 6954-6981. DOI:10.1039/C4CS00114A |
| [12] |
Zhang ZJ, Li SZ, Deng Y, et al. Recent progress in studies of synthetic microbial community and its applications in bioengineering[J]. Chinese Journal of Applied & Environmental Biology, 2015, 21(6): 981-986. (in Chinese) 张照婧, 厉舒祯, 邓晔, 等. 合成微生物群落及其生物处理应用研究新进展[J]. 应用与环境生物学报, 2015, 21(6): 981-986. |
| [13] |
Yan L, Gao YM, Wang YJ, et al. Diversity of a mesophilic lignocellulolytic microbial consortium which is useful for enhancement of biogas production[J]. Bioresource Technology, 2012, 111: 49-54. DOI:10.1016/j.biortech.2012.01.173 |
| [14] |
Wang HP, Li JW, Zhao YQ, et al. Establishing practical strategies to run high loading corn stover anaerobic digestion:methane production performance and microbial responses[J]. Bioresource Technology, 2020, 310: 123364. DOI:10.1016/j.biortech.2020.123364 |
| [15] |
Weiland P. Biogas production:current state and perspectives[J]. Applied Microbiology and Biotechnology, 2010, 85(4): 849-860. DOI:10.1007/s00253-009-2246-7 |
| [16] |
Zheng XM, Zheng P, Sun JB. Systems biology for industrial biotechnology[J]. Chinese Journal of Biotechnology, 2019, 35(10): 1955-1973. (in Chinese) 郑小梅, 郑平, 孙际宾. 面向工业生物技术的系统生物学[J]. 生物工程学报, 2019, 35(10): 1955-1973. |
| [17] |
Konopka A, Lindemann S, Fredrickson J. Dynamics in microbial communities:unraveling mechanisms to identify principles[J]. The ISME Journal, 2015, 9(7): 1488-1495. DOI:10.1038/ismej.2014.251 |
| [18] |
Borenstein E, Kupiec M, Feldman MW, et al. Large-scale reconstruction and phylogenetic analysis of metabolic environments[J]. Proceedings of the National Academy of Sciences of the United States of America, 2008, 105(38): 14482-14487. DOI:10.1073/pnas.0806162105 |
| [19] |
Li JB, Li CN, Kou YP, et al. Distinct mechanisms shape soil bacterial and fungal co-occurrence networks in a mountain ecosystem[J]. FEMS Microbiology Ecology, 2020, 96(4): fiaa030. DOI:10.1093/femsec/fiaa030 |
| [20] |
Klitgord N, Segrè D. Environments that induce synthetic microbial ecosystems[J]. PLoS Computational Biology, 2010, 6(11): e1001002. DOI:10.1371/journal.pcbi.1001002 |
| [21] |
Zhao YQ, Xu CF, Ai SQ, et al. Biological pretreatment enhances the activity of functional microorganisms and the ability of methanogenesis during anaerobic digestion[J]. Bioresource Technology, 2019, 290: 121660. DOI:10.1016/j.biortech.2019.121660 |
| [22] |
Freilich S, Zarecki R, Eilam O, et al. Competitive and cooperative metabolic interactions in bacterial communities[J]. Nature Communications, 2011, 2: 589. DOI:10.1038/ncomms1597 |
| [23] |
Minty JJ, Lin XN.Engineering synthetic microbial consortia for consolidated bioprocessing of ligonocellulosic biomass into valuable fuels and chemicals[A]//Himmel ME.Direct Microbial Conversion of Biomass to Advanced Biofuels[M].Amsterdam: Elsevier, 2015: 365-381
|
| [24] |
Shong J, Diaz MRJ, Collins CH. Towards synthetic microbial consortia for bioprocessing[J]. Current Opinion in Biotechnology, 2012, 23(5): 798-802. DOI:10.1016/j.copbio.2012.02.001 |
| [25] |
Tri CL, Kamei I. Butanol production from cellulosic material by anaerobic co-culture of white-rot fungus Phlebia and bacterium Clostridium in consolidated bioprocessing[J]. Bioresource Technology, 2020, 305: 123065. DOI:10.1016/j.biortech.2020.123065 |
| [26] |
Johns NI, Blazejewski T, Gomes ALC, et al. Principles for designing synthetic microbial communities[J]. Current Opinion in Microbiology, 2016, 31: 146-153. DOI:10.1016/j.mib.2016.03.010 |
| [27] |
Kong WT, Meldgin DR, Collins JJ, et al. Designing microbial consortia with defined social interactions[J]. Nature Chemical Biology, 2018, 14(8): 821-829. DOI:10.1038/s41589-018-0091-7 |
