扩展功能
文章信息
- 吴慧君, 宋权威, 郑瑾, 于文赫, 张坤峰, 林双君, 梁如冰
- WU Hui-Jun, SONG Quan-Wei, ZHENG Jin, YU Wen-He, ZHANG Kun-Feng, LIN Shuang-Jun, LIANG Ru-Bing
- 微生物降解石油烃的功能基因研究进展
- Function genes in microorganisms capable of degrading petroleum hydrocarbon
- 微生物学通报, 2020, 47(10): 3355-3368
- Microbiology China, 2020, 47(10): 3355-3368
- DOI: 10.13344/j.microbiol.china.200402
-
文章历史
- 收稿日期: 2020-04-22
- 接受日期: 2020-08-12
- 网络首发日期: 2020-08-20
2. 上海交通大学生命科学技术学院 微生物代谢国家重点实验室 上海 200240
2. State Key Laboratory of Microbial Metabolism, School of Life Sciences and Biotechnology, Shanghai Jiao Tong University, Shanghai 200240, China
随着工业与社会的快速发展,石油使用量与开采量剧增,导致环境的石油污染问题日益严重。石油成分复杂,其中富含的苯系物和多环芳烃物质结构稳定、不易挥发、难于降解、环境持留时间长、三致效应高[1]。如何有效防治石油烃的污染已成为科学研究与社会发展所重点关注的问题。环境中的石油烃可经历风化作用逐步分解,包括物理作用(扩散)、生理生化作用(蒸发,溶解,吸附,解析)、化学作用(光氧化,自氧化)及生物作用(植物和微生物的代谢)[2]。其中,微生物降解可将石油烃最终完全转化为无毒无害的CO2和H2O,是石油烃自然衰减的主要作用。微生物降解石油烃是微生物利用石油烃作为其自身代谢的碳源和能源,进行转化后供其生长。影响微生物代谢石油烃的主要因素包括微生物自身特性(跨膜运输系统和产表面活性剂等)[3]、污染物的浓度和生化特性[4]以及电子受体(氧气、硝酸盐和Fe3+等)[5]。环境条件如盐度、湿度、营养元素和土壤化学等会影响石油烃的性质或微生物的生长,也成为微生物降解石油烃的制约因素。石油污染发生后,大量微生物无法存活,土壤中微生物多样性和生物量显著降低,仅石油烃耐受菌和石油烃降解菌可存活下来并利用石油烃作为能量来源大量繁殖,其比例可由通常的1%提高到10%,在群落中占据主导地位[6-9];随着时间的延长,石油烃整体毒性降低,微生物群落的数量和多样性又会增加[10]。
石油烃结构的复杂性决定了微生物降解菌的多样性。目前已发现数百种微生物,包括细菌、真菌和酵母菌可降解一种或多种石油烃类物质[11]。在石油烃物质存在时,微生物会表达特定功能基因,产生特异酶来进行石油烃代谢;功能基因的表达是微生物降解石油烃的关键。近年来,多组学技术包括基因组、转录组和蛋白质组等新兴技术快速发展,为微生物降解石油烃功能基因的快速测序和鉴定提供了有力的技术支撑。采用与已知功能基因进行序列比对、基因转录水平分析、功能酶表达水平等研究方法,可有效分析污染环境中降解微生物的功能基因多样性和丰度。Laczi等[12]通过分析Rhodococcus erythropolis PR4的基因序列,鉴定了多个氧化酶基因,并利用转录组学技术分析和鉴定了降解柴油和其他石油烃的关键基因。红球菌是研究最多的石油烃降解微生物之一,近年来越来越多的研究利用多组学技术对该菌属的降解潜力展开研究[13]。功能基因不仅是研究石油降解微生物的群落结构特征、固有降解菌和降解潜力的基础,也是构建基因工程菌进行生物修复的依据。如将来源于Acidovorax sp. CHX100的环己烷降解功能基因:细胞色素P450、铁氧还蛋白还原酶和铁氧还蛋白,在菌株Pseudomonas taiwanensis VLB120中进行过量表达,可极大地提高菌株对环己烷的氧化降解效能[14]。
石油烃污染源可分为石油开采的原油、炼化厂的减压渣油和加油站的成品油。尽管不同地区原油组成和性质各不相同,但是烷烃和芳香烃的比例高达80%[15]。其中链烷烃在原油中的比例约为50%-70%[16],芳香烃约为10%-30%;仅有极少数原油中链烷烃低于10%-15%[16]。原油在炼化厂经过提炼后产生减压渣油,其主要组分为烷烃和芳香烃,其余组分为难降解的胶质和沥青质。与原油相比,减压渣油中烷烃比例减少,芳香烃比例增加。我国部分地区的原油和减压渣油中烷烃和芳香烃比例见表 1[17-18]。成品油组分则几乎全部为烷烃。由此可见,烷烃和芳香烃是石油烃污染微生物降解与修复的主要目标污染物。微生物主要通过羟化作用、脱氢作用、过氧化作用等方式降解石油烃,包括好氧降解和厌氧降解。好氧降解是好氧微生物或兼性厌氧微生物以石油烃作为底物,分子氧作为最终电子受体,将石油烃转化为H2O、CO2和NH3的过程[11]。厌氧降解则是厌氧微生物或兼性好氧微生物在无分子氧的条件下,以硝酸盐、硫酸盐、Mn4+、Fe3+、二氧化碳作为最终电子受体[19-20],将石油烃有机物转化为CH4等。好氧降解比厌氧降解的速率更快,所需时间更短[21]。本文系统梳理了微生物降解烷烃和芳香烃的好氧和厌氧降解途径,总结了有氧和无氧条件下微生物代谢烷烃和芳香烃的功能基因,为降解石油烃的微生物资源挖掘和基因工程菌的构建提供参考(表 2)。
| 油品 Oil |
地点 Site |
烷烃 Alkane |
芳香烃 Aromatic hydrocarbon |
| 原油 Crude oil |
四川马岭 Maling, Sichuan |
64.34 | 15.58 |
| 四川华庆 Huaqing, Sichuan |
54.29 | 10.72 | |
| 减压渣油 Vacuum residuum |
黑龙江大庆 Daqing, Heilongjiang |
40.80 | 32.20 |
| 新疆白克 Baike, Xinjiang |
47.30 | 25.20 | |
| 新疆九区 Nine districts in Xinjiang |
28.20 | 26.90 | |
| 辽宁欢喜岭 Huanxiling, Liaoning |
28.70 | 35.00 | |
| 天津大港 Dagang, Tianjin |
30.60 | 31.60 | |
| 中原 Zhongyuan |
23.60 | 31.60 |
| 降解基因 Degrading genes |
降解底物 Substrates |
参考菌株(基因序列号) Reference strains (GenBank accession number) |
菌株来源 Sources |
参考文献 References |
| alkB | C5-C12 alkane | Pseudomonas putida GPo1 (CAB54064) | Machine shop cutting oil | [22] |
| almA | C10-C40 alkane | Acinetobacter sp. DSM 17874 (ABQ18224) | Unsterilized oil | [23] |
| ladA | C15-C36 alkane | Geobacillus thermodenitrificans NG80-2 (ABO68832) | Deep oil reservoir | [24] |
| alkM | C12-C36 alkane | Acinetobacter sp. ADP1 (CAA05333) | Soil | [25] |
| Methane monooxygenase, MMOs | C1-C8 alkane, alkene, halogenated hydrocarbon, cycloalkane | Methylocapsa spp. | Forest soil, acid sphagnum peat bog, etc | [26] |
| Cytochrome P450 family, CYP153 | Alkane, cycloalkane, aromatic hydrocarbon | Dietzia sp. DQ12-45-1b (AFY63004) | Oil production water of a deep subterranean oil reservoir | [27] |
| assA | C13-C18 alkane, pentadecene, hexadecene | Desulfatibacillum alkenivorans AK-01 (ABH11461) | Oil-contaminated site | [28] |
| masD | C6-C8 alkane, alkylbenzene | Aromatoleum sp. HxN1 (CAO03074) | Sediment samples from ditches | [29] |
| akbD | Benzene, toluene, ortho- xylene, ethylbenzene, phenol | Rhodococcus sp. DK17 (AAR90134) | Oil-contaminated site | [30] |
| xylM | Toluene | P. putida mt-2 (CAC86827) | Field soil | [31] |
| todE | Toluene | P. putida F1 (AAA26010) | Soil | [32-33] |
| tmoA | Toluene | Pseudomonas mendocina KR1 (AAA25999) | Algal-bacterial mat | [34] |
| tbuC | Toluene | Ralstonia pickettii PKO1 (AAB09623) | Benzene-, toluene-, ethylbenzene- and xylene(s)- contaminated aquifer | [35] |
| dmpL | Phenol | P. putida CF600 (AAA25940) | Unknown | [36-37] |
| bnzA1 | Benzene | Rhodococcus opacus B4 (BAD95523) | Gasoline-contaminated soil | [38] |
| nahAc | Naphthalene | P. putida G7 (BAE92156) | Soil | [39-40] |
| narAa | Naphthalene | Rhodococcus sp. NCIMB12038 (AAD28100) | Garden soil with carbaryl | [41-42] |
| ndoB | Naphthalene | P. putida NCIB9816 (P0A110) | Garden soil | [43] |
| doxB | Naphthalene | Pseudomonas sp. C18 (P0A111) | Soil | [44] |
| nagAc | Naphthalene | Ralstonia sp. U2 (AAD12610) | Oil-contaminated soil | [45-46] |
| pahC | Naphthalene, phenanthrene | P. putida OUS82 (BAA20395) | PAH-contaminated soil | [47] |
| phnAc | Naphthalene, phenanthrene, anthracene | Burkholderia sp. RP007 (AAD09872) | PAH-contaminated site | [48] |
