扩展功能
文章信息
- FENG Xin-Qian, HAN A-Xiang, HUANG Jie-Peng, YANG Qiong-Ying, LU Jin-Fang, ZHOU Yan, KANG Yu-Tong, ZHENG Lai-Bao, LOU Yong-Liang, GUAN Wan-Chun
- 丰新倩, 韩阿祥, 黄杰鹏, 杨琼莹, 卢金芳, 周燕, 康雨童, 郑来宝, 楼永良, 关万春
- Metagenomic analysis of the microbial community and antibiotic resistance genes in an urban recreational lake, Jiushan Lake
- 宏基因组分析城市休闲水域九山湖的微生物群落结构和抗性基因
- Microbiology China, 2020, 47(10): 3102-3113
- 微生物学通报, 2020, 47(10): 3102-3113
- DOI: 10.13344/j.microbiol.china.200339
-
文章历史
- Received: April 03, 2020
- Accepted: July 22, 2020
- Published online: August 07, 2020
Emerging antimicrobial resistance is considered by the World Health Organization as a global public health crisis that must be managed with the utmost urgency[1]. Part of the problem is the contamination of aquatic ecosystems with the antibiotics that we use for the treatment or prevention of bacterial infection, as well as in agriculture and aquaculture[2-3], which is attributed to the treatment of wastewater often being poor or even non-existent[4]. The input of antibiotics may challenge the bacterial communities[5] and accelerate the appearance and spread of antibiotic resistance genes (ARGs)[6]. ARGs can be transferred horizontally between different microbial communities, increasing the risk of ARG dissemination[7]. Therefore, ARGs are considered emerging contaminants and raise increasing public concern[8], especially with respect to food safety as they have been detected in bacteria inside edible aquatic species[9].
ARG contamination has been investigated in various aquatic environments, such as lakes[10-11], sewage[12-13], aquacultures[3, 14], oceans[15], and even drinking water[16-17]. Several past publications indicated that urban water systems around the world are highly contaminated by ARGs[18-20]. However, few studies have examined the occurrence and abundance of ARGs in urban recreational waters, despite the latter being perceived as aesthetically and ecologically beneficial elements of urban environments[21]. Moreover, pathogenic bacteria and ARGs may find their way to recreational waters because of the input of sewage discharge and animal excrement[22]. The intensity of anthropogenic activities has been shown to affect the abundance of ARGs[6]. Recreational water contaminated with ARGs may pose a risk for public health, as citizens often come in contact with it through swimming and fishing. Thus, it is necessary to understand the occurrence of ARGs in urban recreational lakes.
To date, studies of ARGs in aquatic environments, including recreational waters, have been using traditional approaches of molecular biology, such as PCR which only allowed them to focus on a specific gene[3, 23-24]. The bias deriving from the very selection process of the genes to amplify, as well as limitations related to how well the designed primers work, shed doubt on whether these studies reflect the real composition of ARG and microbes (especially uncultured microbes) in recreational waters[25]. In recent years, the advent of next-generation sequencing and the development of bioinformatics gave rise to metagenomics, a powerful tool for exploring the complex microbial diversities and creating broad-spectrum profiles of ARGs in samples from various aquatic environments[26-28].
This study aimed to utilize a high-throughput sequencing-based metagenomic approach to characterize the composition and abundance of the microbes and ARGs in Jiushan Lake, a recreational lake located in the city of Wenzhou, and estimate the potential public health risk. Moreover, because the function of urban recreational lake waters as potential reservoirs of ARGs and bacteria is subject to the temporal variation of environmental conditions, we chose to take the temporal factor into account. This work should provide useful information for a better understanding of the ecosystem structure and function of urban recreational lakes.
1 Materials and Methods 1.1 Main reagents and equipmentPowerWater DNA Isolation Kit, Qiagen company; Portable pH meter, Mettler Toledo company; Portable dissolved oxygen meter, YSI company; Polycarbonate filters, Millipore company; Microspectrophotometer, DeNovix company; GF/F glass fiber filters, GE Healthcare company; Illumina HiSeq platform, Illumina company.
1.2 Site information and sampling methodWenzhou, a Chinese city located in southeastern Zhejiang province, is close to the East China Sea and has abundant water resources. Jiushan Lake is a shallow body of water situated in the central city district that serves as a very popular natural swimming pool for locals. Water samples were collected at two different time points, in August and December 2018, representing summer and winter, respectively. Three different sampling sites (locations with frequent citizen activities) were selected, and three surface water samples (to be exact, from a depth of about 0.5 m) were collected for each season. The samples were stored in sterilized Nalgene 1-liter polycarbonate bottles and transported to the laboratory in an icebox within 1 hour from collection. Each sample was divided into two parts, one for physicochemical analysis and the other for analysis of microbial genomes.
1.3 Physicochemical parametersWater temperature and pH were measured on-site using an pH meter. Dissolved oxygen (DO) concentration was also measured on-site with a YSI 550 portable dissolved oxygen meter. After filtration through GF/F glass fiber filters, Chlorophyll-a (Chl-a) was extracted from the filters with pure methanol for 24 h, centrifuged at 3 000 r/min for 10 min, and then spectrophotometrically quantified[29]. Soluble reactive phosphorus (SRP) was measured using the phosphorus molybdenum blue method, ammonium nitrogen (N-NH4) with the sodium hypobromite oxidation method, nitrate-nitrogen (N-NO3) with the Zinc cadmium reduction-azo colorimetric method, and nitrite-nitrogen (N-NO2) using the diazo colorimetric method[30].
