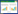扩展功能
文章信息
- 曹敏, 何健, 倪海燕
- CAO Min, HE Jian, NI Hai-Yan
- 二硝基苯胺类除草剂微生物降解研究进展
- Research progress on microbial degradation of dinitroaniline herbicides
- 微生物学通报, 2020, 47(1): 282-294
- Microbiology China, 2020, 47(1): 282-294
- DOI: 10.13344/j.microbiol.china.190213
-
文章历史
- 收稿日期: 2019-03-17
- 接受日期: 2019-06-28
- 网络首发日期: 2019-07-16
2. 南京农业大学生命科学学院 农业部农业环境微生物工程重点开放实验室 江苏 南京 210095
2. Key Laboratory of Microbiological Engineering Agricultural Environment, Ministry of Agriculture; College of Life Sciences, Nanjing Agricultural University, Nanjing, Jiangsu 210095, China
我国是农业生产和农产品出口贸易大国,农业的可持续发展关乎国民经济的健康、持续发展。在保证粮食增产的前提下,除草剂的使用量也随着大幅度增高。二硝基苯胺类除草剂主要包括氟乐灵、二甲戊灵、仲丁灵、安磺灵(图 1)等,是一类广谱、高效的选择性芽前土壤处理剂,具有高效低毒、除草谱广和持效期长等优点,在化学除草剂应用中占很大比重[1]。该类除草剂被广泛应用于棉花、花生、大豆、小麦、水稻、玉米等多种作物农田中,防除一年生禾本科杂草和部分一年生阔叶杂草[2-3],其除草机理是通过抑制细胞的有丝分裂与分化,破坏细胞的正常分裂,从而造成杂草的死亡[4]。二硝基苯胺类除草剂的纯品多为橘黄色晶体,结构通式为2, 6-二硝基苯胺,在我国使用的代表产品是二甲戊灵、氟乐灵和仲丁灵。

|
| 图 1 二硝基苯胺类除草剂主要品种的化学结构式 Figure 1 Chemical structures of the major products of dinitroaniline herbicides |
|
|
二硝基苯胺类除草剂化学性质稳定,残留期长[5-8],易挥发,易进入环境中,已成为一类环境污染物[9-12],对生态环境和人类健康造成严重威胁。二硝基苯胺类除草剂对水生生物和无脊椎动物高毒[5, 13-19],因含二硝基苯胺结构可形成致癌物质——亚硝胺,对人体健康也有潜在危害[20-21]。此外,氟乐灵对某些后茬作物存在危害[17],造成内分泌紊乱[17];对哺乳动物细胞有基因毒性,其致癌性被认定为C类,即可疑的人类致癌剂[19, 22-25]。美国国家环境保护局(US Environmental Protection Agency,USEPA)已将二甲戊灵和氟乐灵归类为持久性生物积聚有毒物质[12]。欧洲一些国家已对氟乐灵采取禁用或限用的措施[22, 26-29],但由于性价比较高,它们在中国仍是首选的除草剂。因此,二硝基苯胺类除草剂在生态环境中迁移、转化、降解以及生态安全性评估已引起了广泛关注。
二硝基苯胺类除草剂在环境中的消解方式主要有挥发、光解和微生物降解[30-32],大量研究表明微生物的降解代谢在其中起主要作用[33-36]。本文简述目前国内外关于二硝基苯胺类除草剂微生物降解菌株、降解代谢途径以及降解基因和酶方面的研究进展,旨在为二硝基苯胺类除草剂残留的微生物降解修复提供理论依据和优良的降解菌株、基因及酶资源。
1 二硝基苯胺类除草剂微生物降解菌株大量研究表明微生物的降解代谢作用是环境中二硝基苯胺除草剂消解的主要因素,目前国内外已筛选到很多能降解该类除草剂的纯品系菌株,这些降解菌株涉及真菌、细菌和放线菌,种属分布非常广泛,具有丰富的多样性(表 1)。目前分离筛选到的二硝基苯胺除草剂微生物降解菌株绝大多数是降解二甲戊灵和氟乐灵的菌株,而已报道的仲丁灵和安磺灵的纯品系降解菌株非常少;具有降解活性的细菌主要来源于芽孢杆菌属(Bacillus sp.)和假单胞菌属(Pseudomonas sp.),具有降解活性的真菌主要是曲霉属(Aspergillus sp.)菌株和尖孢镰刀菌(Fusarium oxysporum),具有降解活性的放线菌目前只有二甲戊灵降解菌株Actinomycetes sp. 42。
| 降解菌株 Degrading strains |
降解底物 Degrading substrates |
来源 References |
|
| 细菌Bacteria | Pseudomonas sp. | Trifluralin | [37-38] |
| Bacteroides ruminicola subsp. brevis GA-33 | Trifluralin | [39] | |
| Azotobacter vinelandii | Pendimethalin | [40] | |
| Alcaligenes sp. T-a Moraxella sp. T-b |
Trifluralin | [41] | |
| Azotobacter chroococcum | Pendimethalin | [42] | |
| Bacillus megaterium | Pendimethalin | [43] | |
| Pseudomonas sp. | |||
| Rhizoctinia sp. | |||
| Rhizoctinia bataticola | |||
| Pyricularia oryzae Cav | |||
| Herbaspirillum sp. Bacillus sp. Klebsiella sp. |
Trifluralin | [44] | |
| Bacillus sp. HB-7 | Pendimethalin | [45] | |
| Bacillus circulans | Pendimethalin | [46] | |
| Pseudomonas aeruginosa | Pendimethalin | [47] | |
| Bacillus mycoides | |||
| Bacillus cereus | |||
| Bacillus megaterium E22 | Pendimethalin | [48] | |
| Pseudomonas putida E15 | Pendimethalin | [49] | |
| Bacillus lehensis XJU | Pendimethalin | [50] | |
| Shewanella marisflavi EP1 | Pendimethalin | [51] | |
| Leucobacter sp. FJ-01 | Trifluralin | [52] | |
| Bacillus subtilis Y3 | Pendimethalin, Trifluralin, Butralin, Oryzalin | [53-54] | |
| Pseudomonas resinovorans E20 | Pendimethalin | [55] | |
| Bacillus simplex Bacillus muralis Micrococcus luteus Micrococcus yunnanensis Clostridium tetani |
Trifluralin | [34] | |
| Bacillus safensis FO-36b | Pendimethalin | [56] | |
| Bacullus subilis subsp. inaquosorum KCTC13429 | |||
| Bacillus cereus ATCC14579 | |||
| Bacillus sp. D8 | Trifluralin | [57] | |