| [28] |
Li Y, Dong GJ, Fang X, et al. Dynamics of microbial community in process of cellulose decomposed by simplified bacterial community F1[J]. Environmental Science & Technology, 2016, 39(8): 35-39,76. (in Chinese) 李莹, 董桂军, 方旭, 等. 简化的纤维素分解复合菌系F1的菌株组成动态[J]. 环境科学与技术, 2016, 39(8): 35-39,76. |
| [29] |
Jiang LL, Zhou JJ, Quan CS, et al. Advances in industrial microbiome based on microbial consortium for biorefinery[J]. Bioresources and Bioprocessing, 2017, 4: 11. DOI:10.1186/s40643-017-0141-0 |
| [30] |
Shou WY, Ram S, Vilar JMG. Synthetic cooperation in engineered yeast populations[J]. Proceedings of the National Academy of Sciences of the United States of America, 2007, 104(6): 1877-1882. DOI:10.1073/pnas.0610575104 |
| [31] |
Bayer TS, Widmaier DM, Temme K, et al. Synthesis of methyl halides from biomass using engineered microbes[J]. Journal of the American Chemical Society, 2009, 131(18): 6508-6515. DOI:10.1021/ja809461u |
| [32] |
Minty JJ, Singer ME, Scholz SA, et al. Design and characterization of synthetic fungal-bacterial consortia for direct production of isobutanol from cellulosic biomass[J]. Proceedings of the National Academy of Sciences of the United States of America, 2013, 110(36): 14592-14597. DOI:10.1073/pnas.1218447110 |
| [33] |
Mukherjee S, Bassler BL. Bacterial quorum sensing in complex and dynamically changing environments[J]. Nature Reviews Microbiology, 2019, 17(6): 371-382. DOI:10.1038/s41579-019-0186-5 |
| [34] |
Papenfort K, Bassler BL. Quorum sensing signal-response systems in Gram-negative bacteria[J]. Nature Reviews Microbiology, 2016, 14(9): 576-588. DOI:10.1038/nrmicro.2016.89 |
| [35] |
Ge C, Sheng HK, Chen X, et al. Quorum sensing system used as a tool in metabolic engineering[J]. Biotechnology Journal, 2020, 15(6): 1900360. DOI:10.1002/biot.201900360 |
| [36] |
GroẞA, Rödel G, Ostermann K. Application of the yeast pheromone system for controlled cell-cell communication and signal amplification[J]. Letters in Applied Microbiology, 2011, 52(5): 521-526. DOI:10.1111/j.1472-765X.2011.03035.x |
| [37] |
Weber W, Daoud-El Baba M, Fussenegger M. Synthetic ecosystems based on airborne inter-and intrakingdom communication[J]. Proceedings of the National Academy of Sciences of the United States of America, 2007, 104(25): 10435-10440. DOI:10.1073/pnas.0701382104 |
| [38] |
Kylilis N, Tuza ZA, Stan GB, et al. Tools for engineering coordinated system behaviour in synthetic microbial consortia[J]. Nature Communications, 2018, 9: 2677. DOI:10.1038/s41467-018-05046-2 |
| [39] |
Alnahhas RN, Winkle JJ, Hirning AJ, et al. Spatiotemporal dynamics of synthetic microbial consortia in microfluidic devices[J]. ACS Synthetic Biology, 2019, 8(9): 2051-2058. DOI:10.1021/acssynbio.9b00146 |
| [40] |
Balagaddé FK, You L, Hansen CL, et al. Long-term monitoring of bacteria undergoing programmed population control in a microchemostat[J]. Science, 2005, 309(5731): 137-140. DOI:10.1126/science.1109173 |
| [41] |
D'Souza G, Shitut S, Preussger D, et al. Ecology and evolution of metabolic cross-feeding interactions in bacteria[J]. Natural Product Reports, 2018, 35(5): 455-488. DOI:10.1039/C8NP00009C |
| [42] |
Zhou J, Ma Q, Yi H, et al. Metabolome profiling reveals metabolic cooperation between Bacillus megaterium and Ketogulonicigenium vulgare during induced swarm motility[J]. Applied and Environmental Microbiology, 2011, 77(19): 7023-7030. DOI:10.1128/AEM.05123-11 |
| [43] |
Ma Q, Bi YH, Wang EX, et al. Integrated proteomic and metabolomic analysis of a reconstructed three-species microbial consortium for one-step fermentation of 2-keto-L-gulonic acid, the precursor of vitamin C[J]. Journal of Industrial Microbiology & Biotechnology, 2019, 46: 21-31. |