1.4 DNA extraction and shotgun metagenomic sequencingHalf of each water sample, i.e., 500 mL, was pre-filtered through 5 μm pore-size polycarbonate filters to remove impurities. Then, each filtrate was filtered again using a 0.22 μm membrane filter to capture and concentrate microbes. The filters were cut into strips using sterile scissors, put into 5 mL sterile tubes, and stored at -20 ℃ until DNA extraction. DNA was extracted using the PowerWater DNA Isolation Kit according to the manufacturer's instructions. The DNA quality was assessed using a microspectrophotometer. All samples had OD260/280 ratios between 1.8 and 2.0. DNA degradation degree and potential contamination were electrophoretically examined on 1% agarose gels. Qualified DNA samples were sent to Novogene (Beijing, China) for Illumina shotgun high-throughput sequencing using the PE 150 (Paired-end sequencing, 150-bp reads) sequencing strategy. A total amount of 1 μg DNA per sample was used as input material for library construction with an insert size of 350 bp, followed by high-throughput sequencing on an Illumina HiSeq platform.
1.5 BioinformaticsThe raw sequencing data were pre-processed for quality control using Readfq V8 (public domain) as follows: a) Removal of reads that contained more than 26.7% (40/150) low-quality bases (the default quality threshold value was set at 38); b) Removal of reads with more than 6.7% (10/150) undetermined bases (N), and; c) Removal of reads that shared an overlap with the adapter sequence longer than 15 bp. Q20, Q30, and GC-content were also calculated. The sequencing output of each water sample was about 12 Gb with more than 85% clean-read bases, i.e., having quality scores higher than 30. The base sequencing error rate was 0.001. A total of 505 097 533 clean reads were generated from 6 samples with an average of 84 182 922 reads per sample. The filtered high-quality sequences were used for subsequent analysis.
To investigate the microbial composition of the water, genes were aligned to the NR database (Version: 2018-01-02) of NCBI using DIAMOND V0.9.9 software (free domain)[31] with an E-value≤10-5. The abundance of a taxonomic group was calculated by summing the abundance of genes annotated to a feature. The identification of human pathogenic bacteria referred to the list of human pathogenic bacteria (51 species) provided in a previous study[32]. All values are averages from three different sequencing runs.
To explore the diversity and abundance of ARGs, genes were aligned to the CARD database using Resistance Gene Identifier (RGI) software[33-35] for annotation and BLASTp comparison (E-value≤10-30). The alignment results were used to calculate both the total and the relative abundance of ARGs. The proportions of different ARG sequences in "total detected ARG sequences" and "total metagenome sequences" were defined as " relative percentage" (%) and "relative abundance" (ppm (parts per million), one hit in one million aligned sequencing genes), respectively[36-37]. All values are averages from three different sequencing runs.
1.6 Statistical analysisDiversity index (Shannon, Inverse Simpson) was calculated based on the genera profiles. Histograms were performed at OriginPro 8.5. Redundancy analysis (RDA) was performed to investigate the correlation between the top 10 most abundant genera and environmental factors. Canonical correspondence analysis (CCA) was used to investigate the relationships between environmental factors, human pathogenic bacteria, and the distribution of ARGs. RDA and CCA were conducted in CANOCO V5 (Biometris, Wageningen, the Netherlands) for windows. One-way ANOVA (LSD test), performed with IBM SPSS Statistics V22 (IBM, Armonk, NY, USA), was used to assess the statistical differences between data sets. The significance level was set at P=0.05.
Data are available at the NCBI Sequence Read Archive under project No. PRJNA613875.
2 Results 2.1 Environmental parametersThe physical and chemical properties of the water samples are shown in Table 1. The pH and dissolved oxygen (DO) values did not display significant seasonal differences between the two seasons. The temperature was 34 ℃ in the summer and 18 ℃ in the winter. Chlorophyll-a concentration was 37.6 μg/L and 17.1 μg/L in the summer and winter, respectively, i.e., the summer concentration was 2.2 times higher than the winter concentration. With respect to nutrients, SRP was 1.5 times higher in the summer than that in the winter water samples. In contrast, the concentration of dissolved inorganic nitrogen, i.e., the sum of N-NO2-, N-NH4+, and N-NO3-, in the summer samples was only 30% of the one in winter samples.