| phdA | Naphthalene, phenanthrene | Nocardioides sp. KP7 (BAA84712) | Ocean | [49] |
| nidA | Toluene, meta-xylene, naphthalene, benzo[a]anthracene, benzo[a]pyrene | Mycobacterium vanbaalenii PYR-1 (AAT51751) | Oil-contaminated sediment | [50-51] |
| C23O | Catechol | P. putida ND6 (AAP44220) | Industrial wastewater | [52] |
| catA/C12O | Catechol | P. putida ND6 (AAP44248) | Industrial wastewater | [52] |
| benA | Benzoate | Halomonas organivorans (CBR26855) | Saline environments | [53] |
| bssA | Benzene, toluene, ethylbenzene, xylene | Thauera aromatica K172 (CAA05052) | Activated sludge | [54] |
| nmsA | Naphthalene | Deltaproteobacteria sp. NaphS2 (CAO72219) | Marine sediment | [55] |
| bclA | Benzoyl-CoA | Magnetospirillum sp. LM-5 (CAA7611436) | Activated sludge | [56] |
| badA | Benzoyl-CoA | Rhodopseudomonas palustris (CAJ18317) | Waste water | [56] |
| bzdA | Benzoyl-CoA | Azoarcus sp. CIB (AAQ08820) | Diesel fuel-contaminated aquifer | [56] |
| bamY | Benzoyl-CoA | Geobacter metallireducen GS-15 (ABB32372) | Freshwater sediment | [56] |
直链烷烃的降解方式主要有末端氧化、次末端氧化、β-氧化和ω-氧化[11, 57-58]。在微生物好氧降解烷烃体系中,加氧酶的羟化作用是主要限速步骤。短链烷烃的单加氧酶主要为甲烷单加氧酶(methane monooxygenase,MMOs),MMOs分为可溶性甲烷单加氧酶(sMMO)和颗粒性甲烷单加氧酶(pMMO),sMMO能氧化C1-C8烷烃、卤代烃、烯烃、环烷烃和芳香烃,pMMO能氧化C1-C5烷烃、卤代烃和烯烃[26, 59]。AlkB烷烃单加氧酶家族主要负责催化短链与中等链长烷烃的氧化[22],该家族广泛存在于变形菌和放线菌中,是研究得最透彻的烷烃单加氧酶。P. putida GPo1菌株中AlkB单加氧酶可氧化丙烷、丁烷和C5-C12短链烷烃;alkB基因位于OCT质粒上的alkBFGHJKL基因簇,这一基因簇表达的一系列酶将烷烃转化为乙酰辅酶A[22];alkST是这一基因簇的转录调控因子[59]。在P. putida GPo1菌株AlkB的同源蛋白中,很多能氧化链长超过C10的烷烃。细胞色素P450s单加氧酶(cytochrome P450,CYPs)是一类硫醇盐蛋白超家族,可催化很多化合物的氧化反应。基于氨基酸序列相似性,CYPs超家族可细分为不同的家族,其中氧化C6-C10烷烃的单加氧酶属于细菌CYP153家族[60]。CYP153家族的底物除了烷烃,还包括柠檬烯、环己烯、苯乙烯和辛烯等[61-62]。
碳链超过C18的烷烃以固体形式存在,通常将固体石蜡作为固体长链烷烃降解的研究对象。固体石蜡是烃类混合物,碳链长为C20-C40[63]。Lazar等[64]于1999年首次提出利用微生物降解固体长链烷烃中的石蜡组分。但是能以固体石蜡作为降解底物的微生物种类不多,目前发现的菌属有Pseudomonas、Bacillus、Acinetobacter、Gordonia、Geobacillus和Rhodococcus等[63]。微生物与固体烷烃的接触机制有两种:一种是与石蜡直接接触,比如Rhodococcus erythropolis能在石蜡表面形成厚厚的生物被膜,同时伴随着细胞电势降低和细胞膜脂肪酸组分变化[63];另一种是产生表面活性剂使石蜡乳化[65]。固体长链烷烃的降解酶有烷烃羟化酶和细胞色素P450s,这两种酶的降解方式都为末端氧化。Acinetobacter baylyi ADP1能降解C12-C36烷烃,其烷烃羟化酶为AlkM[25],是AlkB的同源蛋白。Acinetobacter sp. M-1中有2个alkM的同源基因,命名为alkMa和alkMb,催化C20-C44烷烃降解[66]。细胞色素P450s家族中CYP116B5酶使Acinetobacter radioresistens菌株能够以C14-C36为唯一碳源生长[67]。此外,降解固体长链烷烃还有2个代表性基因,分别为almA和ladA。Acinetobacter sp. DSM 17874菌株产生的almA是首个克隆出来的能够特异性降解碳链超过C30的基因[23],随后在其他菌属中都发现了almA的同源基因,但是该基因的降解机制尚不明晰。嗜热酶LadA是通过基因组学和蛋白质组学技术在Geobacillus thermodenitrificans NG80-2菌株中被发现,能够对C15-C36烷烃进行末端氧化[24]。
环烃类化合物通常很难降解,以环己烷为例,只有少数微生物能以环己烷为唯一碳源进行生长[68]。环己烷降解需要两种氧化酶的作用,一种氧化酶将环己烷氧化为环己醇,脱氢形成环己酮;另一种氧化酶则氧化环己酮为己内酯后开环继续降解,最终生成CO2和H2O[11]。研究表明,环己烷单加氧酶为细胞色素P450蛋白家族,如Bacillus megaterium菌株利用细胞色素P450BM3 (CYP102A1)氧化环己烷,菌株Acidovorax sp. CHX100利用细胞色素P450单加氧酶(CYP450chx)将环己烷氧化为环己醇[68-69]。
1.2 芳香烃的好氧微生物降解芳香烃因含有苯环结构,相比于烷烃较难降解。在芳香烃的好氧降解过程中,芳环结构在加氧酶的作用下生成中心中间产物,主要包括邻苯二酚、原儿茶酸、龙胆酸盐和尿黑酸等,而后开环进一步降解,产物最终进入三羧酸循环[2, 11, 70-71]。
图 1以甲苯降解为例来阐述苯系物(苯、甲苯、乙苯、二甲苯)好氧降解途径中所需的酶。P. putida mt-2菌株是研究甲苯降解的模式菌株,其质粒pWW0上有2个xyl操纵子,分别编码甲苯降解上游途径和下游途径的酶。上游途径是将甲苯转化为苯甲酸,由操纵子xylUWCMABN完成;下游途径是将苯甲酸转化为三羧酸循环的中间产物,由操纵子xylXYZLTEGFJQKIH完成[72-73]。单加氧酶XylMA催化甲苯降解的初始反应,将甲基基团羟基化[35]。XylE为邻苯二酚2, 3-双加氧酶(C23O),可将邻苯二酚间位开环;该酶在不动杆菌属、考克氏菌属和假单胞菌属中均有发现[74]。XylR和XylS则是转录正调控因子,分别激活上游途径和下游途径[35]。值得关注的是,Kim等从土壤中分离出的Rhodococcus sp. DK17菌株具有两条不同的芳烃开环途径,能降解多种单环芳烃,如苯、甲苯、乙苯、邻二甲苯、苯酚等,其降解基因簇是akbBCDEF,akbS和akbT作为双组分调控系统调控这一基因簇的表达[30, 75-76]。
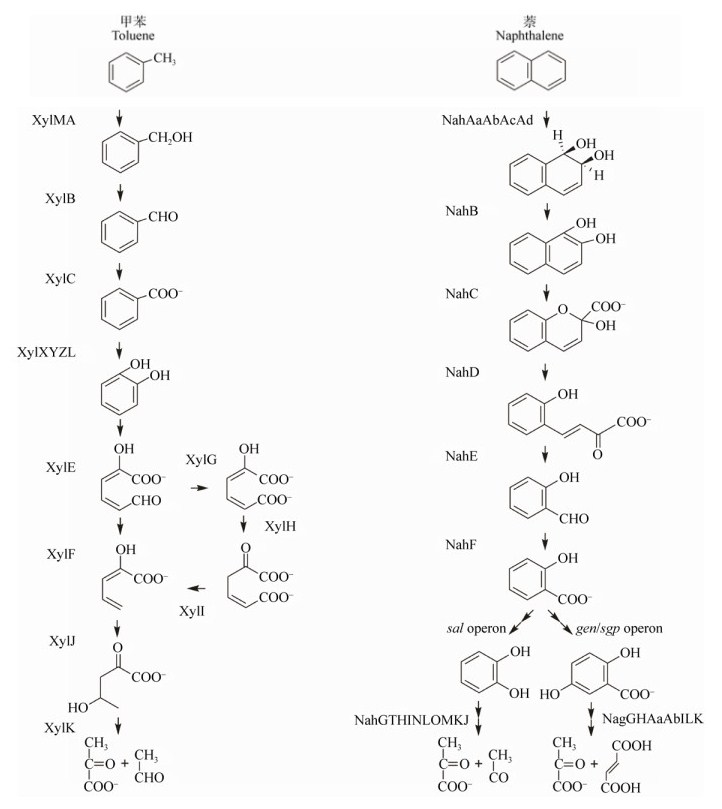
|
| 图 1 甲苯和萘的好氧降解途径 Figure 1 Aerobic degradation pathways of methylbenzene and naphthalene |
|
|
作为最简单的多环芳烃,萘已经成为研究芳香烃代谢的模式物质。研究显示,萘降解途径也分为上游途径和下游途径。上游途径是将萘转化为水杨酸的过程,由操纵子nahAaAbAcAdBFCED完成,其中NahAaAbAcAd为双加氧酶,将萘转化为顺- 1, 2-二氢-1, 2-萘二醇。下游途径中,操纵子sal或gen/sgp分别将水杨酸转化为邻苯二酚或龙胆酸[71]。邻苯二酚由操纵子nahGTHINLOMKJ通过间位开环途径最终转化为丙酮酸和乙醛;龙胆酸由操纵子nagGHAaAbILK转化为丙酮酸和富马酸[47, 77-78]。萘的降解途径在不同菌株中呈现多样性,可能是降解基因发生基因水平转移所致[47, 71]。以部分假单胞菌中nah样基因为例,P. putida NCIB9816菌株ndo基因与nah基因功能相同[40, 79];Pseudomonas sp. C18菌株dox基因的功能是氧化二苯并噻吩[44];P. putida OUS82和P. aeruginosa PaK1菌株中pah基因参与多环芳烃的降解[80-82],上述菌株都可利用萘作为底物生长。尽管不同菌株的萘代谢途径各有不同,但是萘降解调控基因在不同菌株中高度保守,可作为功能基因标签[83]。萘双加氧酶基因nahAc是表征萘好氧降解的基因标签;同样地,其他芳香烃的双加氧酶基因也能作为功能基因标签[84],如吡喃双加氧酶基因nidA[50-51, 85]、邻苯二酚2, 3-双加氧酶基因C23O[86-88]和邻苯二酚1, 2-双加氧酶基因catA (或C12O基因)[89]。此外,还有一些在细菌中普遍存在的芳环羟基化双加氧酶基因可作为功能基因标签的参考,如nag、nar、phn和bph[90-91],以及苯酚羟基化单加氧酶基因phe[92]。
2 微生物厌氧降解石油烃的关键酶与功能基因 2.1 烷烃的厌氧微生物降解相较于好氧降解研究,厌氧降解的酶系统研究较少。延胡索酸加成是厌氧降解的主要途径之一,由甘氨酰基自由基酶催化,如烷基琥珀酸合酶(alkylsuccinate synthase,ASS)或甲烷基琥珀酸合酶(methylalkylsuccinate synthase,MAS)[58]。硫酸盐还原菌Desulfatibacillum alkenivorans AK-01和脱氮菌Aromatoleum HxN1分别是研究ASS和MAS的模式菌株[28-29]。硫酸盐还原菌D. alkenivorans AK-01能在C13–C18烷烃、十五碳烯和十六碳烯中生长,其基因组中包含2个不同的ass操纵子,参与底物激活、辅酶A连接、碳链重排和脱羧反应等烷烃代谢过程[29, 93-94]。ASS/MAS酶通常参与短链和中链烷烃的延胡索酸加成反应[95-96],有研究显示ASS也能介导固态石蜡的延胡索酸加成:Wawrik等[97]通过基因组学分析结合assA转录分析,发现海洋沉积物中的微生物能将固体石蜡C25–C50转化为甲烷。assA和masD基因分别是ASS酶和MAS酶的主要编码基因,已成为鉴定厌氧烷烃降解微生物的基因标签[29, 98]。
2.2 芳香烃的厌氧微生物降解芳香烃类厌氧降解是在合酶、羧酶和脱氢酶的作用下生成中心中间产物苯甲酰辅酶A及其衍生物(如间苯二酚、间苯三酚、羟氢醌),而后在还原酶作用下进一步降解为乙酰辅酶A[70, 99-100]。通常认为,无取代基芳香烃降解途径的初始反应由羧化作用激活,甲基化芳香烃降解途径的初始反应则由延胡索酸加成反应激活[100] (图 2)。

|
| 图 2 烷烃、苯、甲苯、萘和1-甲基萘的厌氧降解途径 Figure 2 Anaerobic degradation pathways of alkane, benzene, toluene, naphthalene and 1-methylnaphthalene |
|
|
