扩展功能
文章信息
- 郑慧娟, 白晓晔, 高旭, 孙志宏, 张和平
- ZHENG Hui-Juan, BAI Xiao-Ye, GAO Xu, SUN Zhi-Hong, ZHANG He-Ping
- 双歧杆菌属特异性测序引物筛选及优化
- Screening and optimization of Bifidobacterium-specific sequencing primers
- 微生物学通报, 2019, 46(7): 1736-1747
- Microbiology China, 2019, 46(7): 1736-1747
- DOI: 10.13344/j.microbiol.china.180610
-
文章历史
- 收稿日期: 2018-08-04
- 接受日期: 2018-11-13
- 网络首发日期: 2018-12-24
双歧杆菌(Bifidobacterium)是无芽孢革兰氏阳性菌,定殖于哺乳动物的胃肠道、口腔、阴道等位置,是哺乳动物肠道菌群的主要菌属之一[1-2]。1899年,Tissier第一次从母乳喂养婴儿的粪便中分离出双歧杆菌,并因其末端常见有分叉将其正式命名为双歧杆菌[3]。1984年,第8版《伯杰细菌鉴定手册》首次将其列为一个独立的属——双歧杆菌属[4]。有研究发现双歧杆菌具有提高机体免疫力、改善营养元素代谢、改善乳糖不耐症、缓解便秘、保护肝脏、预防肠道疾病、抗氧化和增强机体免疫等一系列益生功能[5-8]。同时,越来越多的研究发现患病人群的肠道菌群存在以双歧杆菌为代表的有益菌降低而有害菌升高的规律[9]。Mizuno等认为供体粪便中双歧杆菌的高丰富度是粪便微生物移植(Fecal microbiota transplantation,FMT)成功的一个积极预测因子[10],并且有临床研究证实双歧杆菌可以减少人体菌群失调,进而预防细菌感染[11]。
鉴于双歧杆菌具有众多益生特性和潜在作为生物标签的功能,双歧杆菌多样性及其变化规律逐渐成为研究热点。对样品中双歧杆菌多样性进行分析的传统方法主要包括平板计数与菌落形态特征观察、选择性培养基和细胞质蛋白模式分析等[12-14]。传统生物学手段虽可以在一定程度上揭示样品中双歧杆菌的组成,但这些方法往往耗时耗力且难以充分展现样品中双歧杆菌的多样性,而且据估计60%−80%的人类肠道微生物群可能是不可培养的[15]。现代分子生物学手段则无需培养,更适合双歧杆菌的多样性分析。常见鉴定双歧杆菌的分子生物学手段包括限制性酶切片段长度多态性分析、DNA分子杂交和PCR-DGGE等[16-18],其精密性差与对检测分子要求高等特点使其不适于更精密、快速的多样性分析。
分子生物学方法在近些年得到了长足发展。三代测序技术主要包括单分子实时测序、单分子DNA测序和纳米孔单分子测序。2013年,PacBio RS Ⅱ测序平台面世,为样品中微生物的精准分析带来了新的技术手段[19]。单分子实时测序具有“长读长”的特点,并且采用循环多次测定模式,以保证较高的准确度[20]。而对低丰度物种信息的检测则易受到测序深度的影响[21],且其信息量小难以支持深入的分析。有研究表明肠道中双歧杆菌的含量仅占肠道中厌氧菌的0−0.8%[22],这使其易被肠道菌群中其他优势物种的信息掩盖,很难通过常规检测手段进行深入的分析。特异性引物则可以放大低丰度物种的信息量,目前已有能检测双歧杆菌种水平的引物多为定量引物和多重引物,前者扩增长度为300 bp左右,不能发挥三代测序“长读长”的优势,后者在后续处理时不易去除[23-24]。同时,通常使用的细菌通用测序引物虽然可以展现复杂样品的微生物多样性,但是对低丰度双歧杆菌缺乏足够的特异性。
本研究根据已有基于16S rRNA基因序列的双歧杆菌特异性引物,筛选优化适用于粪便样品中低丰度双歧杆菌的特异性引物,为研究肠道菌群中低丰度双歧杆菌的多样性奠定基础。
1 材料与方法 1.1 实验菌株实验菌株由内蒙古农业大学乳品生物技术与工程教育部重点实验室提供,其中双歧杆菌2株,乳酸菌20株,共计22株菌,具体如表 1所示。
| 菌株名称Strains name | 菌株编号Strains number |
| Bifidobacterium gallicum | DSM 20093T |