| Season | T (℃) | pH | DO (mg/L) | Chl-a (μg/L) | SRP (μmol/L) | DIN (μmol/L) | N/P |
| Summer | 34 (0)a | 7.37 (0.02) | 5 (0.7) | 37.6 (1.34)a | 2.15 (0.02)a | 22.32 (1.04)b | 10.39 (0.38)b |
| Winter | 18 (0.1)b | 7.28 (0.045) | 4.73 (0.87) | 17.1 (0.63)b | 1.43 (0)b | 69.28 (3.22) a | 48.37 (2.25)a |
| Note: n=3, T: Temperature; DO: Dissolved oxygen; Chl-a: Chlorophyll a; SRP: Soluble reactive phosphorus; DIN: Total inorganic nitrogen; N/P: Ratio of total inorganic nitrogen and SRP. One-way ANOVA (LSD test) results among the different seasonal samples are indicated by superscript letters. The data with identical superscript letters indicate that the mean values are not significantly different. 注:n=3, T:温度; DO:溶解氧; Chl-a:叶绿素a; SRP:可溶性活性磷; DIN:总无机氮; N/P:总无机氮与SRP的比值.不同季节样品间的单因素方差分析(LSD test)结果用上标字母表示.上标字母相同的数据表示平均值没有显著差异. | |||||||
At the kingdom level, in summer water samples, 54% of genes were aligned to Bacteria, 9% of genes to Viruses, 0.4% to Archaea, and 0.04% to Eukaryota, while about 36% of the genes could not be annotated using the NR database. In winter samples, 64% of genes were aligned to Bacteria, 5.9% to Viruses, 0.3% to Archaea, and 0.04% to Eukaryota, while 29.41% of the genes were not annotated. Notably, the relative abundance of Bacteria in the winter was higher than that in the summer.
This study identified 148 phyla in the summer and 152 phyla in the winter samples (including Bacteria, Eukaryota, and Archaea), respectively. As seen in Figure 1, the phyla with the highest relative abundance greater than 1% in both seasons were Proteobacteria, Actinobacteria, Bacteroidetes, Cyanobacteria, and Verrucomicrobia. Several important observations can be made. Proteobacteria and Actinobacteria dominated samples from both seasons, together accounting for nearly 35% and 46% of the microbial community in summer and winter samples, respectively. Next in order of abundance in the summer water samples were Cyanobacteria and Bacteroidetes, while the corresponding positions in winter samples were occupied by Bacteroidetes and Verrucomicrobia. Proteobacteria, Actinobacteria, and Bacteroidetes had significantly higher abundance in winter samples than that in summer samples (P < 0.05), while both Cyanobacteria and Verrucomicrobia were more abundant in summer than those in winter (P < 0.01).

|
| Figure 1 Microbial community structure in the summer and winter water samples at the phylum level 图 1 夏季和冬季水样在门水平上的微生物群落结构 |
|
|
At the genus level, bacteria with high relative abundance in both seasons were those belonging to the genera Limnohabitans (a genus of Comamonadaceae family, Burkholderiales order, Betaproteobacteria class, Proteobacteria phylum), Synechococcus (a genus of Chroococcales family, Cyanobacteria class, Cyanobacteria phylum), Candidatus Planktophila (a genus of Candidatus Nanopelagicales order, Actinobacteria class, Actinobacteria phylum), Polynucleobacter (a genus of Burkholderiaceae family, Burkholderiales order, Betaproteobacteria class, Proteobacteria phylum), and Candidatus Methylopumilus (a genus of Methylophilaceae family, Nitrosomonadales order, Betaproteobacteria class, Proteobacteria phylum) (Figure 2). Synechococcus was the most common genus in summer samples (2.78%) and its abundance was significantly higher in summer samples than that in winter (P < 0.01), while Limnohabitans was significantly higher in winter than that in summer (P < 0.05). RDA was performed to investigate the correlation between the top 10 most abundant genera and environmental factors. The results (Figure 3) demonstrated that NH4+-N, NO2--N, and temperature were found to be significantly correlated with the distribution of the main planktonic bacteria (P < 0.05).

|
| Figure 2 Microbial community structure in the summer and winter water samples at the genus level 图 2 夏季和冬季水体样本属水平的微生物群落结构 |
|
|

|
| Figure 3 Redundancy analysis (RDA) of environmental factors and the 10 major genera 图 3 环境因子和10个主要菌属的冗余分析 |
|
|
In the diversity analysis, the trend of Shannon and Simpson index was consistent, and the results showed that a high α diversity was observed in summer than that in winter (Table 2). Of particular interest was the presence among the identified species in this study associated with human opportunistic infections, such as Pseudomonas aeruginosa, Escherichia coli, Chlamydia trachomatis, Clostridium botulinum, and Bacillus cereus. Amongst, P. aeruginosa (a species of Pseudomonas genus, Pseudomonadaceae family, Pseudomonadales order, Gammaproteobacteria class, Proteobacteria phylum) (0.013 3%/0.020 8%) had the highest abundance in all samples.