相较于甲基化芳香烃,无取代基芳香烃的降解研究较少。Meckenstock等总结了无取代基芳香烃降解微生物、初始羧化反应及可能参与降解的酶[101]。Zhang等通过稳定同位素标记分析出严格厌氧条件下萘降解的主要代谢产物萘甲酸,证实了萘降解途径的第一步反应是羧化作用[102]。因打开苯环中的C–H键是一个高耗能的过程,因此,苯的直接羧化作用研究在基因组学研究出现后才逐渐清晰。Abu Laban等[103]通过聚丙烯酰胺凝胶电泳发现可能参与苯羧化作用的厌氧苯羧化酶AbcA,Luo等[104]通过转录组学分析也发现了类似基因。此外,苯厌氧降解的初始反应也能通过羟化作用实现。Zhang等[105]发现Geobacter metallireducens在厌氧条件下将苯转化为苯酚后再进行下一步降解。氨基酸序列比对发现催化萘和苯的羧化酶属于UbiD酶家族[100]。尽管参与菲厌氧降解的酶尚属未知,但由于菲的主要代谢产物为菲甲酸,因而菲厌氧降解途径的初始反应被认为也是羧化作用[102, 106]。目前,关于无取代基芳香烃羧化作用的证据几乎都是来自代谢产物分析和基因分析,具体酶催化的分子机制尚不明晰。
与烷烃厌氧降解的延胡索酸加成反应相似,甲基苯和甲基萘的延胡索酸加成途径需要琥珀酸苄基合酶(benzylsuccinate synthase,BSS)和萘甲基琥珀酸合酶(naphthylmethylsuccinate synthase,NMS)的参与[107] (图 2)。BSS和NMS的α亚基编码基因分别为bssA和nmsA[108],已被用作基因标签来鉴定芳香烃厌氧降解微生物的存在[107, 109]。苯系物经由BssA参与的延胡索酸加成反应,转化为核心中间产物苯甲酰CoA,而后再由两类酶催化苯甲酰CoA实现初始去芳化[20, 110]。一类酶为依赖于ATP的苯甲酰CoA还原酶BcrCBAD/BzdNOPQ/ BadDEFG,分布于Thauera aromatica、Azoarcus spp.、Aromatoleum spp.和Rhodopseudomonas palustris等兼性厌氧菌中;另一类为不依赖ATP的苯甲酰CoA还原酶BamBCDEFGHI,分布于严格厌氧菌中,如Geobacter metallireducen[107]。去芳化后的产物由水解酶BamA/BzdY/Oah逐步氧化,最终生成乙酰CoA和CO2[56, 107]。与此类似,甲基萘经由NmsA参与的延胡索酸加成反应,生成中心中间产物萘甲酰CoA,而后由萘甲酰CoA还原酶Ncr催化其初始的去芳化(图 2)。尽管萘甲酰CoA还原酶(Ncr)在一些硫酸盐还原菌中保守存在[111],但仍需要提供其他类型的降解条件(如铁还原)或更多降解物(如菲)来共同确定ncr是否能作为基因标签[100]。
3 结论与展望自然界中的微生物参与营养物质的循环和有机物的降解,与动植物产生互利共生关系,在维持生态系统平衡中发挥了重要作用。石油烃污染区域的微生物群落特征通常与未污染区域不同,通过深入研究污染物类型、环境条件和微生物群落等的各自特征与相互作用,有利于快速精准地实施高效的污染物微生物修复。随着高通量测序技术的快速发展,该技术在微生物降解与环境修复领域中得到了广泛应用,已成为石油降解微生物研究的重要手段。综合利用多种组学技术,挖掘与鉴定核心功能基因、解析关键代谢途径已成为研究降解基因多样性、自然衰减潜力与改造和构建基因工程菌等研究的重要基础。然而,由于功能基因的发现是建立在酶学研究的基础上,对石油烃的微生物代谢途径与关键限速酶的解析与鉴定是发现新功能基因的主要限制因素。尽管一些石油烃降解功能基因被发现,但由于石油烃物质的多样性与复杂性,仍然难以满足科学研究与实际应用的需要。如由于复杂多环芳烃的微生物代谢途径尚不清楚,其关键酶的鉴定仍很匮乏,功能基因的研究鲜有报道,在一定程度上限制了此类污染物的微生物降解研究与环境修复进程。因此,后续研究应注重相关降解限速酶的功能研究与新代谢途径的明晰,进一步丰富和完善功能基因的数据库。
当利用功能基因研究微生物群落结构和功能时,在微生物降解底物的多样性和功能基因的特异性扩增方面需要关注。一种石油烃污染物通常能被多种微生物降解,一种微生物也可以同时降解多种石油烃污染物。Rojo[22]将石油烃降解微生物分为两类:一类为专一降解微生物,另一类为广谱降解微生物,后者能够同时催化降解饱和脂肪烃和芳香烃。之前的研究大多集中于单个降解微生物的分离鉴定和对单一石油烃类(如低分子量的PAHs和烷烃)的降解特征研究,对广谱性降解微生物的分类研究很少。随着高通量测序技术的快速发展,基因组与转录组相结合的功能基因鉴定方法已成为研究广谱石油烃降解菌的重要技术手段。Whyte等[112]发现Pseudomonas sp. BI7含有烷烃和芳香烃降解基因,能同时降解烷烃和芳香烃,首次证明了广谱降解微生物的存在。此后,关于广谱降解微生物的研究报道逐渐增多,包括分枝杆菌属(Mycobacterium)、红球菌属(Rhodococcus)、假单胞菌属(Pseudomonas)、变形杆菌属(Firmicutes)等微生物,其中分枝杆菌属和红球菌属在混合石油烃降解中最常报道。Mycobacterium vanbaalenii PYR-1菌株中单加氧酶NidAB和双加氧酶NidA3B3参与多种底物的代谢,不仅包括甲苯、间二甲苯、萘、苯并[a]蒽和苯并[a]芘,还包括咔唑和二苯并噻吩这些非烃类物质[113]。菌株P. aeruginosa SJTD-1利用2个AlkB单加氧酶、2个P450单加氧酶和1个AlmA单加氧酶降解C12–C24烷烃[114-115]。利用多种底物进行生长是微生物进化的趋势,因此,在基于功能基因研究微生物降解底物时应重点考虑广谱降解菌的存在与特征[116]。
针对功能基因的特异性扩增和基因定量,在石油烃降解微生物的研究中有着重要的应用价值和科研意义。由于不同生物分类的功能基因其序列差异较大,因此往往需要设计简并引物对环境样品的功能基因进行扩增。Kloos等[117]根据烷烃降解功能基因alkB的全长序列,与GenBank中已知的烷烃降解微生物的序列进行比对,设计了能够检测大多数alkB基因的简并引物,并对土壤样品DNA进行扩增和测序。Lueders等[118]利用延胡索酸加成功能基因(bssA、assA、bamA、bcrA等)的简并引物,通过PCR扩增、末端限制性片段长度多态性(T-RFLP)分析和扩增子测序,建立了一套直接快速检测石油烃厌氧降解固有菌群和降解潜力的方法。Shahsavari等[84]则利用简并引物,通过实时荧光定量PCR (qPCR)对石油烃污染土壤中降解多环芳烃的功能基因进行定量。需要注意的是,由于石油污染物成分复杂而降解基因多样性高,直接借鉴已有的简并引物可能会导致微生物群落功能研究有所疏漏。因此,可通过设计代谢通路中多个关键基因的简并引物,实现全面研究不同环境样品的功能基因多样性。
| [1] |
O'Brien PL, Desutter TM, Casey FXM, et al. Evaluation of soil function following remediation of petroleum hydrocarbons-A review of current remediation techniques[J]. Current Pollution Reports, 2017, 3(3): 192-205. DOI:10.1007/s40726-017-0063-7 |
| [2] |
Truskewycz A, Gundry TD, Khudur LS, et al. Petroleum hydrocarbon contamination in terrestrial ecosystems-fate and microbial responses[J]. Molecules, 2019, 24(18): 3400. DOI:10.3390/molecules24183400 |
| [3] |
Hua F, Wang HQ. Uptake and trans-membrane transport of petroleum hydrocarbons by microorganisms[J]. Biotechnology & Biotechnological Equipment, 2014, 28(2): 165-175. |
| [4] |
Johnsen AR, Wick LY, Harms H. Principles of microbial PAH-degradation in soil[J]. Environmental Pollution, 2005, 133(1): 71-84. |
| [5] |
Varjani SJ, Upasani VN. A new look on factors affecting microbial degradation of petroleum hydrocarbon pollutants[J]. International Biodeterioration & Biodegradation, 2017, 120: 71-83. |
| [6] |
Chikere CB, Okpokwasili GC, Chikere BO. Monitoring of microbial hydrocarbon remediation in the soil[J]. 3 Biotech, 2011, 1(3): 117-138. DOI:10.1007/s13205-011-0014-8 |
| [7] |
Tang JC, Lü HH, Liu QL, et al. Recent review on the microbial molecular ecology during contamination and remediation of petroleum hydrocarbons[J]. Microbiology China, 2015, 42(5): 944-955. (in Chinese) 唐景春, 吕宏虹, 刘庆龙, 等. 石油烃污染及修复过程中的微生物分子生态学研究进展[J]. 微生物学通报, 2015, 42(5): 944-955. |
| [8] |
Khudur LS, Shahsavari E, Miranda AF, et al. Evaluating the efficacy of bioremediating a diesel-contaminated soil using ecotoxicological and bacterial community indices[J]. Environmental Science and Pollution Research, 2015, 22(19): 14809-14819. DOI:10.1007/s11356-015-4624-2 |
| [9] |
van Dorst J, Siciliano SD, Winsley T, et al. Bacterial targets as potential indicators of diesel fuel toxicity in subantarctic soils[J]. Applied and Environmental Microbiology, 2014, 80(13): 4021-4033. DOI:10.1128/AEM.03939-13 |
| [10] |
Yan LJ, Sinkko H, Penttinen P, et al. Characterization of successional changes in bacterial community composition during bioremediation of used motor oil-contaminated soil in a boreal climate[J]. Science of the Total Environment, 2016, 542: 817-825. DOI:10.1016/j.scitotenv.2015.10.144 |
| [11] |