| Serratia sp. JY-2 | Pendimethalin | [58-59] | |
| Pseudomonas sp. JY-5 | |||
| Bacillus cereus | Pendimethalin | [60] | |
| Asaccharospora irregularis | |||
| Paracoccum sp. P13 | Pendimethalin | [61] | |
| 放线菌Actinomycetes | Actinomycetes sp. 42 | Pendimethalin | [43] |
| 真菌Fungi | Aspergillus niger | Trifluralin | [62] |
| Lachnospira multiparus D-32 | Trifluralin | [39] | |
| Paecilomyces sp. | Trifluralin, Butralin | [63-64] | |
| Candida sp. | Trifluralin | [65] | |
| Aspergillus carneus Fusarium oxysporum Trichoderma viride |
Trifluralin | [66] | |
| Aspergillus flavus Aspergillus terreus Fusarium solani Fusarium oxysporum Penicillium citrinum Penicillium simplicissimum |
Pendimethalin | [67] | |
| Fusarium oxysporum | Pendimethalin | [68] | |
| Paecilomyces variotii | |||
| Rhizoctonia bataticola | |||
| Aspergillus niger 2012 | Pendimethalin | [43] | |
| Fusarium oxysporum | |||
| Paecilomyces variotii | |||
| Aspergillus terreus YF-209 | Pendimethalin | [69] | |
| Monilochaetes sp. YF-212 | |||
| Aspergillus furnigatus YF-216 | |||
| Fusarium oxysporum Aspergillus oryzae Lentinula edodes Penicillium brevicompactum Lecanicillium saksenae |
Pendimethalin | [70] | |
| Phanerochaete chrysosporium | Pendimethalin | [71] | |
| Aspergillus tubingensis Qsun-6 | Pendimethalin | [72] | |
| Penicillium thrichoderma Penicillium simplicissimum Penicillium talaromyces Metacordyceps chlamydosporia Stachybotrys chartarum Alternia alternata |
Trifluralin | [34] | |
| Lentinula edodes EL1 | Pendimethalin | [35] | |
| Clavispora lusitaniae YC2 | Pendimethalin | [73] |
在表 1的二硝基苯胺除草剂降解细菌菌株中,朱鲁生等以二甲戊灵为唯一碳源选择性富集筛选二甲戊灵降解菌株,并在转接培养的同时不断提高培养基中的二甲戊灵浓度加强筛选效果,最终得到一株二甲戊灵降解菌株HB-7,鉴定为芽孢杆菌属(Bacillus sp.);不同的培养条件会影响菌株的生长和降解效率,在最适pH 7.0和30 ℃的条件下,再外源添加少量葡萄糖(0.1%)作为碳源可以提高菌株降解二甲戊灵的效率,24 h内能降解90.0%以上的100 mg/L二甲戊灵[45]。Ni等从活性污泥中通过选择富集的方法分离筛选到一株二甲戊灵降解菌株——枯草芽孢杆菌(Bacillus subtilis) Y3,并通过定时取样的方法对该菌降解二甲戊灵的效果进行监测,该菌能在60 h内降解100 mg/L二甲戊灵,也能在相同时间内降解100 mg/L的仲丁灵和安磺灵以及60 mg/L氟乐灵[53-54]。陈贝贝等以氟乐灵为唯一碳源从连作15年以上的棉田土壤中选择分离到一株氟乐灵降解菌株D8,鉴定为芽孢杆菌(Bacillus sp.),并通过单因素实验对菌株D8降解氟乐灵的降解特性进行研究,菌株D8在氟乐灵初始浓度为50 mg/L、接种量4%、外加氮源(酵母浸粉) 0.1%、pH 5.0、培养温度37 ℃的条件下,3 d对氟乐灵的降解率达到59.9%以上[57]。胡佳月等采用以蛋白胨为氮源、二甲戊灵和少量葡萄糖为碳源的富集培养基,从长期施用二甲戊灵的棉田土壤中富集筛选能够降解二甲戊灵的菌株,并通过不断提高培养基中的二甲戊灵浓度来转接培养进一步选择筛选,最终得到2株二甲戊灵降解菌株——嗜线虫沙雷氏菌(Serratia sp.) JY-2和假单胞菌(Pseudomonas sp.) JY-5,对这2株菌的降解特性进行了研究,结果表明这2株菌在最佳条件(即外加碳源量0.5%、菌株接种量10%、pH 7.0、培养温度30 ℃、初始二甲戊灵浓度200 mg/L)下作用3 d对二甲戊灵的降解率能分别达到83.3%和82.8%[58-59]。Archana等用以二甲戊灵为唯一碳源的选择培养基从受农药污染的土壤中富集筛选到的2株二甲戊灵降解细菌,通过菌株形态、生理生化特征和16S rRNA基因序列将这2株菌鉴定为蜡样芽孢杆菌(Bacillus cereus)和不规则梭菌(Asaccharospora irregularis),这2株细菌能在二甲戊灵初始浓度为500 mg/L的条件下生长,每隔7 d对这2株菌降解二甲戊灵的情况进行测定,结果表明这2株细菌在28 d内分别能降解408 mg/L和422 mg/L二甲戊灵[60]。Ni等通过富集培养的方法从果园土壤中分离到的一株二甲戊灵降解菌株——副球菌属(Pacacoccus sp.) P13,它能以二甲戊灵为唯一碳源和能源生长,2 d能降解100 mg/L二甲戊灵,5 d能降解200 mg/L二甲戊灵[61]。
在已公开报道的二硝基苯胺类除草剂降解真菌菌株中,Zayed等测定了埃及土壤中3个主要真菌类群肉色曲霉(Aspergillus carneus)、尖孢镰刀菌(Fusarium oxysporum)和绿色木霉(Trichoderma virida)降解转化氟乐灵的能力,并对氟乐灵的转化产物进行了鉴定,这3株菌10 d内对初始浓度200 mg/L氟乐灵的转化率均高于90.0%,其中绿色木霉(T. virida)对氟乐灵的转化率最高,高达97.0%[66]。林爱军等通过以二甲戊灵为唯一碳源的无机盐培养基对污泥中的微生物进行富集培养,并不断提高二甲戊灵的浓度进行转接培养,最终分离筛选到多株二甲戊灵降解真菌菌株,选取了其中降解效率较高且对二甲戊灵耐受性较强的3株真菌——土生曲霉(Aspergillus terreus) YF-209、长梗串孢霉属(Monilochaetes sp.) YF-212和烟色曲霉(Aspergillus furnigatus) YF-216进行了降解特性的测定,结果表明不同的培养条件对微生物降解二甲戊灵的效果有较大影响,但这3株真菌在外加蔗糖浓度为0.5%-1.0%、二甲戊灵初始浓度为100 mg/L、pH 6.0-8.0、温度20-30 ℃的条件下,菌株的生长量和对二甲戊灵的降解量都能达到最大,这3株真菌YF-209、YF-212和YF-216在5 d内对100 mg/L二甲戊灵的降解率分别为66.5%、78.4%和71.3%[69]。宫宏琨等以加入适量二甲戊灵的察氏培养基为富集培养基,以富集培养基中加入100 mg/L二甲戊灵开始富集筛选二甲戊灵降解菌株,并在转接培养过程中逐步提高富集培养基中二甲戊灵的浓度,最终达到5 000 mg/L,稀释涂布获取单菌落,并将各个不同的单菌落转接到以二甲戊灵为唯一碳源的选择培养基中进一步纯化,同时在转接培养时也不断提高二甲戊灵的浓度,最终结合菌株降解实验得到一株二甲戊灵降解真菌Qsun-6,被鉴定为塔宾曲霉(Aspergillus tubingensis),该菌株在5 d内对100 mg/L二甲戊灵的降解率能达到75.2%,对菌株Qsun-6降解二甲戊灵的特性进行研究,发现在外源添加适量速效碳源(如1%蔗糖)、二甲戊灵浓度为100 mg/L、pH 6.0和30 ℃的条件下,菌株的生长量和对二甲戊灵的降解率都能达到最大[72]。Han等以二甲戊灵为碳源的Czapek培养基为富集培养基,从连续15年施用二甲戊灵的棉田土中分离筛选二甲戊灵降解菌株,同样在富集的过程中不断提高培养基中的二甲戊灵浓度,最终得到一株二甲戊灵降解菌株YC2,被鉴定为葡萄牙棒孢酵母(Clavispora lusitaniae),该酵母在5 d内能降解60.0%以上的200 mg/L二甲戊灵,8 d的降解率能达到74.0%[73]。