| Sample | Shannon | Inverse Simpson |
| Summer.1 | 4.813 | 29.026 |
| Summer.2 | 4.794 | 28.113 |
| Summer.3 | 4.777 | 29.970 |
| Winter.1 | 4.147 | 12.633 |
| Winter.2 | 4.080 | 11.677 |
| Winter.3 | 4.533 | 23.019 |
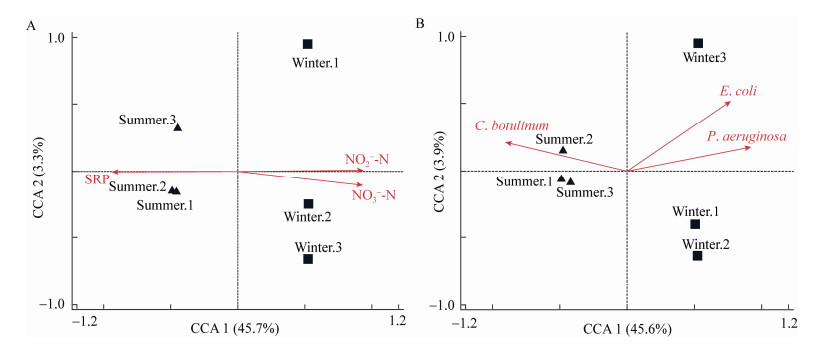
A total of 449 ARGs were detected in this lake, 370 in summer and 383 in winter water samples, respectively. More than half of the ARGs (304 ARGs) were shared, 66 were summer-exclusive, and 79 winter-exclusive (Figure 4). The relative abundance of ARGs in the summer samples (359.02±7.0 ppm) was higher than that in the winter samples (318.94±11.6 ppm) (Figure 5A), with the difference being statistically significant (P < 0.05). The ARGs with the relative percentage were shown in Figure 5B. The predominant resistance gene in the summer samples was MCR-1.2 (polymyxin E resistance gene) (4.66%), followed by LsaC (aminoglycoside resistance gene) (4.46%), APH6-Ia (aminoglycoside resistance gene) (3.16%), and LpeB (macrolide resistance gene) (2.40%). In winter samples, BcI (β-lactam resistance gene) (7.35%) was the predominant ARG, followed by LRA-13 (cephalosporin resistance gene) (3.73%), acrD (aminoglycoside resistance gene) (3.53%), and SHV-180 (β-lactam resistance gene) (2.60%). The canonical correspondence analysis (CCA) (Figure 6) indicated that the environmental variables (NO3--N, NO2--N, and SRP) and human pathogenic bacteria (P. aeruginosa, E. coli, and C. botulinum) showed significant relationships with the distribution of ARGs (Monte Carlo test, P < 0.05).

|
| Figure 4 The number of shared and unique ARGs in summer and winter water samples 图 4 夏季和冬季水样中共有的和特有的抗性基因数目 |
|
|

|
| Figure 5 Relative abundance of all ARGs (A) and relative percentage of different ARGs (B) 图 5 所有抗性基因的相对丰度(A)和不同抗性基因的相对百分比(B) Note: Relative abundance was defined as the ratio of the number of sequences of ARGs to the total number of metagenome sequences. The unit of "ppm" was defined as one hit in one million sequences. 注:相对丰度定义为抗性基因序列数与宏基因组序列总数的比值. “ppm”的单位被定义为一百万条序列中的一条. |
|
|
|
| Figure 6 Canonical correspondence analysis (CCA) shows the relationships between ARGs and environmental variables (A) or pathogenic species (top 10) (B) 图 6 典型对应分析(CCA)显示了ARGs与环境变量(A)或致病菌(前10) (B)之间的关系 |
|
|
Figure 7 shows the percentage of resistance mechanisms represented in the ARGs repertoire of summer and winter water samples. In both seasons, three resistance mechanisms were dominant, accounting for more than 90%: antibiotic efflux, antibiotic inactivation, and antibiotic target alteration. However, their relative representation depended on the season. Specifically, antibiotic inactivation had a much higher representation in winter than in summer, whereas the percentages of the other two major mechanisms showed the opposite trend. In both seasons, antibiotic efflux and inactivation were the largest groups, with a collective percentage of 73.94% in summer and 80.37% in winter.

|
| Figure 7 The percentages of different resistant mechanisms 图 7 不同抗性机制的百分比 |
|
|
In this study, we used Illumina high-throughput shotgun metagenomic sequencing to investigate the seasonal composition and abundance of microbial communities and ARGs in Jiushan Lake. Proteobacteria and Actinobacteria were the predominant phyla in all water samples, which was consistent with previous reports on other freshwater environments, such as general freshwater lakes[38-39] and freshwater aquacultures[40-41]. At the genus level, the relative abundance of Synechococcus spp. was much higher in summer than that in winter, while the opposite was true for Limnohabitans spp. and Candidatus Planktophila spp. This seasonal difference in bacterial community composition may be caused by the seasonal variation in physical and chemical properties such as nitrogen availability, temperature, pH, and total phosphorus[42-44]. For example, inorganic nutrients are often the main limiting factor for microbial growth[45]. Working with samples from five lakes with different trophic status and humic content, Lindström showed that the quantity and community structure of bacteria vary in different habitats and nutrient conditions[46]. Synechococcus spp. are photoautotroph organisms belonging to Cyanobacteria[47], the result of RDA demonstrated that Synechococcus was positively correlated with temperature, thus the higher summer temperature may provide favorable conditions for its growth[48]. On the other hand, Limnohabitans spp. belongs to βetaproteobacteria, which are nitrogen cycle-related organisms with a wide distribution in freshwater systems[42]. Total nitrogen plays an important role in the growth and reproduction of this genus[49]. Consistent with these reports, our study shows a positive correlation between NH4+-N, NO2--N and Limnohabitans (Figure 3).
Extremely worrisome is our detection of species typically associated with human opportunistic infections, such as P. aeruginosa, E. coli, C. trachomatis, C. botulinum, and B. cereus, as these findings suggest a clear health hazard to citizens coming into contact with the water of the lake during recreational activities. P. aeruginosa was the most abundant potential pathogen in all water samples. It has been reported that the infection of human external ear and skin in recreational waters is related to the presence of P. aeruginosa[50] and it has attracted more attention because of the multidrug resistance of the widely specific multi-drug efflux system[51]. B. cereus primarily causes food poisoning and severe eye infections[52], Resistance of B. cereus to beta-lactam and carbapenem has also been reported[53]. Drug-resistant pathogens can cause intestinal problems in swimmers by swallowing in lake water, and opportunistic pathogens found in recreational waters can pose a considerable potential threat to public health.