Hua T, Li SN, Di ZH, et al. Review on mechanism and application of microbe degrading petroleum pollutants[J]. Biotechnology Bulletin, 2018, 34(10): 26-34. (in Chinese) 华涛, 李胜男, 邸志珲, 等. 微生物降解石油污染物机制研究进展[J]. 生物技术通报, 2018, 34(10): 26-34. |
| [12] |
Laczi K, Kis Á, Horváth B, et al. Metabolic responses of Rhodococcus erythropolis PR4 grown on diesel oil and various hydrocarbons[J]. Applied Microbiology and Biotechnology, 2015, 99(22): 9745-9759. DOI:10.1007/s00253-015-6936-z |
| [13] |
Zampolli J, Zeaiter Z, di Canito A, et al. Genome analysis and -omics approaches provide new insights into the biodegradation potential of Rhodococcus[J]. Applied Microbiology and Biotechnology, 2019, 103(3): 1069-1080. DOI:10.1007/s00253-018-9539-7 |
| [14] |
Karande R, Debor L, Salamanca D, et al. Continuous cyclohexane oxidation to cyclohexanol using a novel cytochrome P450 monooxygenase from Acidovorax sp. CHX100 in recombinant P. taiwanensis VLB120 biofilms[J]. Biotechnology and Bioengineering, 2016, 113(1): 52-61. DOI:10.1002/bit.25696 |
| [15] |
Widdel F, Rabus R. Anaerobic biodegradation of saturated and aromatic hydrocarbons[J]. Current Opinion in Biotechnology, 2001, 12(3): 259-276. DOI:10.1016/S0958-1669(00)00209-3 |
| [16] |
Mi ZT. Chemical and Process Engineering[M]. 2nd ed. Beijing: Chemical Industry Press, 2006: 13. (in Chinese) 米镇涛. 化学工艺学[M]. 2版. 北京: 化学工业出版社, 2006: 13. |
| [17] |
Mao HX, Zhang HL, Yang Q. The application research on oily sludge treatment by microbiological technology in longdong oilfield[J]. Environmental Protection of Oil & Gas Fields, 2012, 22(5): 8-10. (in Chinese) 毛怀新, 张海玲, 杨琴. 陇东油田井场油泥微生物处理应用[J]. 油气田环境保护, 2012, 22(5): 8-10. |
| [18] |
Shen BX. Petroleum Refining Technology[M]. 2nd ed. Beijing: China Petrochemical Press, 2017: 18. (in Chinese) 沈本贤. 石油炼制工艺学[M]. 2版. 北京: 中国石化出版社, 2017: 18. |
| [19] |
Vogt C, Kleinsteuber S, Richnow HH. Anaerobic benzene degradation by bacteria[J]. Microbial Biotechnology, 2011, 4(6): 710-724. DOI:10.1111/j.1751-7915.2011.00260.x |
| [20] |
Fuchs G, Boll M, Heider J. Microbial degradation of aromatic compounds-from one strategy to four[J]. Nature Reviews Microbiology, 2011, 9(11): 803-816. DOI:10.1038/nrmicro2652 |
| [21] |
Varjani SJ. Microbial degradation of petroleum hydrocarbons[J]. Bioresource Technology, 2017, 223: 277-286. DOI:10.1016/j.biortech.2016.10.037 |
| [22] |
Rojo F. Degradation of alkanes by bacteria[J]. Environmental Microbiology, 2009, 11(10): 2477-2490. DOI:10.1111/j.1462-2920.2009.01948.x |
| [23] |
Throne-Holst M, Wentzel A, Ellingsen TE, et al. Identification of novel genes involved in long-chain n-alkane degradation by Acinetobacter sp. strain DSM 17874[J]. Applied and Environmental Microbiology, 2007, 73(10): 3327-3332. DOI:10.1128/AEM.00064-07 |
| [24] |
Li L, Liu XQ, Yang W, et al. Crystal structure of long-chain alkane monooxygenase (LadA) in complex with coenzyme FMN: unveiling the long-chain alkane hydroxylase[J]. Journal of Molecular Biology, 2008, 376(2): 453-465. DOI:10.1016/j.jmb.2007.11.069 |
| [25] |
Ratajczak A, Geißdörfer W, Hillen W. Alkane hydroxylase from Acinetobacter sp. strain ADP1 is encoded by alkM and belongs to a new family of bacterial integral-membrane hydrocarbon hydroxylases[J]. Applied and Environmental Microbiology, 1998, 64(4): 1175-1179. DOI:10.1128/AEM.64.4.1175-1179.1998 |
| [26] |
Singh JS, Singh D. Agro-Environmental Sustainability[M]. Switzerland: Springer, 2017: 1-18.
|
| [27] |
Wang XB, Chi CQ, Nie Y, et al. Degradation of petroleum hydrocarbons (C6-C40) and crude oil by a novel Dietzia strain[J]. Bioresource Technology, 2011, 102(17): 7755-7761. DOI:10.1016/j.biortech.2011.06.009 |
| [28] |
Callaghan AV, Morris BEL, Pereira IAC, et al. The genome sequence of Desulfatibacillum alkenivorans AK-01: a blueprint for anaerobic alkane oxidation[J]. Environmental Microbiology, 2012, 14(1): 101-113. DOI:10.1111/j.1462-2920.2011.02516.x |
| [29] |
Callaghan AV. Enzymes involved in the anaerobic oxidation of n-alkanes: from methane to long-chain paraffins[J]. Frontiers in Microbiology, 2013, 4: 89. |
| [30] |
Kim D, Chae JC, Zylstra GJ, et al. Identification of a novel dioxygenase involved in metabolism of o-xylene, toluene, and ethylbenzene by Rhodococcus sp. strain DK17[J]. Applied and Environmental Microbiology, 2004, 70(12): 7086-7092. DOI:10.1128/AEM.70.12.7086-7092.2004 |
| [31] |
Greated A, Lambertsen L, Williams PA, et al. Complete sequence of the IncP-9 TOL plasmid pWW0 from Pseudomonas putida[J]. Environmental Microbiology, 2002, 4(12): 856-871. DOI:10.1046/j.1462-2920.2002.00305.x |
| [32] |
Zylstra GJ, Gibson DT. Aromatic hydrocarbon degradation: a molecular approach[A]//Setlow JK. Genetic Engineering[M]. Boston: Springer, 1991: 183-203
|
| [33] |
Busch A, Lacal J, Silva-Jímenez H, et al. Catabolite repression of the TodS/TodT two-component system and effector-dependent transphosphorylation of TodT as the basis for toluene dioxygenase catabolic pathway control[J]. Journal of Bacteriology, 2010, 192(16): 4246-4250. DOI:10.1128/JB.00379-10 |
| [34] |
Kukor JJ, Olsen RH. Genetic organization and regulation of a meta cleavage pathway for catechols produced from catabolism of toluene, benzene, phenol, and cresols by Pseudomonas pickettii PKO1[J]. Journal of Bacteriology, 1991, 173(15): 4587-4594. DOI:10.1128/JB.173.15.4587-4594.1991 |