2 二硝基苯胺类除草剂的微生物降解途径目前国内外分离到许多株二硝基苯胺类除草剂降解菌株,研究了菌株的降解特性,通过液相色谱、气相色谱、质谱和核磁共振等技术鉴定了降解过程中产生的代谢产物,并根据鉴定到的代谢产物结构分析了微生物降解该类除草剂的代谢途径。研究表明各种微生物菌株降解二甲戊灵、氟乐灵、仲丁灵和安磺灵的方式主要包括硝基还原、氧化N端脱烷基、环化生成苯并咪唑、芳甲基氧化等[39, 41-43, 46-47, 50, 53-54, 62, 65-68],各个微生物菌株利用二硝基苯胺类除草剂的方式不尽相同,这也造成了二甲戊灵、氟乐灵等除草剂的代谢产物多种多样。纵观现有公开发表的研究成果,二硝基苯胺除草剂的微生物降解途径研究往往停滞于降解过程的前3步(即降解的上游代谢途径),而完整的矿化途径尚无研究报道。
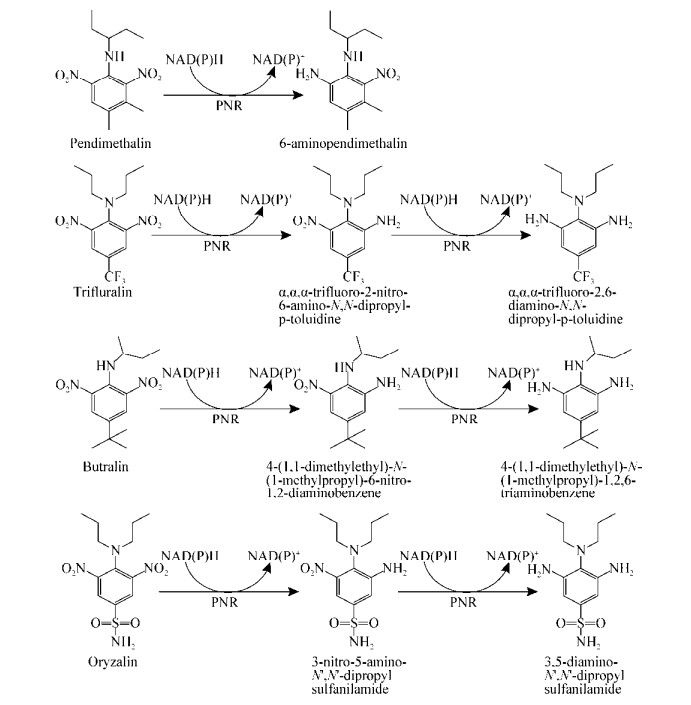
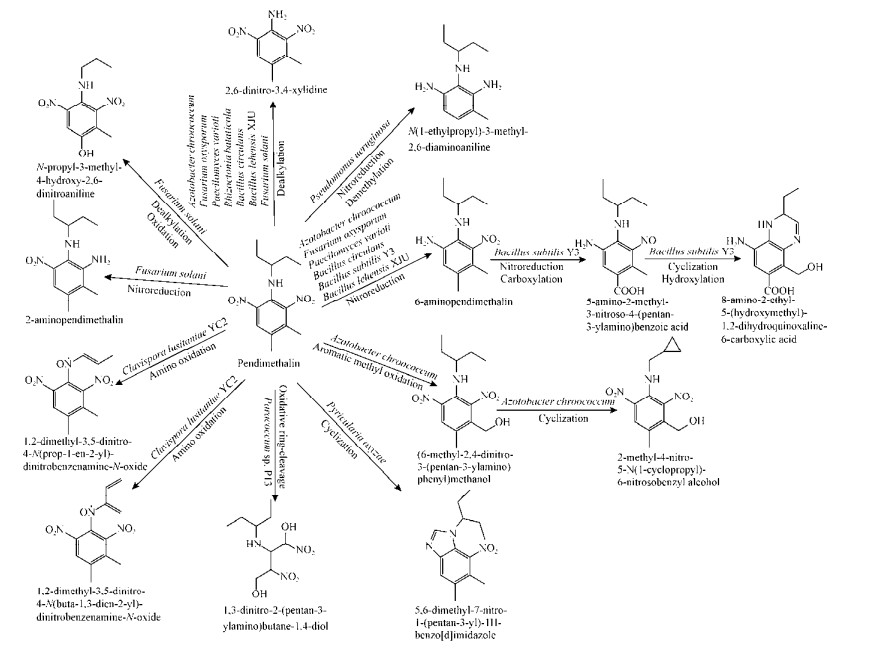
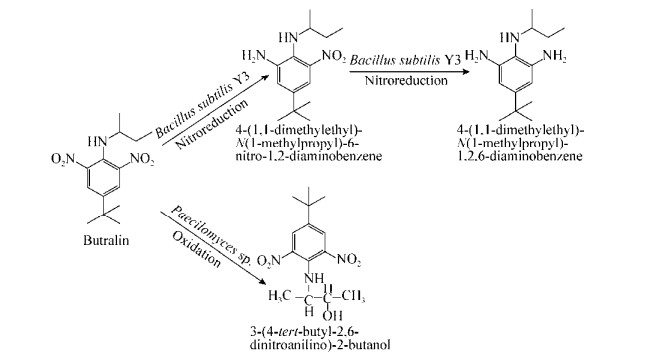
已公开报道的微生物降解二甲戊灵、氟乐灵、仲丁灵和安磺灵的部分代谢途径分别见图 2-5。其中,B. subtilis Y3能以硝基还原的方式起始降解二甲戊灵生成6-氨基二甲戊灵[N-(1-乙基丙基)-2-硝基-6-氨基-3, 4-二甲苯胺],进而又通过硝基还原作用和羧化作用生成5-氨基-2-甲基-3-亚硝基-4-(戊烷基-3-氨基)-苯甲酸,随后发生环化和羟基化作用得到8-氨基-2-乙基-5-(羟甲基)-1, 2-二氢喹喔啉-6-羧酸[53]。菌株Y3还能连续催化还原氟乐灵、仲丁灵和安磺灵的2个对称的硝基基团[54]。此外,褐球固氮菌(Azotobacter chroococcum)、尖孢镰刀菌(F. oxysporum)、环状芽孢杆菌(B. circulans)和芽孢杆菌(Bacillus lehensis) XJU均能分别以N端氧化脱烷基和硝基还原的方式开始降解二甲戊灵[42-43, 46, 50]。在氟乐灵的微生物降解中,栖瘤胃拟杆菌短亚种(Bacteroides ruminicola subsp. brevis) GA-33和多对毛螺菌(Lachnospira multiparus) D-32也能分别通过N端氧化脱烷基和硝基还原两种方式开始降解氟乐灵[39]。此外,在产碱杆菌属菌株(Alcaligenes sp.) T-a和莫拉菌属菌株(Moraxella sp.) T-b降解氟乐灵的过程中,不仅检测到了硝基还原产物和脱烷基产物,也检测到了环化产物苯并咪唑,说明这2株菌可以通过多种途径代谢氟乐灵[41]。
|
| 图 2 推测的二甲戊灵微生物降解途径 Figure 2 Proposed metabolic pathways of pendimethalin by pure isolates |
|
|

|
| 图 3 推测的氟乐灵微生物降解途径 Figure 3 Proposed metabolic pathways of trifluralin by pure isolates |
|
|
|
| 图 4 推测的仲丁灵微生物降解途径 Figure 4 Proposed metabolic pathways of butralin by pure isolates |
|
|

|
| 图 5 推测的B. subtilis Y3降解安磺灵的部分代谢途径 Figure 5 Proposed partial metabolic pathway of oryzalin by B. subtilis Y3 |
|
|
二硝基苯胺类除草剂的微生物降解菌株资源在不断地丰富,这也促进了微生物降解该类除草剂分子机制的研究,但目前该类除草剂的降解基因和酶方面的研究报道还是很少。2004年,朱鲁生等采用碱裂解法从二甲戊灵降解菌株Bacillus sp. HB-7中提取到一个大小约为20 kb的质粒,该质粒可以产生能够降解二甲戊灵的酶系[45];同年,Bellinaso等通过PCR和DNA探针杂交技术在4株氟乐灵降解菌株(Klebsiella oxytoca、Herbaspirillum seropedicae和2株Bacillus megaterium)中均克隆到了nodB (编码萘1, 2-双加氧酶)相似基因,说明在这4株菌中都可能有一个双加氧酶参与氟乐灵的代谢,但是还需要进一步的测序和蛋白质组学等实验证明[74]。2015年,More等通过蛋白纯化技术从环状芽孢杆菌(B. circulans)中纯化到了二甲戊灵硝基还原酶,该酶单体分子量为27 kD,需要NADPH作为辅因子,能催化还原二甲戊灵、氟乐灵、地乐胺等二硝基苯胺除草剂,也能还原对硝基苯胺、2, 4, 6-三硝基甲苯(TNT)、2, 4-二硝基甲苯和2-硝基苯甲酸等硝基芳烃化合物,但对二甲戊灵和氟乐灵具有更高的催化活性[75]。2016年,Ni等通过降解酶蛋白纯化、肽指纹图谱分析和基因组学分析等方法从菌株B. subtilis Y3中克隆到负责二甲戊灵第一步降解的硝基还原酶基因pnr,并通过降解酶催化产物纯化和核磁共振技术鉴定到PNR还原二甲戊灵的产物为6-氨基二甲戊灵,PNR还能连续催化还原氟乐灵、仲丁灵和安磺灵的2个对称的硝基基团(图 6)[54]。
二硝基苯胺类除草剂是一类广谱高效且广泛使用的选择性除草剂,其在环境中的残留对生态环境和人类健康都具有潜在危害,微生物因其数量、种类和代谢类型的多样性以及适应性强等特性而在二硝基苯胺类除草剂的降解中占据至关重要的位置。目前国内外已经积累了许多二硝基苯胺类除草剂的降解资源,但是当前对该类除草剂的微生物降解研究还比较浅。例如:(1)菌株的降解特性研究较少;(2)高效的降解菌株资源较少;(3)现有的除草剂降解途径绝大多数是基于中间代谢产物的鉴定分析推测而来的,而且降解途径的研究往往止步于降解转化的前3步,该类除草剂在环境的矿化并没有研究清楚;(4)虽然脱烷基和硝基还原是该类除草剂微生物降解的主要方式,但是研究发现还存在很多其他的降解方式(图 2-3),对于这些降解过程中得到的降解产物是不是具有生态毒性还需要进一步验证;(5)降解基因和酶的研究进展缓慢,缺乏对菌株降解机制的深入和全面研究等,从而限制了对该类除草剂残留污染修复的降解菌剂、降解基因和酶资源的开发和利用。
除草剂降解/脱毒酶基因在抗除草剂转基因工程领域具有潜在的应用价值,而我国目前还缺乏具有自主知识产权、性能优良的除草剂降解/脱毒酶基因。抗除草剂转基因工程可以极有效地控制农田杂草,其中最为成功的是抗草甘膦转基因作物,已经大量种植。但长期使用草甘膦带来严重的杂草抗性问题,急需发掘其他除草剂降解/脱毒酶基因,为研发新的抗除草剂转基因作物提供基因资源。二硝基苯胺类除草剂是一类广泛使用、杀草谱较广的除草剂,可作为抗除草剂转基因工程的靶标除草剂,因此它的降解/脱毒酶基因在构建抗除草剂作物中具有良好的应用前景,而要构建具备完善降解体系的抗二硝基苯胺类除草剂转基因作物,就迫切需要进一步探索除草剂的矿化过程,全面深入解析其降解机制。