With respect to ARGs, our study is consistent with previous reports according to which urban aquatic environments, such as rivers and lakes (including recreational waters), may serve as ARG reservoirs[23, 26], as we were able to detect hundreds of ARGs in Jiushan Lake. Most of the detected ARGs endowed resistance to aminoglycoside, β-lactam antibiotics, or macrolide. Moreover, we observed a significant difference in the abundance and composition of ARGs between summer and winter water samples. Human pathogenic bacteria (P. aeruginosa, E. coli, and C. botulinum) showed significant relationships with the distribution of ARGs, implying the threatening risk of antibiotic resistance to the public health. The relative abundance of ARGs was higher in summer than that in winter. With respect to the cause, a seasonal study of ARGs in a peri-urban river denoted temperature as a potential factor driving the shift of ARG profiles[54]. However, other factors may also be at play, such as the increased human presence in the lake during summer.
MCR-1.2 was the predominant ARG in summer water samples. It is a novel variant of MCR-1 (mobilized colistin resistance gene -1), a plasmid-mediated resistance gene for polymyxin E (also called colistin, hence the gene's name)[55-56]. Polymyxin E is a polypeptide antibiotic produced by Bacillus polymyxavar and is figuratively called the "last line of defense" against gram-negative bacterial infection[57-58]. To date, MCR-1.2 has been found in Klebsiella pneumonia and E. coli[55, 59], both of which were detected in this study. As ARGs can be transferred between different bacteria via plasmids during conjugation, MCR-1.2 will ultimately find its way to opportunistic human pathogens, at which point it will pose a hard challenge to clinical medication and a significant threat to human health. The detection of MCR-1.2 in Jiushan Lake, whose waters come in contact with a large number of swimming enthusiasts, should arouse the public's concern.
BcI was the preponderant ARG in winter water samples. It codes for Bacillus cereus beta-lactamase, which is a zinc metallo-beta-lactamase that hydrolyzes a large number of penicillins and cephalosporins[60]. β-lactam antibiotics are the most widely used antibiotics[61], which might explain why ARGs against them are widely distributed in aquatic environments. An example is the detection of the other β-lactamase-coding gene, bla-TEM, in recreational lakes in parks of Beijing[23].
The predominant resistance mechanisms detected in Jiushan Lake were antibiotic efflux and antibiotic inactivation, which were also found to be the chief mechanism in paddy soils from South China[27] and the receiving surface water of a wastewater treatment plant[62]. Antibiotic inactivation is mainly associated with resistance to common anthropogenic antibiotics, such as aminoglycosides, β-lactams, and macrolides[36]. Efflux pumps can transfer not only antibiotics but also heavy metals and other toxins from the cell to the external environment and are involved in various processes such as detoxification of metabolic intermediates, virulence, and signal trafficking[63]. Thus, antibiotic extrusion by efflux pumps is the most effective method to resist the different pressures of diverse environments[64-65].
4 ConclusionThis study provides a description of the microbial community and ARG composition in an urban recreational lake as assessed by metagenomic analysis. The microbial diversity and relative abundance of ARG were higher in summer than in winter. MCR-1.2 and BcI were the predominant ARGs in the summer and winter water samples, respectively. An expanded study with samples from several months should help to understand the composition and dynamic change of bacteria and ARGs more clearly. More importantly, as the high abundance of observed ARGs may pose a public health risk, our results suggest that more attention should be paid to the ARG contamination of urban recreational waters.