| [35] |
Parales RE, Parales JV, Pelletier DA, et al. Chapter 1 Diversity of microbial toluene degradation pathways[J]. Advances in Applied Microbiology, 2008, 64: 1-73. DOI:10.1016/S0065-2164(08)00401-2 |
| [36] |
van der Meer JR. Evolution of novel metabolic pathways for the degradation of chloroaromatic compounds[J]. Antonie Van Leeuwenhoek, 1997, 71(1/2): 159-178. DOI:10.1023/A:1000166400935 |
| [37] |
Suenaga H, Koyama Y, Miyakoshi M, et al. Novel organization of aromatic degradation pathway genes in a microbial community as revealed by metagenomic analysis[J]. The ISME Journal, 2009, 3(12): 1335-1348. DOI:10.1038/ismej.2009.76 |
| [38] |
Na KS, Kuroda A, Takiguchi N, et al. Isolation and characterization of benzene-tolerant Rhodococcus opacus strains[J]. Journal of Bioscience and Bioengineering, 2005, 99(4): 378-382. DOI:10.1263/jbb.99.378 |
| [39] |
Menn FM, Applegate BM, Sayler GS. NAH plasmid-mediated catabolism of anthracene and phenanthrene to naphthoic acids[J]. Applied and Environmental Microbiology, 1993, 59(6): 1938-1942. DOI:10.1128/AEM.59.6.1938-1942.1993 |
| [40] |
Simon MJ, Osslund TD, Saunders R, et al. Sequences of genes encoding naphthalene dioxygenase in Pseudomonas putida strains G7 and NCIB 9816-4[J]. Gene, 1993, 127(1): 31-37. |
| [41] |
Larkin MJ, Allen CCR, Kulakov LA, et al. Purification and characterization of a novel naphthalene dioxygenase from Rhodococcus sp. strain NCIMB12038[J]. Journal of Bacteriology, 1999, 181(19): 6200-6204. DOI:10.1128/JB.181.19.6200-6204.1999 |
| [42] |
Kulakov LA, Allen CCR, Lipscomb DA, et al. Cloning and characterization of a novel cis-naphthalene dihydrodiol dehydrogenase gene (narB) from Rhodococcus sp. NCIMB12038[J]. FEMS Microbiology Letters, 2000, 182(2): 327-331. DOI:10.1111/j.1574-6968.2000.tb08916.x |
| [43] |
Peng RH, Xiong AS, Xue Y, et al. Microbial biodegradation of polyaromatic hydrocarbons[J]. FEMS Microbiology Reviews, 2008, 32(6): 927-955. DOI:10.1111/j.1574-6976.2008.00127.x |
| [44] |
Denome SA, Stanley DC, Olson ES, et al. Metabolism of dibenzothiophene and naphthalene in Pseudomonas strains: complete DNA sequence of an upper naphthalene catabolic pathway[J]. Journal of Bacteriology, 1993, 175(21): 6890-6901. DOI:10.1128/JB.175.21.6890-6901.1993 |
| [45] |
Fuenmayor SL, Wild M, Boyes AL, et al. A gene cluster encoding steps in conversion of naphthalene to gentisate in Pseudomonas sp. strain U2[J]. Journal of Bacteriology, 1998, 180(9): 2522-2530. DOI:10.1128/JB.180.9.2522-2530.1998 |
| [46] |
Zhou NY, Fuenmayor SL, Williams PA. Nag genes of Ralstonia (formerly Pseudomonas) sp. strain U2 encoding enzymes for gentisate catabolism[J]. Journal of Bacteriology, 2001, 183(2): 700-708. DOI:10.1128/JB.183.2.700-708.2001 |
| [47] |
Habe H, Omori T. Genetics of polycyclic aromatic hydrocarbon metabolism in diverse aerobic bacteria[J]. Bioscience, Biotechnology, and Biochemistry, 2003, 67(2): 225-243. DOI:10.1271/bbb.67.225 |
| [48] |
Laurie AD, Lloyd-Jones G. The phn genes of Burkholderia sp. strain RP007 constitute a divergent gene cluster for polycyclic aromatic hydrocarbon catabolism[J]. Journal of Bacteriology, 1999, 181(2): 531-540. DOI:10.1128/JB.181.2.531-540.1999 |
| [49] |
Saito A, Iwabuchi T, Harayama S. A novel phenanthrene dioxygenase from Nocardioides sp. strain KP7: expression in Escherichia coli[J]. Journal of Bacteriology, 2000, 182(8): 2134-2141. DOI:10.1128/JB.182.8.2134-2141.2000 |
| [50] |
Peng JJ, Cai C, Qiao M, et al. Dynamic changes in functional gene copy numbers and microbial communities during degradation of pyrene in soils[J]. Environmental Pollution, 2010, 158(9): 2872-2879. DOI:10.1016/j.envpol.2010.06.020 |
| [51] |
DeBruyn JM, Chewning CS, Sayler GS. Comparative quantitative prevalence of Mycobacteria and functionally abundant nidA, nahAc, and nagAc dioxygenase genes in coal tar contaminated sediments[J]. Environmental Science & Technology, 2007, 41(15): 5426-5432. |
| [52] |
Jiang Y, Yang XL, Liu B, et al. Catechol 2, 3-dioxygenase from Pseudomonas sp. strain ND6: gene sequence and enzyme characterization[J]. Bioscience, Biotechnology, and Biochemistry, 2004, 68(8): 1798-1800. DOI:10.1271/bbb.68.1798 |
| [53] |
de Lourdes Moreno M, Sánchez-Porro C, Piubeli F, et al. Cloning, characterization and analysis of cat and ben genes from the phenol degrading halophilic bacterium Halomonas organivorans[J]. PLoS One, 2011, 6(6): e21049. DOI:10.1371/journal.pone.0021049 |
| [54] |
Leuthner B, Leutwein C, Schulz H, et al. Biochemical and genetic characterization of benzylsuccinate synthase from Thauera aromatica: a new glycyl radical enzyme catalysing the first step in anaerobic toluene metabolism[J]. Molecular Microbiology, 1998, 28(3): 615-628. DOI:10.1046/j.1365-2958.1998.00826.x |
| [55] |
Musat F, Galushko A, Jacob J, et al. Anaerobic degradation of naphthalene and 2-methylnaphthalene by strains of marine sulfate-reducing bacteria[J]. Environmental Microbiology, 2009, 11(1): 209-219. DOI:10.1111/j.1462-2920.2008.01756.x |
| [56] |