| Bifidobacterium longum | DSM 20088T |
| Enterococcus asini | DSM 11492T |
| Enterococcus faecium | ATCC 19434T |
| Enterococcus italicus | DSM 15952T |
| Escherichia coli | ATCC 11775T |
| Lactobacillus helveticus | DSM 20075T |
| Lactobacillus casei | ATCC 334T |
| Leuconostoc pseudomesenteroides | DSM 20193T |
| Lactobacillus salivarius | DSM 20555T |
| Lactococcus plantarum | DSM 20686T |
| Listeria monocytogenes | ATCC 19115T |
| Pediococcus acidilactici | JCM 2014T |
| Pediococcus pentosaceus | DSM 20336T |
| Pseudomonas aeruginosa | ATCC 10145T |
| Salmonella typhi | ATCC 19430T |
| Streptococcus thermophilus | NM-81-2 |
| Shigella flexneri | ATCC 29903T |
| Staphylococcus aureus | ATCC 25923T |
| Weissella beninensis | DSM 22752T |
| Weissella confusa | IMAU 10245 |
| Weissella kandleri | DSM 20593T |
细菌基因组DNA提取试剂盒,天根生化科技(北京)有限公司;QIAamp Fast DNA Stool Mini Kit,Qiagen公司;引物由上海桑尼生物科技有限公司合成。PCR仪,赛默飞世尔科技公司;电泳仪,北京六一生物科技有限公司;全自动凝胶成像分析系统,北京赛智创业科技有限公司;PacBio SMRT测序平台,Pacific Biosciences公司。
1.3 DNA提取依照细菌基因组DNA提取试剂盒说明书对表 1中菌株提取DNA。A1为1份自然分娩并母乳喂养健康婴儿(汉族)的胎便样品,本文引用了其细菌通用引物的扩增序列[25],原文中编号为IF8。A2和A3为2份健康母亲(汉族)的粪便样品,分两批采集自内蒙古医科大学附属医院。3份样品采集后均贮藏于−80 ℃。依照QIAamp Fast DNA Stool Mini Kit说明书提取粪便中微生物DNA,均保存于−20 ℃备用。
1.4 PCR扩增PCR体系:10×PCR buffer (Mg2+ plus) 2 µL,dNTPs (2.5 mmol/L) 1.6 µL,DNA模板(10 ng/µL) 1 µL,上、下游引物(100 µmol/L)各0.5 µL,Taq酶(5 U/µL) 0.2 µL,蒸馏水14.2 µL。PCR反应条件参照Rinttilä等的方法[26]。
1.5 引物的理论筛选查阅文献搜索基于双歧杆菌16S rRNA基因序列设计的引物。依照LPSN (http://www.bacterio.net/)确定双歧杆菌属模式菌株,并下载相应16S rRNA基因序列。使用MEGA 6.0计算上述引物与序列的匹配率,并确定其在双歧杆菌模式菌株B. longum subsp. infantis 16S rRNA基因序列(NCBI登录号为ATCC 15697)的相对位置。
1.6 引物的重组与优化通过上述步骤筛选高匹配率的引物,并根据其相对位置进行重组优化,筛选扩增片段长度大于800 bp的引物,以增加可以检测到双歧杆菌属种水平的几率。双歧杆菌与乳酸菌属在肠道中均较常见,并且比对发现两者16S rRNA结构具有相似性,因此将重组引物通过MEGA与包含乳酸菌7个属[肠球菌属(54)、链球菌属(101)、明串珠菌属(19)、片球菌属(13)、乳球菌属(15)、乳酸杆菌属(219)、魏斯氏菌属(22),括号内为各菌属包含序列数] 16S rRNA基因序列的本地数据库进行BLAST比对,得到只对双歧杆菌属匹配率高的引物进行后续实验验证。