| [1] |
Gong WW, Liu XH, Xia SH, et al. Abiotic reduction of trifluralin and pendimethalin by sulfides in black-carbon-amended coastal sediments[J]. Journal of Hazardous Materials, 2016, 310: 125-134. DOI:10.1016/j.jhazmat.2016.02.022 |
| [2] |
Muñoz A, Vera T, Ródenas M, et al. Gas-phase degradation of the herbicide ethalfluralin under atmospheric conditions[J]. Chemosphere, 2014, 95: 395-401. DOI:10.1016/j.chemosphere.2013.09.053 |
| [3] |
Epp JB, Schmitzer PR, Crouse GD. Fifty years of herbicide research: comparing the discovery of trifluralin and halauxifen-methyl[J]. Pest Management Science, 2018, 74(1): 9-16. DOI:10.1002/ps.4657 |
| [4] |
Tomlin CDS. The Pesticide Manual[M]. 13th ed. Alton, UK: British Crop Protection Council, 2003: 752-753
|
| [5] |
Helling CS. Dinitroaniline herbicides in soils[J]. Journal of Environmental Quality, 1976, 5(1): 1-15. |
| [6] |
Wheeler WB, Stratton GD, Twilley RR, et al. Trifluralin degradation and binding in soil[J]. Journal of Agricultural and Food Chemistry, 1979, 27(4): 702-706. DOI:10.1021/jf60224a019 |
| [7] |
Huang B, Li J, Fang WS, et al. Effect of soil fumigation on degradation of pendimethalin and oxyfluorfen in laboratory and ginger field studies[J]. Journal of Agricultural and Food Chemistry, 2016, 64(46): 8710-8721. DOI:10.1021/acs.jafc.6b01437 |
| [8] |
Rodríguez-Liébana JA, ElGouzi S, Peña A. Laboratory persistence in soil of thiacloprid, pendimethalin and fenarimol incubated with treated wastewater and dissolved organic matter solutions. Contribution of soil biota[J]. Chemosphere, 2017, 181: 508-517. DOI:10.1016/j.chemosphere.2017.04.111 |
| [9] |
Barbash JE, Resek EA. Pesticides in Ground Water: Distribution, Trends, and Governing Factors[M]. Chelsea, Michigan: Ann Arbor Press, 1996.
|
| [10] |
Warren N, Allan IJ, Carter JE, et al. Pesticides and other micro-organic contaminants in freshwater sedimentary environments — a review[J]. Applied Geochemistry, 2003, 18(2): 159-194. DOI:10.1016/S0883-2927(02)00159-2 |
| [11] |
Konstantinou IK, Hela DG, Albanis TA. The status of pesticide pollution in surface waters (rivers and lakes) of Greece. Part Ⅰ. Review on occurrence and levels[J]. Environmental Pollution, 2006, 141(3): 555-570. DOI:10.1016/j.envpol.2005.07.024 |
| [12] |
Palma P, Kuster M, Alvarenga P, et al. Risk assessment of representative and priority pesticides, in surface water of the Alqueva reservoir (South of Portugal) using on-line solid phase extraction-liquid chromatography-tandem mass spectrometry[J]. Environment International, 2009, 35(3): 545-551. DOI:10.1016/j.envint.2008.09.015 |
| [13] |
Fliedner A. Ecotoxicity of poorly water-soluble substances[J]. Chemosphere, 1997, 35(1/2): 295-305. |
| [14] |
Hou LF, Lee WJ, Rusiecki J, et al. Pendimethalin exposure and cancer risk among pesticide applicators: a report from the U.S.-based agricultural health study[J]. Annals of Epidemiology, 2004, 14(8): 608. |
| [15] |
Emmerson J, Pierce E, McGrath J. The chronic toxicity of compound 36352 (trifluralin) given as a component of the diet to Fischer 344 rats for two years[R]. Studies R-87 and R-97. Indianapolis, IN: Elanco Products Co., Eli Lilly and Co., 1980
|
| [16] |
Naqvi SM, Leung TS. Trifluralin and oryzalin herbicides toxicities to juvenile crawfish (Procambarus clarkii) and mosquitofish (Gambusia affinis)[J]. Bulletin of Environmental Contamination and Toxicology, 1983, 31(3): 304-308. DOI:10.1007/BF01608703 |
| [17] |