Carmona M, Zamarro MT, Blázquez B, et al. Anaerobic catabolism of aromatic compounds: a genetic and genomic view[J]. Microbiology and Molecular Biology Reviews, 2009, 73(1): 71-133. |
| [57] |
Abbasian F, Lockington R, Mallavarapu M, et al. A comprehensive review of aliphatic hydrocarbon biodegradation by bacteria[J]. Applied Biochemistry and Biotechnology, 2015, 176(3): 670-699. DOI:10.1007/s12010-015-1603-5 |
| [58] |
Wilkes H, Buckel W, Golding BT, et al. Metabolism of hydrocarbons in n-alkane-utilizing anaerobic bacteria[J]. Journal of Molecular Microbiology and Biotechnology, 2016, 26(1/3): 138-151. |
| [59] |
van Beilen JB, Funhoff EG. Alkane hydroxylases involved in microbial alkane degradation[J]. Applied Microbiology and Biotechnology, 2007, 74(1): 13-21. |
| [60] |
Funhoff EG, Bauer U, García-Rubio I, et al. CYP153A6, a soluble P450 oxygenase catalyzing terminal-alkane hydroxylation[J]. Journal of Bacteriology, 2006, 188(14): 5220-5227. DOI:10.1128/JB.00286-06 |
| [61] |
Li QS, Ogawa J, Schmid RD, et al. Engineering cytochrome P450 BM-3 for oxidation of polycyclic aromatic hydrocarbons[J]. Applied and Environmental Microbiology, 2001, 67(12): 5735-5739. DOI:10.1128/AEM.67.12.5735-5739.2001 |
| [62] |
Kubota M, Nodate M, Yasumoto-Hirose M, et al. Isolation and functional analysis of cytochrome P450 CYP153A genes from various environments[J]. Bioscience, Biotechnology, and Biochemistry, 2005, 69(12): 2421-2430. DOI:10.1271/bbb.69.2421 |
| [63] |
Rodrigues CJC, de Carvalho CCCR. Phenotypic adaptations help Rhodococcus erythropolis cells during the degradation of paraffin wax[J]. Biotechnology Journal, 2019, 14(8): 1800598. DOI:10.1002/biot.201800598 |
| [64] |
Lazar I, Voicu A, Nicolescu C, et al. The use of naturally occurring selectively isolated bacteria for inhibiting paraffin deposition[J]. Journal of Petroleum Science and Engineering, 1999, 22(1/3): 161-169. |
| [65] |
Singh A, van Hamme JD, Ward OP. Surfactants in microbiology and biotechnology: part 2. Application aspects[J]. Biotechnology Advances, 2007, 25(1): 99-121. |
| [66] |
Tani A, Ishige T, Sakai Y, et al. Gene structures and regulation of the alkane hydroxylase complex in Acinetobacter sp. strain M-1[J]. Journal of Bacteriology, 2001, 183(5): 1819-1823. DOI:10.1128/JB.183.5.1819-1823.2001 |
| [67] |
Minerdi D, Sadeghi SJ, Di Nardo G, et al. CYP116B5: a new class Ⅶ catalytically self-sufficient cytochrome P450 from Acinetobacter radioresistens that enables growth on alkanes[J]. Molecular Microbiology, 2015, 95(3): 539-554. |
| [68] |
Salamanca D, Karande R, Schmid A, et al. Novel cyclohexane monooxygenase from Acidovorax sp. CHX100[J]. Applied Microbiology and Biotechnology, 2015, 99(16): 6889-6897. DOI:10.1007/s00253-015-6599-9 |
| [69] |
Kawakami N, Shoji O, Watanabe Y. Use of perfluorocarboxylic acids to trick cytochrome P450BM3 into initiating the hydroxylation of gaseous alkanes[J]. Angewandte Chemie International Edition, 2011, 50(23): 5315-5318. DOI:10.1002/anie.201007975 |
| [70] |
Ladino-Orjuela G, Gomes E, da Silva R, et al. Metabolic pathways for degradation of aromatic hydrocarbons by bacteria[J]. Reviews of Environmental Contamination and Toxicology, 2016, 237: 105-121. |
| [71] |
Phale PS, Shah BA, Malhotra H. Variability in assembly of degradation operons for naphthalene and its derivative, carbaryl, suggests mobilization through horizontal gene transfer[J]. Genes, 2019, 10(8): 569. DOI:10.3390/genes10080569 |
| [72] |
Ramos JL, Marqués S, Timmis KN. Transcriptional control of the Pseudomonas TOL plasmid catabolic operons is achieved through an interplay of host factors and plasmid-encoded regulators[J]. Annual Review of Microbiology, 1997, 51: 341-373. DOI:10.1146/annurev.micro.51.1.341 |
| [73] |
Assinder SJ, Williams PA. The TOL plasmids: determinants of the catabolism of toluene and the xylenes[J]. Advances in Microbial Physiology, 1990, 31: 1-69. DOI:10.1016/S0065-2911(08)60119-8 |
| [74] |
Méndez V, Fuentes S, Hernández M, et al. Isolation of hydrocarbon-degrading heavy-metal-resistant bacteria from crude oil-contaminated soil in central chile[J]. Journal of Biotechnology, 2010, 150(Suppl 1): 287. |
| [75] |
Kim D, Kim YS, Kim SK, et al. Monocyclic aromatic hydrocarbon degradation by Rhodococcus sp. strain DK17[J]. Applied and Environmental Microbiology, 2002, 68(7): 3270-3278. DOI:10.1128/AEM.68.7.3270-3278.2002 |
| [76] |
Kim D, Chae JC, Zylstra GJ, et al. Identification of two-component regulatory genes involved in o-xylene degradation by Rhodococcus sp. strain DK17[J]. Journal of Microbiology (Seoul, Korea), 2005, 43(1): 49-53. |
| [77] |
Goyal AK, Zylstra GJ. Genetics of naphthalene and phenanthrene degradation by Comamonas testosteroni[J]. Journal of Industrial Microbiology and Biotechnology, 1997, 19(5/6): 401-407. |
| [78] |
Fuentes S, Méndez V, Aguila P, et al. Bioremediation of petroleum hydrocarbons: catabolic genes, microbial communities, and applications[J]. Applied Microbiology and Biotechnology, 2014, 98(11): 4781-4794. DOI:10.1007/s00253-014-5684-9 |
| [79] |