| [44] |
Chignell J, Reardon KF, Park S.Label-free, strain-resolved, shotgun proteomics of a defined bacterial co-culture[A]//Proceedings of 2013 AIChE Annual Meeting[C].San Francisco, CA: 2013
|
| [45] |
Yin J, Ma AZ, Song MY, et al. Research progress in synthetic microbial systems[J]. Microbiology China, 2020, 47(2): 583-593. (in Chinese) 尹珺, 马安周, 宋茂勇, 等. 合成微生物体系研究进展[J]. 微生物学通报, 2020, 47(2): 583-593. |
| [46] |
Klitgord N, Segrè D. Environments that induce synthetic microbial ecosystems[J]. PLoS Computational Biology, 2010, 6(11): e1001002. DOI:10.1371/journal.pcbi.1001002 |
| [47] |
Alkhnbashi OS, Meier T, Mitrofanov A, et al. CRISPR-Cas bioinformatics[J]. Methods, 2020, 172: 3-11. DOI:10.1016/j.ymeth.2019.07.013 |
| [48] |
Andreou AI, Nakayama N. Mobius assembly:a versatile golden-gate framework towards universal DNA assembly[J]. PLoS One, 2018, 13(1): e0189892. DOI:10.1371/journal.pone.0189892 |
| [49] |
Yasen A, Aini A, Wang H, et al. Progress and applications of single-cell sequencing techniques[J]. Infection, Genetics and Evolution, 2020, 80: 104198. DOI:10.1016/j.meegid.2020.104198 |
| [50] |
Dusny C, Grünberger A. Microfluidic single-cell analysis in biotechnology:from monitoring towards understanding[J]. Current Opinion in Biotechnology, 2020, 63: 26-33. DOI:10.1016/j.copbio.2019.11.001 |
| [51] |
Choi KR, Jang WD, Yang D, et al. Systems metabolic engineering strategies:integrating systems and synthetic biology with metabolic engineering[J]. Trends in Biotechnology, 2019, 37(8): 817-837. DOI:10.1016/j.tibtech.2019.01.003 |
| [52] |
McCarty NS, Ledesma-Amaro R. Synthetic biology tools to engineer microbial communities for biotechnology[J]. Trends in Biotechnology, 2019, 37(2): 181-197. DOI:10.1016/j.tibtech.2018.11.002 |
| [53] |
Chen Y, Li C, Zhou ZX, et al. Enhanced biodegradation of alkane hydrocarbons and crude oil by mixed strains and bacterial community analysis[J]. Applied Biochemistry and Biotechnology, 2014, 172(7): 3433-3447. DOI:10.1007/s12010-014-0777-6 |
| [54] |
Zhou K, Qiao KJ, Edgar S, et al. Distributing a metabolic pathway among a microbial consortium enhances production of natural products[J]. Nature Biotechnology, 2015, 33(4): 377-383. DOI:10.1038/nbt.3095 |
| [55] |
Zuroff TR, Xiques SB, Curtis WR. Consortia-mediated bioprocessing of cellulose to ethanol with a symbiotic Clostridium phytofermentans/yeast co-culture[J]. Biotechnology for Biofuels, 2013, 6: 59. DOI:10.1186/1754-6834-6-59 |
| [56] |
Park EY, Naruse K, Kato T. One-pot bioethanol production from cellulose by co-culture of Acremonium cellulolyticus and Saccharomyces cerevisiae[J]. Biotechnology for Biofuels, 2012, 5: 64. DOI:10.1186/1754-6834-5-64 |
| [57] |
Szambelan K, Nowak J, Czarnecki Z. Use of Zymomonas mobilis and Saccharomyces cerevisiae mixed with Kluyveromyces fragilis for improved ethanol production from Jerusalem artichoke tubers[J]. Biotechnology Letters, 2004, 26(10): 845-848. DOI:10.1023/B:BILE.0000025889.25364.4b |
| [58] |
Nakayama S, Kiyoshi K, Kadokura T, et al. Butanol production from crystalline cellulose by cocultured Clostridium thermocellum and Clostridium saccharoperbutylacetonicum N1-4[J]. Applied and Environmental Microbiology, 2011, 77(18): 6470-6475. DOI:10.1128/AEM.00706-11 |
| [59] |
Wang ZY, Cao GL, Zheng J, et al. Developing a mesophilic co-culture for direct conversion of cellulose to butanol in consolidated bioprocess[J]. Biotechnology for Biofuels, 2015, 8: 84. DOI:10.1186/s13068-015-0266-3 |
| [60] |
Wen ZQ, Wu MB, Lin YJ, et al. Artificial symbiosis for acetone-butanol-ethanol (ABE) fermentation from alkali extracted deshelled corn cobs by co-culture of Clostridium beijerinckii and Clostridium cellulovorans[J]. Microbial Cell Factories, 2014, 13: 92. DOI:10.1186/s12934-014-0092-5 |