U.S. EPA (U.S. Environmental Protection Agency). Persistence, bioaccumulative, and toxic (PBT) profiler model suite[R]. 2006. Available: http://www.epa.gov/oppt/pbtprofiler
|
| [18] |
Székács A, Trummer N, Adányi N, et al. Development of a non-labeled immunosensor for the herbicide trifluralin via optical waveguide lightmode spectroscopic detection[J]. Analytica Chimica Acta, 2003, 487(1): 31-42. DOI:10.1016/S0003-2670(03)00302-7 |
| [19] |
Könen S, Çavaş T. Genotoxicity testing of the herbicide trifluralin and its commercial formulation Treflan using the piscine micronucleus test[J]. Environmental and Molecular Mutagenesis, 2008, 49(6): 434-438. DOI:10.1002/em.20401 |
| [20] |
Ritter L, Solomon KR, Forget J, et al. An assessment report on: DDT-Aldrin-Dieldrin-Endrin-Chlordane heptachlor- hexachlorobenzene mirex-toxaphene polychlorinated biphenyls dioxins and furans[R]//International Programme on Chemical Safety (IPCS)[M]. IOMC, 1995: 1-43
|
| [21] |
Dimitrov BD, Gadeva PG, Benova DK, et al. Comparative genotoxicity of the herbicides Roundup, Stomp and Reglone in plant and mammalian test systems[J]. Mutagenesis, 2006, 21(6): 375-382. DOI:10.1093/mutage/gel044 |
| [22] |
Chen QY, Zheng WJ, Xiao YB, et al. Determination of three dinitroaniline herbicide in fruits and vegetables using MSPD-GC-MS/MS[J]. Food Research and Development, 2011, 32(9): 155-158. (in Chinese) 陈其勇, 郑文杰, 肖亚兵, 等. 果蔬中二甲戊乐灵、氟乐灵、双丁乐灵残留测定[J]. 食品研究与开发, 2011, 32(9): 155-158. DOI:10.3969/j.issn.1005-6521.2011.09.047 |
| [23] |
Ribas G, Frenzilli G, Barale R, et al. Herbicide-induced DNA damage in human lymphocytes evaluated by the single-cell gel electrophoresis (SCGE) assay[J]. Mutation Research/Genetic Toxicology, 1995, 344(1/2): 41-54. |
| [24] |
Greene SA, Pohanish RP. Sittig's Handbook of Pesticides and Agricultural Chemicals[M]. 2nd edition. Norwich: William Andrew Publishing, N.Y., USA, 20052005
|
| [25] |
Saghir SA, Charles GD, Bartels MJ, et al. Mechanism of trifluralin-induced thyroid tumors in rats[J]. Toxicology Letters, 2008, 180(1): 38-45. DOI:10.1016/j.toxlet.2008.05.019 |
| [26] |
Du LT, Wu HQ, Yang XF, et al. The effects of trifluralin of liver and renal microsomal enzymes[J]. Chinese Occupational Medicine, 2000, 27(5): 16-18. (in Chinese) 杜柳涛, 邬惠琼, 杨杏芬, 等. 氟乐灵对大鼠肝、肾微粒体酶的影响[J]. 中国职业医学, 2000, 27(5): 16-18. DOI:10.3969/j.issn.1000-6486.2000.05.007 |
| [27] |
Zhang ZC, Zeng M, Li ZY, et al. A matrix solid-phase dispersion method using AgNO3-loaded basic alumina as solid-phase for determination of pendimethalin in garlic[J]. Chinese Journal of Pesticide Science, 2005, 7(3): 264-268. (in Chinese) 张智超, 曾敏, 李朝阳, 等. 以涂敷硝酸银的碱性氧化铝为分散剂的基质固相分散法测定大蒜中二甲戊灵残留[J]. 农药学学报, 2005, 7(3): 264-268. DOI:10.3321/j.issn:1008-7303.2005.03.013 |
| [28] |
Huo JL, Li J, Ge YQ, et al. Research progress on determination methodology for dinitroaniline herbicide residues[J]. Agrochemicals, 2006, 45(4): 222-226. (in Chinese) 霍江莲, 李军, 葛毅强, 等. 二硝基苯胺类除草剂残留检测技术的研究进展[J]. 农药, 2006, 45(4): 222-226. DOI:10.3969/j.issn.1006-0413.2006.04.002 |
| [29] |
Alshallash KS. Effect of pendimethalin, trifluralin and terbutryn on Lolium multiflorum growing with barley during pre-emergence stage[J]. Annals of Agricultural Sciences, 2014, 59(2): 239-242. DOI:10.1016/j.aoas.2014.11.012 |
| [30] |
Behin J, Farhadian N. Response surface methodology for ozonation of trifluralin using advanced oxidation processes in an airlift photoreactor[J]. Applied Water Science, 2017, 7(6): 3103-3112. DOI:10.1007/s13201-016-0443-y |
| [31] |