Eaton RW. Organization and evolution of naphthalene catabolic pathways: sequence of the DNA encoding 2-hydroxychromene-2-carboxylate isomerase and trans-o-hydroxybenzylidenepyruvate hydratase-aldolase from the NAH7 plasmid[J]. Journal of Bacteriology, 1994, 176(24): 7757-7762. DOI:10.1128/JB.176.24.7757-7762.1994 |
| [80] |
Kiyohara H, Torigoe S, Kaida N, et al. Cloning and characterization of a chromosomal gene cluster, pah, that encodes the upper pathway for phenanthrene and naphthalene utilization by Pseudomonas putida OUS82[J]. Journal of Bacteriology, 1994, 176(8): 2439-2443. DOI:10.1128/JB.176.8.2439-2443.1994 |
| [81] |
Takizawa N, Kaida N, Torigoe S, et al. Identification and characterization of genes encoding polycyclic aromatic hydrocarbon dioxygenase and polycyclic aromatic hydrocarbon dihydrodiol dehydrogenase in Pseudomonas putida OUS82[J]. Journal of Bacteriology, 1994, 176(8): 2444-2449. DOI:10.1128/JB.176.8.2444-2449.1994 |
| [82] |
Takizawa N, Iida T, Sawada T, et al. Nucleotide sequences and characterization of genes encoding naphthalene upper pathway of Pseudomonas aeruginosa PaK1 and Pseudomonas putida OUS82[J]. Journal of Bioscience and Bioengineering, 1999, 87(6): 721-731. DOI:10.1016/S1389-1723(99)80144-3 |
| [83] |
Park W, Padmanabhan P, Padmanabhan S, et al. nahR, encoding a LysR-type transcriptional regulator, is highly conserved among naphthalene-degrading bacteria isolated from a coal tar waste-contaminated site and in extracted community DNA[J]. Microbiology, 2002, 148(8): 2319-2329. DOI:10.1099/00221287-148-8-2319 |
| [84] |
Shahsavari E, Aburto-Medina A, Taha M, et al. A quantitative PCR approach for quantification of functional genes involved in the degradation of polycyclic aromatic hydrocarbons in contaminated soils[J]. MethodsX, 2016, 3: 205-211. DOI:10.1016/j.mex.2016.02.005 |
| [85] |
Klankeo P, Nopcharoenkul W, Pinyakong O. Two novel pyrene-degrading Diaphorobacter sp. and Pseudoxanthomonas sp. isolated from soil[J]. Journal of Bioscience and Bioengineering, 2009, 108(6): 488-495. DOI:10.1016/j.jbiosc.2009.05.016 |
| [86] |
Sei K, Asano KI, Tateishi N, et al. Design of PCR primers and gene probes for the general detection of bacterial populations capable of degrading aromatic compounds via catechol cleavage pathways[J]. Journal of Bioscience and Bioengineering, 1999, 88(5): 542-550. DOI:10.1016/S1389-1723(00)87673-2 |
| [87] |
Merimaa M, Heinaru E, Liivak M, et al. Grouping of phenol hydroxylase and catechol 2, 3-dioxygenase genes among phenol-and p-cresol-degrading Pseudomonas species and biotypes[J]. Archives of Microbiology, 2006, 186(4): 287-296. DOI:10.1007/s00203-006-0143-3 |
| [88] |
Kasuga I, Nakajima F, Furumai H. Diversity of catechol 2, 3-dioxygenase genes of bacteria responding to dissolved organic matter derived from different sources in a eutrophic lake[J]. FEMS Microbiology Ecology, 2007, 61(3): 449-458. DOI:10.1111/j.1574-6941.2007.00347.x |
| [89] |
El Azhari N, Devers-Lamrani M, Chatagnier G, et al. Molecular analysis of the catechol-degrading bacterial community in a coal wasteland heavily contaminated with PAHs[J]. Journal of Hazardous Materials, 2010, 177(1/3): 593-601. |
| [90] |
Cébron A, Norini MP, Beguiristain T, et al. Real-Time PCR quantification of PAH-ring hydroxylating dioxygenase (PAH-RHDα) genes from Gram positive and Gram negative bacteria in soil and sediment samples[J]. Journal of Microbiological Methods, 2008, 73(2): 148-159. DOI:10.1016/j.mimet.2008.01.009 |
| [91] |
Jurelevicius D, Alvarez VM, Peixoto R, et al. Bacterial polycyclic aromatic hydrocarbon ring-hydroxylating dioxygenases (PAH-RHD) encoding genes in different soils from King George Bay, Antarctic Peninsula[J]. Applied Soil Ecology, 2012, 55: 1-9. DOI:10.1016/j.apsoil.2011.12.008 |
| [92] |
Peng AP, Liu J, Ling WT, et al. Diversity and distribution of 16S rRNA and phenol monooxygenase genes in the rhizosphere and endophytic bacteria isolated from PAH-contaminated sites[J]. Scientific Reports, 2015, 5(1): 12173. DOI:10.1038/srep12173 |
| [93] |
Callaghan AV, Gieg LM, Kropp KG, et al. Comparison of mechanisms of alkane metabolism under sulfate-reducing conditions among two bacterial isolates and a bacterial consortium[J]. Applied and Environmental Microbiology, 2006, 72(6): 4274-4282. DOI:10.1128/AEM.02896-05 |
| [94] |
Callaghan AV, Wawrik B, Chadhain SMN, et al. Anaerobic alkane-degrading strain AK-01 contains two alkylsuccinate synthase genes[J]. Biochemical and Biophysical Research Communications, 2008, 366(1): 142-148. DOI:10.1016/j.bbrc.2007.11.094 |
| [95] |
Kniemeyer O, Musat F, Sievert SM, et al. Anaerobic oxidation of short-chain hydrocarbons by marine sulphate-reducing bacteria[J]. Nature, 2007, 449(7164): 898-901. DOI:10.1038/nature06200 |
| [96] |
Savage KN, Krumholz LR, Gieg LM, et al. Biodegradation of low-molecular-weight alkanes under mesophilic, sulfate-reducing conditions: metabolic intermediates and community patterns[J]. FEMS Microbiology Ecology, 2010, 72(3): 485-495. DOI:10.1111/j.1574-6941.2010.00866.x |
| [97] |
Wawrik B, Marks CR, Davidova IA, et al. Methanogenic paraffin degradation proceeds via alkane addition to fumarate by 'Smithella' spp. mediated by a syntrophic coupling with hydrogenotrophic methanogens[J]. Environmental Microbiology, 2016, 18(8): 2604-2619. DOI:10.1111/1462-2920.13374 |