| [1] |
Eurosurveillance Editorial Team. WHO member states adopt global action plan on antimicrobial resistance[J]. Euro Surveillance, 2015, 20(21): 21137. |
| [2] |
Dolliver H, Gupta S. Antibiotic losses in leaching and surface runoff from manure-amended agricultural land[J]. Journal of Environmental Quality, 2008, 37(3): 1227-1237. DOI:10.2134/jeq2007.0392 |
| [3] |
Chen CQ, Zheng L, Zhou JL, et al. Persistence and risk of antibiotic residues and antibiotic resistance genes in major mariculture sites in Southeast China[J]. Science of the Total Environment, 2017, 580: 1175-1184. DOI:10.1016/j.scitotenv.2016.12.075 |
| [4] |
Gücker B, Brauns M, Solimini AG, et al. Urban stressors alter the trophic basis of secondary production in an agricultural stream[J]. Canadian Journal of Fisheries and Aquatic Sciences, 2011, 68(1): 74-88. DOI:10.1139/F10-126 |
| [5] |
Watkinson AJ, Murby EJ, Costanzo SD. Removal of antibiotics in conventional and advanced wastewater treatment: Implications for environmental discharge and wastewater recycling[J]. Water Research, 2007, 41(18): 4164-4176. DOI:10.1016/j.watres.2007.04.005 |
| [6] |
Jiang HY, Zhou RJ, Yang Y, et al. Characterizing the antibiotic resistance genes in a river catchment: influence of anthropogenic activities[J]. Journal of Environmental Sciences, 2018, 69: 125-132. DOI:10.1016/j.jes.2017.08.009 |
| [7] |
Sanderson H, Fricker C, Brown RS, et al. Antibiotic resistance genes as an emerging environmental contaminant[J]. Environmental Reviews, 2016, 24(2): 205-218. DOI:10.1139/er-2015-0069 |
| [8] |
Pruden A, Pei RT, Storteboom H, et al. Antibiotic resistance genes as emerging contaminants: studies in northern Colorado[J]. Environmental Science & Technology, 2006, 40(23): 7445-7450. |
| [9] |
Zhao YT, Zhang XX, Zhao ZH, et al. Metagenomic analysis revealed the prevalence of antibiotic resistance genes in the gut and living environment of freshwater shrimp[J]. Journal of Hazardous Materials, 2018, 350: 10-18. DOI:10.1016/j.jhazmat.2018.02.004 |
| [10] |
Devarajan N, Laffite A, Graham ND, et al. Accumulation of clinically relevant antibiotic-resistance genes, bacterial load, and metals in freshwater lake sediments in Central Europe[J]. Environmental Science & Technology, 2015, 49(11): 6528-6537. |
| [11] |
Wang Z, Han MZ, Li EH, et al. Distribution of antibiotic resistance genes in an agriculturally disturbed lake in China: Their links with microbial communities, antibiotics, and water quality[J]. Journal of Hazardous Materials, 2020, 393: 122426. DOI:10.1016/j.jhazmat.2020.122426 |
| [12] |
Novo A, André S, Viana P, et al. Antibiotic resistance, antimicrobial residues and bacterial community composition in urban wastewater[J]. Water Research, 2013, 47(5): 1875-1887. DOI:10.1016/j.watres.2013.01.010 |
| [13] |
Cacace D, Fatta-Kassinos D, Manaia CM, et al. Antibiotic resistance genes in treated wastewater and in the receiving water bodies: a pan-European survey of urban settings[J]. Water Research, 2019, 162: 320-330. DOI:10.1016/j.watres.2019.06.039 |
| [14] |
Gao PP, Mao DQ, Luo Y, et al. Occurrence of sulfonamide and tetracycline-resistant bacteria and resistance genes in aquaculture environment[J]. Water Research, 2012, 46(7): 2355-2364. DOI:10.1016/j.watres.2012.02.004 |
| [15] |
Niu ZG, Zhang K, Zhang Y. Occurrence and distribution of antibiotic resistance genes in the coastal area of the Bohai Bay, China[J]. Marine Pollution Bulletin, 2016, 107(1): 245-250. |
| [16] |
Jiang L, Hu XL, Xu T, et al. Prevalence of antibiotic resistance genes and their relationship with antibiotics in the Huangpu River and the drinking water sources, Shanghai, China[J]. Science of the Total Environment, 2013, 458-460: 267-272. DOI:10.1016/j.scitotenv.2013.04.038 |
| [17] |
Bai XH, Ma XL, Xu FM, et al. The drinking water treatment process as a potential source of affecting the bacterial antibiotic resistance[J]. Science of the Total Environment, 2015, 533: 24-31. DOI:10.1016/j.scitotenv.2015.06.082 |
| [18] |
Wu D, Su YL, Xi H, et al. Urban and agriculturally influenced water contribute differently to the spread of antibiotic resistance genes in a mega-city river network[J]. Water Research, 2019, 158: 11-21. DOI:10.1016/j.watres.2019.03.010 |
| [19] |
Fresia P, Antelo V, Salazar C, et al. Urban metagenomics uncover antibiotic resistance reservoirs in coastal beach and sewage waters[J]. Microbiome, 2019, 7(1): 35. DOI:10.1186/s40168-019-0648-z |
| [20] |
Chen HY, Bai XM, Jing LJ, et al. Characterization of antibiotic resistance genes in the sediments of an urban river revealed by comparative metagenomics analysis[J]. Science of the Total Environment, 2019, 653: 1513-1521. DOI:10.1016/j.scitotenv.2018.11.052 |
| [21] |
Sales-Ortells H, Agostini G, Medema G. Quantification of waterborne pathogens and associated health risks in urban water[J]. Environmental Science & Technology, 2015, 49(11): 6943-6952. |