Pramanik SK, Joarder S, Das S, et al. Photodegradation of oryzalin in aqueous isopropanol and acetonitrile[J]. Journal of Environmental Science and Health, Part B, 2016, 51(5): 287-297. DOI:10.1080/03601234.2015.1128741 |
| [32] |
Chelme-Ayala P, El-Din MG, Smith DW. Degradation of bromoxynil and trifluralin in natural water by direct photolysis and UV plus H2O2 advanced oxidation process[J]. Water Research, 2010, 44(7): 2221-2228. DOI:10.1016/j.watres.2009.12.045 |
| [33] |
Mohan SV, Krishna MR, Muralikrishna P, et al. Solid phase bioremediation of pendimethalin in contaminated soil and evaluation of leaching potential[J]. Bioresource Technology, 2007, 98(15): 2905-2910. DOI:10.1016/j.biortech.2006.09.049 |
| [34] |
Erguven GO, Bayhan H, Ikizoglu B, et al. The capacity of some newly bacteria and fungi for biodegradation of herbicide trifluralin under agiated culture media[J]. Cellular and Molecular Biology, 2016, 62(6): 74-79. |
| [35] |
Pinto AP, Rodrigues SC, Caldeira AT, et al. Exploring the potential of novel biomixtures and Lentinula edodes fungus for the degradation of selected pesticides. Evaluation for use in biobed systems[J]. Science of the Total Environment, 2016, 541: 1372-1381. DOI:10.1016/j.scitotenv.2015.10.046 |
| [36] |
Huang X, He J, Yan X, et al. Microbial catabolism of chemical herbicides: microbial resources, metabolic pathways and catabolic genes[J]. Pesticide Biochemistry and Physiology, 2017, 143: 272-297. DOI:10.1016/j.pestbp.2016.11.010 |
| [37] |
Hamdi YA, Tewfik MS. Decomposition of the herbicide trifluralin by a pseudomonad[J]. Acta Microbiologica Polonica, Series B: Microbiologia Applicate, 1969, 1(2): 83-84. |
| [38] |
Carter GE, Camper ND. Soil enrichment studies with trifluralin[J]. Weed Science, 1975, 23(1): 71-74. DOI:10.1017/S004317450006255X |
| [39] |
Williams PP, Feil VJ. Identification of trifluralin metabolites from rumen microbial cultures. Effect of trifluralin on bacteria and protozoa[J]. Journal of Agricultural and Food Chemistry, 1971, 19(6): 1198-1204. DOI:10.1021/jf60178a033 |
| [40] |
Saha J, Chowdhury A, Chaudhuri S. Stimulation of heterotrophic dinitrogen fixation in barley root association by the herbicide pendimethalin and its metabolic transformation by Azotobacter SPP[J]. Soil Biology and Biochemistry, 1991, 23(6): 569-573. DOI:10.1016/0038-0717(91)90114-Y |
| [41] |
Sato Y. Degradation of trifluralin by bacteria isolated from soil[J]. Weed Research, Japan, 1992, 37(3): 213-219. |
| [42] |
Kole RK, Saha J, Pal S, et al. Bacterial degradation of the herbicide pendimethalin and activity evaluation of its metabolites[J]. Bulletin of Environmental Contamination and Toxicology, 1994, 52(5): 779-786. |
| [43] |
Kulshrestha G, Singh SB, Lal SP, et al. Effect of long-term field application of pendimethalin: enhanced degradation in soil[J]. Pest Management Science, 2000, 56(2): 202-206. DOI:10.1002/(SICI)1526-4998(200002)56:2<202::AID-PS97>3.0.CO;2-C |
| [44] |
de Lourdes Bellinaso M, Greer CW, do Carmo Peralba M, et al. Biodegradation of the herbicide trifluralin by bacteria isolated from soil[J]. FEMS Microbiology Ecology, 2003, 43(2): 191-194. DOI:10.1111/j.1574-6941.2003.tb01058.x |
| [45] |
Zhu LS, Lin AJ, Wang J, et al. Isolation of pendimethalin degrading bacteria HB-7 and its research of degrading characteristics[J]. Acta Scientiae Circumstantiae, 2004, 24(2): 360-365. (in Chinese) 朱鲁生, 林爱军, 王军, 等. 二甲戊乐灵降解细菌HB-7的分离及降解特性研究[J]. 环境科学学报, 2004, 24(2): 360-365. DOI:10.3321/j.issn:0253-2468.2004.02.033 |
| [46] |