| [98] |
Callaghan AV, Davidova IA, Savage-Ashlock K, et al. Diversity of benzyl-and alkylsuccinate synthase genes in hydrocarbon-impacted environments and enrichment cultures[J]. Environmental Science & Technology, 2010, 44(19): 7287-7294. |
| [99] |
Heider J, Spormann AM, Beller HR, et al. Anaerobic bacterial metabolism of hydrocarbons[J]. FEMS Microbiology Reviews, 1998, 22(5): 459-473. DOI:10.1111/j.1574-6976.1998.tb00381.x |
| [100] |
Meckenstock RU, Boll M, Mouttaki H, et al. Anaerobic degradation of benzene and polycyclic aromatic hydrocarbons[J]. Journal of Molecular Microbiology and Biotechnology, 2016, 26(1/3): 92-118. |
| [101] |
Meckenstock RU, Mouttaki H. Anaerobic degradation of non-substituted aromatic hydrocarbons[J]. Current Opinion in Biotechnology, 2011, 22(3): 406-414. DOI:10.1016/j.copbio.2011.02.009 |
| [102] |
Zhang X, Young LY. Carboxylation as an initial reaction in the anaerobic metabolism of naphthalene and phenanthrene by sulfidogenic consortia[J]. Applied and Environmental Microbiology, 1997, 63(12): 4759-4764. DOI:10.1128/AEM.63.12.4759-4764.1997 |
| [103] |
Abu Laban N, Selesi D, Rattei T, et al. Identification of enzymes involved in anaerobic benzene degradation by a strictly anaerobic iron-reducing enrichment culture[J]. Environmental Microbiology, 2010, 12(10): 2783-2796. |
| [104] |
Luo F, Gitiafroz R, Devine CE, et al. Metatranscriptome of an anaerobic benzene-degrading, nitrate-reducing enrichment culture reveals involvement of carboxylation in benzene ring activation[J]. Applied and Environmental Microbiology, 2014, 80(14): 4095-4107. DOI:10.1128/AEM.00717-14 |
| [105] |
Zhang T, Tremblay PL, Chaurasia AK, et al. Anaerobic benzene oxidation via phenol in Geobacter metallireducens[J]. Applied and Environmental Microbiology, 2013, 79(24): 7800-7806. DOI:10.1128/AEM.03134-13 |
| [106] |
Davidova IA, Gieg LM, Duncan KE, et al. Anaerobic phenanthrene mineralization by a carboxylating sulfate-reducing bacterial enrichment[J]. The ISME Journal, 2007, 1(5): 436-442. DOI:10.1038/ismej.2007.48 |
| [107] |
von Netzer F, Kuntze K, Vogt C, et al. Functional gene markers for fumarate-adding and dearomatizing key enzymes in anaerobic aromatic hydrocarbon degradation in terrestrial environments[J]. Journal of Molecular Microbiology and Biotechnology, 2016, 26(1/3): 180-194. |
| [108] |
von Netzer F, Pilloni G, Kleindienst S, et al. Enhanced gene detection assays for fumarate-adding enzymes allow uncovering of anaerobic hydrocarbon degraders in terrestrial and marine systems[J]. Applied and Environmental Microbiology, 2013, 79(2): 543-552. DOI:10.1128/AEM.02362-12 |
| [109] |
Winderl C, Schaefer S, Lueders T. Detection of anaerobic toluene and hydrocarbon degraders in contaminated aquifers using benzylsuccinate synthase (bssA) genes as a functional marker[J]. Environmental Microbiology, 2007, 9(4): 1035-1046. DOI:10.1111/j.1462-2920.2006.01230.x |
| [110] |
Boll M, Löffler C, Morris BEL, et al. Anaerobic degradation of homocyclic aromatic compounds via arylcarboxyl-coenzyme A esters: organisms, strategies and key enzymes[J]. Environmental Microbiology, 2014, 16(3): 612-627. DOI:10.1111/1462-2920.12328 |
| [111] |
Morris BEL, Gissibl A, Kümmel S, et al. A PCR-based assay for the detection of anaerobic naphthalene degradation[J]. FEMS Microbiology Letters, 2014, 354(1): 55-59. DOI:10.1111/1574-6968.12429 |
| [112] |
Whyte LG, Bourbonniére L, Greer CW. Biodegradation of petroleum hydrocarbons by psychrotrophic Pseudomonas strains possessing both alkane (alk) and naphthalene (nah) catabolic pathways[J]. Applied and Environmental Microbiology, 1997, 63(9): 3719-3723. DOI:10.1128/AEM.63.9.3719-3723.1997 |
| [113] |
Kweon O, Kim SJ, Freeman JP, et al. Substrate specificity and structural characteristics of the novel Rieske nonheme iron aromatic ring-hydroxylating oxygenases NidAB and NidA3B3 from Mycobacterium vanbaalenii PYR-1[J]. mBio, 2010, 1(2): e00135-10. |
| [114] |
Liu H, Xu J, Liang RB, et al. Characterization of the medium- and long-chain n-alkanes degrading Pseudomonas aeruginosa strain SJTD-1 and its alkane hydroxylase genes[J]. PLoS One, 2014, 9(8): e105506. DOI:10.1371/journal.pone.0105506 |
| [115] |
Ji NN, Wang XL, Yin C, et al. CrgA protein represses AlkB2 monooxygenase and regulates the degradation of medium-to-long-chain n-alkanes in Pseudomonas aeruginosa SJTD-1[J]. Frontiers in Microbiology, 2019, 10: 400. DOI:10.3389/fmicb.2019.00400 |
| [116] |
Copley SD. Evolution of a metabolic pathway for degradation of a toxic xenobiotic: the patchwork approach[J]. Trends in Biochemical Sciences, 2000, 25(6): 261-265. DOI:10.1016/S0968-0004(00)01562-0 |
| [117] |
Kloos K, Munch JC, Schloter M. A new method for the detection of alkane-monooxygenase homologous genes (alkB) in soils based on PCR-hybridization[J]. Journal of Microbiological Methods, 2006, 66(3): 486-496. DOI:10.1016/j.mimet.2006.01.014 |
| [118] |
Lueders T, von Netzer F. Functional genes for anaerobic hydrocarbon degrading microbes[A]//McGenity TJ, Timmis KN, Nogales B. Hydrocarbon and Lipid Microbiology Protocols[M]. Berlin: Springer, 2014: 39-55
|
 2020, Vol. 47
2020, Vol. 47