| [22] |
Arnone RD, Walling JP. Waterborne pathogens in urban watersheds[J]. Journal of Water & Health, 2007, 5(1): 149-162. |
| [23] |
Dong PY, Cui QJ, Fang TT, et al. Occurrence of antibiotic resistance genes and bacterial pathogens in water and sediment in urban recreational water[J]. Journal of Environmental Sciences, 2019, 77: 65-74. DOI:10.1016/j.jes.2018.06.011 |
| [24] |
Zhang HH, He HY, Chen SN, et al. Abundance of antibiotic resistance genes and their association with bacterial communities in activated sludge of wastewater treatment plants: Geographical distribution and network analysis[J]. Journal of Environmental Sciences, 2019, 82: 24-38. DOI:10.1016/j.jes.2019.02.023 |
| [25] |
Zhu YG, Johnson TA, Su JQ, et al. Diverse and abundant antibiotic resistance genes in Chinese swine farms[J]. Proceedings of the National Academy of Sciences of the United States of America, 2013, 110(9): 3435-3440. DOI:10.1073/pnas.1222743110 |
| [26] |
Jiang HY, Zhou RJ, Zhang MD, et al. Exploring the differences of antibiotic resistance genes profiles between river surface water and sediments using metagenomic approach[J]. Ecotoxicology and Environmental Safety, 2018, 161: 64-69. DOI:10.1016/j.ecoenv.2018.05.044 |
| [27] |
Xiao KQ, Li B, Ma LP, et al. Metagenomic profiles of antibiotic resistance genes in paddy soils from South China[J]. FEMS Microbiology Ecology, 2016, 92(3): fiw023. DOI:10.1093/femsec/fiw023 |
| [28] |
Su JQ, An XL, Li B, et al. Metagenomics of urban sewage identifies an extensively shared antibiotic resistome in China[J]. Microbiome, 2017, 5(1): 84. DOI:10.1186/s40168-017-0298-y |
| [29] |
Porra RJ. The chequered history of the development and use of simultaneous equations for the accurate determination of chlorophylls a and b[J]. Photosynthesis Research, 2002, 73(1/3): 149-156. DOI:10.1023/A:1020470224740 |
| [30] |
APHA-AWWA-WEF. Standard Methods for the Examination of Water and Wastewater[M]. 22nd ed. Washington, DC: American Public Health Association, 2012
|
| [31] |
Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND[J]. Nature Methods, 2015, 12(1): 59-60. DOI:10.1038/nmeth.3176 |
| [32] |
Ye L, Zhang T. Pathogenic bacteria in sewage treatment plants as revealed by 454 pyrosequencing[J]. Environmental Science & Technology, 2011, 45(17): 7173-7179. |
| [33] |
Martínez JL, Coque TM, Baquero F. What is a resistance gene? Ranking risk in resistomes[J]. Nature Reviews Microbiology, 2015, 13(2): 116-123. DOI:10.1038/nrmicro3399 |
| [34] |
Jia BF, Raphenya AR, Alcock B, et al. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database[J]. Nucleic Acids Research, 2017, 45(D1): D566-D573. DOI:10.1093/nar/gkw1004 |
| [35] |
McArthur AG, Waglechner N, Nizam F, et al. The comprehensive antibiotic resistance database[J]. Antimicrobial Agents and Chemotherapy, 2013, 57(7): 3348-3357. DOI:10.1128/AAC.00419-13 |
| [36] |
Chen BW, Yang Y, Liang XM, et al. Metagenomic profiles of antibiotic resistance genes (ARGs) between human impacted estuary and deep ocean sediments[J]. Environmental Science & Technology, 2013, 47(22): 12753-12760. |
| [37] |
Hu Q, Zhang XX, Jia SY, et al. Metagenomic insights into ultraviolet disinfection effects on antibiotic resistome in biologically treated wastewater[J]. Water Research, 2016, 101: 309-317. DOI:10.1016/j.watres.2016.05.092 |
| [38] |
Wu X, Xi WY, Ye WJ, et al. Bacterial community composition of a shallow hypertrophic freshwater lake in China, revealed by 16S rRNA gene sequences[J]. FEMS Microbiology Ecology, 2007, 61(1): 85-96. DOI:10.1111/j.1574-6941.2007.00326.x |
| [39] |
Wu L, Ge G, Gong SJ, et al. Diversity and composition of the bacterial community of Poyang Lake (China) as determined by 16S rRNA gene sequence analysis[J]. World Journal of Microbiology and Biotechnology, 2012, 28(1): 233-244. |
| [40] |
Xiong WG, Sun YX, Zhang T, et al. Antibiotics, antibiotic resistance genes, and bacterial community composition in fresh water aquaculture environment in China[J]. Microbial Ecology, 2015, 70(2): 425-432. DOI:10.1007/s00248-015-0583-x |
| [41] |
Fang H, Huang KL, Yu JN, et al. Metagenomic analysis of bacterial communities and antibiotic resistance genes in the Eriocheir sinensis freshwater aquaculture environment[J]. Chemosphere, 2019, 224: 202-211. DOI:10.1016/j.chemosphere.2019.02.068 |
| [42] |
Yan QY, Bi YH, Deng Y, et al. Impacts of the Three Gorges Dam on microbial structure and potential function[J]. Scientific Reports, 2015, 5: 8605. DOI:10.1038/srep08605 |
| [43] |
Karner M, Fuks D, Herndl GJ. Bacterial activity along a trophic gradient[J]. Microbial Ecology, 1992, 24(3): 243-257. DOI:10.1007/BF00167784 |
| [44] |