Megadi VB, Tallur PN, Hoskeri RS, et al. Biodegradation of pendimethalin by Bacillus circulans[J]. Indian Journal of Biotechnology, 2010, 9(2): 173-177. |
| [47] |
Sharef IB, Abdelbagi AO, Elsheikh EAE, et al. Biodegradation of pendimethalin by three strains of bacteria isolated from pesticide-polluted soils[J]. University of Khartoum Journal of Agricultural Sciences, 2013, 21(2): 233-252. |
| [48] |
Belal EB, Hassan NE. Dissipation of pendimethalin by Bacillus megaterium[J]. Mansoura Journal of Plant Protection and Pathology, 2013, 5: 463-472. |
| [49] |
Elsayed BB, El-Nady MF. Bioremediation of pendimethalin-contaminated soil[J]. African Journal of Microbiology Research, 2013, 7(21): 2574-2588. DOI:10.5897/AJMR12.1919 |
| [50] |
More VS, Tallur PN, Niyonzima FN, et al. Enhanced degradation of pendimethalin by immobilized cells of Bacillus lehensis XJU[J]. 3 Biotech, 2015, 5(6): 967-974. DOI:10.1007/s13205-015-0299-0 |
| [51] |
Ning GJ, Li FL, Huang JX. Reductive biotransformation of pendimethalin by Shewanella marisflavi EP1 under anaerobic conditions[J]. Environmental Science & Technology, 2015, 38(2): 19-24. (in Chinese) 宁国静, 李非里, 黄杰勋. 厌氧下Shewanella marisflavi EP1还原转化二甲戊乐灵[J]. 环境科学与技术, 2015, 38(2): 19-24. |
| [52] |
Ji L, Wu W. Isolation, identification and degradation characteristics of trifluralin-degrading bacterium[J]. Jiangsu Agricultural Sciences, 2016, 44(3): 390-393, 531. (in Chinese) 季丽, 吴伟. 氟乐灵降解菌株的分离鉴定及降解特性研究[J]. 江苏农业科学, 2016, 44(3): 390-393, 531. |
| [53] |
Ni HY, Yao L, Li N, et al. Biodegradation of pendimethalin by Bacillus subtilis Y3[J]. Journal of Environmental Sciences, 2016, 41: 121-127. DOI:10.1016/j.jes.2015.04.035 |
| [54] |
Ni HY, Wang F, Li N, et al. Pendimethalin nitroreductase is responsible for the initial pendimethalin degradation step in Bacillus subtilis Y3[J]. Applied and Environmental Microbiology, 2016, 82(24): 7052-7062. DOI:10.1128/AEM.01771-16 |
| [55] |
Aboulila AA, Belal EB, Metwaly MM, et al. Degenotoxicity of pendimethalin contaminated clay soil by Pseudomonas resinovorans using anatomical, cytogenetic and biochemical analysis in Vicia faba plants[J]. International Journal of Current Research in Biosciences and Plant Biology, 2016, 3(2): 38-53. DOI:10.20546/ijcrbp.2016.302.005 |
| [56] |
Ishag AESA, Abdelbagi AO, Hammad AMA, et al. Biodegradation of endosulfan and pendimethalin by three strains of bacteria isolated from pesticides-polluted soils in the Sudan[J]. Applied Biological Chemistry, 2017, 60(3): 287-297. DOI:10.1007/s13765-017-0281-0 |
| [57] |
Chen BB, Dong HQ, Cao ZH, et al. Separation, identification and degradation characteristics of trifluralin degrading strain Bacillus sp.[J]. Agrochemicals, 2017, 56(1): 30-35. (in Chinese) 陈贝贝, 董红强, 曹志恒, 等. 氟乐灵降解菌Bacillus sp.的分离鉴定及其降解特性[J]. 农药, 2017, 56(1): 30-35. DOI:10.3969/j.issn.1671-5284.2017.01.007 |
| [58] |
Hu JY, Zhang GQ, Han XQ, et al. Screening, identification and degradation characterization of pendimethalin degrading bacteria[J]. Journal of Henan Agricultural Sciences, 2017, 46(7): 64-70. (in Chinese) 胡佳月, 张国强, 韩小强, 等. 二甲戊灵降解细菌的筛选、鉴定及其降解特性研究[J]. 河南农业科学, 2017, 46(7): 64-70. |
| [59] |
Hu JY, Zhang GQ, Han XQ, et al. Degradation effect of two bacterial strains on soil pendimethalin[J]. Jiangsu Journal of Agricultural Sciences, 2017, 33(3): 585-591. (in Chinese) 胡佳月, 张国强, 韩小强, 等. 2株降解菌对土壤中二甲戊灵的降解效果[J]. 江苏农业学报, 2017, 33(3): 585-591. DOI:10.3969/j.issn.1000-4440.2017.03.015 |
| [60] |