| [61] |
Jiang YJ, Guo D, Lu JS, et al. Consolidated bioprocessing of butanol production from xylan by a thermophilic and butanologenic Thermoanaerobacterium sp.M5[J]. Biotechnology for Biofuel, 2018, 11: 89. DOI:10.1186/s13068-018-1092-1 |
| [62] |
Wen ZQ, Minton NP, Zhang Y, et al. Enhanced solvent production by metabolic engineering of a twin-clostridial consortium[J]. Metabolic Engineering, 2017, 39: 38-48. DOI:10.1016/j.ymben.2016.10.013 |
| [63] |
Brethauer S, Studer MH. Consolidated bioprocessing of lignocellulose by a microbial consortium[J]. Energy & Environmental Science, 2014, 7(4): 1446-1453. |
| [64] |
Wang WD, Cui ZJ, Yang HY, et al. Stability of a composite microbial system WSC-6 with efficient cellulose degrading[J]. China Environmental Science, 2005, 25(5): 567-571. (in Chinese) 王伟东, 崔宗均, 杨洪岩, 等. 高效稳定纤维素分解菌复合系WSC-6的稳定性[J]. 中国环境科学, 2005, 25(5): 567-571. |
| [65] |
Wen X, Fu BR, Wang YJ, et al. Isolation of anaerobic bacterial strains from cellulolytic bacterial community WSC-9[J]. Journal of Northeast Agricultural University, 2013, 44(2): 47-52. (in Chinese) 温雪, 付博锐, 王彦杰, 等. 纤维素分解复合菌系WSC-9中厌氧细菌的分离[J]. 东北农业大学学报, 2013, 44(2): 47-52. |
| [66] |
Ai SQ, Zhao YQ, Sun ZY, et al. Change of bacterial community structure during cellulose degradation by the microbial consortium[J]. Chinese Journal of Biotechnology, 2018, 34(11): 1794-1808. (in Chinese) 艾士奇, 赵一全, 孙志远, 等. 复合菌系降解纤维素过程中微生物群落结构的变化[J]. 生物工程学报, 2018, 34(11): 1794-1808. |
| [67] |
Li ZG, Wen X, Zhang Y, et al. Construction of cellulose decomposing complex microbial system and effects of different component on cellulose degradation[J]. Journal of Heilongjiang Bayi Agricultural University, 2015, 27(1): 78-84. (in Chinese) 李振国, 温雪, 张杨, 等. 人工组合纤维素分解复合菌系的构建及各组分对纤维素分解的影响[J]. 黑龙江八一农垦大学学报, 2015, 27(1): 78-84. |
| [68] |
Shahab RL, Luterbacher JS, Brethauer S, et al. Consolidated bioprocessing of lignocellulosic biomass to lactic acid by a synthetic fungal-bacterial consortium[J]. Biotechnology and Bioengineering, 2018, 115(5): 1207-1215. DOI:10.1002/bit.26541 |
| [69] |
Fu N, Peiris P, Markham J, et al. A novel co-culture process with Zymomonas mobilis and Pichia stipitis for efficient ethanol production on glucose/xylose mixtures[J]. Enzyme and Microbial Technology, 2009, 45(3): 210-217. DOI:10.1016/j.enzmictec.2009.04.006 |
| [70] |
Ben Said S, Tecon R, Borer B, et al. The engineering of spatially linked microbial consortia-potential and perspectives[J]. Current Opinion in Biotechnology, 2020, 62: 137-145. DOI:10.1016/j.copbio.2019.09.015 |
| [71] |
Ben Said S, Or D. Synthetic microbial ecology:engineering habitats for modular consortia[J]. Frontiers in Microbiology, 2017, 8: 1125. DOI:10.3389/fmicb.2017.01125 |
| [72] |
Kalscheuer R, Stölting T, Steinbüchel A. Microdiesel:Escherichia coli engineered for fuel production[J]. Microbiology, 2006, 152(9): 2529-2536. DOI:10.1099/mic.0.29028-0 |
| [73] |
Lin H, Wang Q, Shen Q, et al. Genetic engineering of microorganisms for biodiesel production[J]. Bioengineered, 2013, 4(5): 292-304. DOI:10.4161/bioe.23114 |
| [74] |
Zheng XM, Zheng P, Zhang K, et al. 5S rRNA promoter for guide RNA expression enabled highly efficient CRISPR/Cas9 genome editing in Aspergillus niger[J]. ACS Synthetic Biology, 2019, 8(7): 1568-1574. DOI:10.1021/acssynbio.7b00456 |
| [75] |
Sun DH, Chen JZ, Wang Y, et al. Metabolic engineering of Corynebacterium glutamicum by synthetic small regulatory RNAs[J]. Journal of Industrial Microbiology & Biotechnology, 2019, 46(2): 203-208. |
 2020, Vol. 47
2020, Vol. 47