Jing XY, Gou HL, Gong YH, et al. Seasonal dynamics of the coastal bacterioplankton at intensive fish-farming areas of the Yellow Sea, China revealed by high-throughput sequencing[J]. Marine Pollution Bulletin, 2019, 139: 366-375. DOI:10.1016/j.marpolbul.2018.12.052 |
| [45] |
Chrzanowski TH, Sterner RW, Elser JJ. Nutrient enrichment and nutrient regeneration stimulate bacterioplankton growth[J]. Microbial Ecology, 1995, 29(3): 221-230. DOI:10.1007/BF00164886 |
| [46] |
Lindström ES. Bacterioplankton community composition in five lakes differing in trophic status and humic content[J]. Microbial Ecology, 2000, 40(2): 104-113. DOI:10.1007/s002480000036 |
| [47] |
Moore LR, Post AF, Rocap G, et al. Utilization of different nitrogen sources by the marine cyanobacteria Prochlorococcus and Synechococcus[J]. Limnology and Oceanography, 2002, 47(4): 989-996. DOI:10.4319/lo.2002.47.4.0989 |
| [48] |
Mackey KRM, Paytan A, Caldeira K, et al. Effect of temperature on photosynthesis and growth in marine Synechococcus spp[J]. Plant Physiology, 2013, 163(2): 815-829. |
| [49] |
Salcher MM. Same same but different: ecological niche partitioning of planktonic freshwater prokaryotes[J]. Journal of Limnology, 2014, 73(S1): 74-87. |
| [50] |
Ratnam S, Hogan K, March SB, et al. Whirlpool-associated folliculitis caused by Pseudomonas aeruginosa: report of an outbreak and review[J]. Journal of Clinical Microbiology, 1986, 23(3): 655-659. DOI:10.1128/JCM.23.3.655-659.1986 |
| [51] |
Poole K. Multidrug efflux pumps and antimicrobial resistance in Pseudomonas aeruginosa and related organisms[J]. Journal of Molecular Microbiology and Biotechnology, 2001, 3(2): 255-264. |
| [52] |
Bottone EJ. Bacillus cereus, a volatile human pathogen[J]. Clinical Microbiology Reviews, 2010, 23(2): 382-398. DOI:10.1128/CMR.00073-09 |
| [53] |
Torkar KG, Bedenić B. Antimicrobial susceptibility and characterization of metallo-β-lactamases, extended- spectrum β-lactamases, and carbapenemases of Bacillus cereus isolates[J]. Microbial Pathogenesis, 2018, 118: 140-145. DOI:10.1016/j.micpath.2018.03.026 |
| [54] |
Zheng J, Zhou ZC, Wei YY, et al. High-throughput profiling of seasonal variations of antibiotic resistance gene transport in a peri-urban river[J]. Environment International, 2018, 114: 87-94. DOI:10.1016/j.envint.2018.02.039 |
| [55] |
di Pilato V, Arena F, Tascini C, et al. mcr-1.2, a new mcr variant carried on a transferable plasmid from a colistin-resistant KPC carbapenemase-producing Klebsiella pneumoniae strain of sequence type 512[J]. Antimicrobial Agents and Chemotherapy, 2016, 60(9): 5612-5615. DOI:10.1128/AAC.01075-16 |
| [56] |
Liu YY, Wang Y, Walsh TR, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study[J]. The Lancet Infectious Diseases, 2016, 16(2): 161-168. DOI:10.1016/S1473-3099(15)00424-7 |
| [57] |
Falagas ME, Rafailidis PI, Matthaiou DK. Resistance to polymyxins: mechanisms, frequency and treatment options[J]. Drug Resistance Updates, 2010, 13(4/5): 132-138. |
| [58] |
Poirel L, Jayol A, Nordmann P. Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes[J]. Clinical Microbiology Reviews, 2017, 30(2): 557-596. DOI:10.1128/CMR.00064-16 |
| [59] |
Caltagirone M, Nucleo E, Spalla M, et al. Occurrence of extended spectrum β-lactamases, KPC-type, and MCR-1.2-producing Enterobacteriaceae from wells, river water, and wastewater treatment plants in oltrepò pavese area, Northern Italy[J]. Frontiers in Microbiology, 2017, 8: 2232. DOI:10.3389/fmicb.2017.02232 |
| [60] |
Carfi A, Pares S, Duée E, et al. The 3-D structure of a zinc metallo-beta-lactamase from Bacillus cereus reveals a new type of protein fold[J]. The EMBO Journal, 1995, 14(20): 4914-4921. DOI:10.1002/j.1460-2075.1995.tb00174.x |
| [61] |
Pitout JDD, Sanders CC, Sanders Jr WE. Antimicrobial resistance with focus on β-lactam resistance in gram-negative bacilli[J]. The American Journal of Medicine, 1997, 103(1): 51-59. DOI:10.1016/S0002-9343(97)00044-2 |
| [62] |
Tang JY, Bu YQ, Zhang XX, et al. Metagenomic analysis of bacterial community composition and antibiotic resistance genes in a wastewater treatment plant and its receiving surface water[J]. Ecotoxicology and Environmental Safety, 2016, 132: 260-269. DOI:10.1016/j.ecoenv.2016.06.016 |
| [63] |
Martínez JL. Antibiotics and antibiotic resistance genes in natural environments[J]. Science, 2008, 321(5887): 365-367. DOI:10.1126/science.1159483 |
| [64] |
Zhang T, Yang Y, Pruden A. Effect of temperature on removal of antibiotic resistance genes by anaerobic digestion of activated sludge revealed by metagenomic approach[J]. Applied Microbiology and Biotechnology, 2015, 99(18): 7771-7779. DOI:10.1007/s00253-015-6688-9 |
| [65] |
Wright GD. The antibiotic resistome: the nexus of chemical and genetic diversity[J]. Nature Reviews Microbiology, 2007, 5(3): 175-186. DOI:10.1038/nrmicro1614 |
 2020, Vol. 47
2020, Vol. 47