Archana C, Saharan N, Rathore G, et al. Isolation and characterization of potential pendimethalin degrading bacteria from pesticides polluted soil[J]. Journal of Entomology and Zoology Studies, 2018, 6(4): 1842-1848. |
| [61] |
Ni HY, Li N, Qiu JG, et al. Biodegradation of pendimethalin by Paracoccus sp. P13[J]. Current Microbiology, 2018, 75(8): 1077-1083. DOI:10.1007/s00284-018-1494-0 |
| [62] |
Funderburk Jr HH, Schultz DP, Negi NS, et al. Metabolism of trifluralin by soil microorganisms and higher plants[A]//Metabolism of trifluralin by soil microorganisms and higher plants[C]. Proceedings-Southern Weed Conference, 1967: 389
|
| [63] |
Laanio TL, Kearney PC, Kaufman DD. Microbial metabolism of dinitramine[J]. Pesticide Biochemistry and Physiology, 1973, 3(3): 271-277. DOI:10.1016/0048-3575(73)90025-4 |
| [64] |
Kearney PC, Plimmer JR, Williams VP, et al. Soil persistence and metabolism of N-sec-butyl-4-tert-butyl-2, 6-dinitroaniline[J]. Journal of Agricultural and Food Chemistry, 1974, 22(5): 856-859. DOI:10.1021/jf60195a046 |
| [65] |
Zeyer J, Kearney PC. Microbial dealkylation of trifluralin in pure culture[J]. Pesticide Biochemistry and Physiology, 1983, 20(1): 10-18. DOI:10.1016/0048-3575(83)90116-5 |
| [66] |
Zayed SMAD, Mostafa IY, Parghaly MM, et al. Microbial degradation of tripluralin by Aspergillus carneus, Fusarium oxysporum and Trichoderma viride[J]. Journal of Environmental Science and Health, Part B, 1983, 18(2): 253-267. DOI:10.1080/03601238309372367 |
| [67] |
Barua AS, Saha J, Chaudhuri S, et al. Degradation of pendimethalin by soil fungi[J]. Pesticide Science, 1990, 29(4): 419-425. DOI:10.1002/ps.2780290406 |
| [68] |
Singh SB, Kulshrestha G. Microbial degradation of pendimethalin[J]. Journal of Environmental Science and Health, Part B, 1991, 26(3): 309-321. DOI:10.1080/03601239109372737 |
| [69] |
Lin AJ, Zhu LS, Wang J, et al. Biodegradation of herbicide pendimethalin by fungi and its characteristics[J]. Chinese Journal of Applied Ecology, 2003, 14(11): 1929-1933. (in Chinese) 林爱军, 朱鲁生, 王军, 等. 除草剂二甲戊灵的真菌降解及其特性研究[J]. 应用生态学报, 2003, 14(11): 1929-1933. DOI:10.3321/j.issn:1001-9332.2003.11.027 |
| [70] |
Pinto AP, Serrano C, Pires T, et al. Degradation of terbuthylazine, difenoconazole and pendimethalin pesticides by selected fungi cultures[J]. Science of the Total Environment, 2012, 435-436: 402-410. DOI:10.1016/j.scitotenv.2012.07.027 |
| [71] |
Belal E, Elkhateeb NM. Biodegradation of pendimethalin residues by P. chrysosporium in aquatic system and soils[J]. Journal of Biological Chemistry and Environmental Sciences, 2014, 9(3): 383-400. |
| [72] |
Gong HK, Sun QY, Liu SS, et al. Isolation and characteristics of pendimethalin degrading fungus strain Qsun-6[J]. Journal of Dalian Polytechnic University, 2014, 33(4): 254-257. (in Chinese) 宫宏琨, 孙庆元, 刘栓栓, 等. 除草剂二甲戊乐灵降解真菌Qsun-6的分离及其特性[J]. 大连工业大学学报, 2014, 33(4): 254-257. |
| [73] |
Han YJ, Tang ZG, Bao HF, et al. Degradation of pendimethalin by the yeast YC2 and determination of its two main metabolites[J]. RSC Advances, 2019, 9(1): 491-497. DOI:10.1039/C8RA07872F |
| [74] |
De L Bellinaso M, Henriques JA, Gaylarde CC, et al. Genes similar to naphthalene dioxygenase genes in trifluralin-degrading bacteria[J]. Pest Management Science, 2004, 60(5): 474-478. DOI:10.1002/ps.835 |
| [75] |
More VS, Tallur PN, Ninnekar HZ, et al. Purification and properties of pendimethalin nitroreductase from Bacillus circulans[J]. Applied Biochemistry and Microbiology, 2015, 51(3): 329-335. DOI:10.1134/S0003683815030138 |
 2020, Vol. 47
2020, Vol. 47