1.7 重组引物的特异性验证首先应用PCR体系,在52−60 ℃之间设置温度梯度,确定最佳退火温度;继而进行特异性对照实验,并通过凝胶电泳筛选能准确区分双歧杆菌的引物。
1.8 PacBio SMRT测序依据QIAamp Fast DNA Stool Mini Kit说明书,分别对3份粪便样品中细菌DNA的扩增产物构建文库并通过PacBio RS Ⅱ测序平台测序。使用SMRT® Portal (V2.7)软件中RS_Readsofinsert.1方案对样品下机数据进行质控,并根据Barcode将序列拆分到不同的样品以进行后续的生物信息学分析。通过QIIME (V1.7.0)平台对序列进行物种多样性分析[27]。以细菌通用引物(27f/1492r)扩增结果为参照,根据上述物种注释的结果在种水平和属水平对新引物的扩增多样性进行分析比较。通过OriginPro 2015绘制饼图、柱状图。通过R语言软件(V3.4.3)的Pheatmap软件包,以热图形式呈现样品扩增结果中双歧杆菌属菌种缺失情况。
2 结果与分析 2.1 已有双歧杆菌属引物特征 2.1.1 基本特征所搜集引物涉及了定性与定量两大用途,多针对复杂环境样品。引物基本特征汇总见表 2。
| 编号 Number |
引物名称 Primers name |
引物序列 Primers sequence (5′→3′) |
退火温度 Annealing temperature (℃) |
参考文献 References |
检测目标 Detection targets |
| 1 | g-Bifid-F g-Bifid-R | CTCCTGGAAACGGGTGG | 55 | [28] | Adult feces |
| GGTGTTCTTCCCGATATCTACA | |||||
| 2 | Bif164-f Bif662-r | GGGTGGTAATGCCGGATG | 66 | [29] | Infant feces |
| CCACCGTTACACCGGGAA | |||||
| 3 | lm26 lm3 | GATTCTGGCTCAGGATGAACG | 57 | [30] | Sour milk |
| CGGGTGCTYCCCACTTTCATG | |||||
| 4 | F R | TCGCGTC(C/T)GGTGTGAAAG | 58 | [26] | Adult feces |
| CCACATCCAGC(A/G)TCCAC | |||||
| 5 | Pbi F1 Pbi R2 | CCGGAATAGCTCC | 50 | [31] | Strains |
| GACCATGCACCACCTGTGAA | |||||
| 6 | Bif-F Bif-R | TTCGGGTTGTAAACCGCTTTT | 62 | [32] | Waste water |
| TACGTATTACCGCGGCTGCT | |||||
| 7 | FP RP | GGGCGTAAAGGGCTCGTA | 53 | [33] | Duck intestinal tract |
| CTTCCCGATATCTACACATTCCA | |||||
| 8 | P175 P674 | GGGTGGTAATGCCGGATG | 56 | [34] | Strains |
| CACCGTTACACCGGAATT | |||||
| 9 | Bif S11 Bif S12 | TCCTCGCTTTGCTCCCGATA | 56 | [35] | Sour milk |
| AATTCCCGTTGTAACGGTGG | |||||
| Note: Y: A/T/C/G. | |||||
通过MEGA计算引物与双歧杆菌属匹配率,结果见图 1。3条引物匹配率较低,其中P674的序列依据相关匹配信息被改进为P674- (5′-TCCA CCGTTACACCGGGAATT-3′)继续后续实验,Bif S11/Bif S12则停止后续实验。
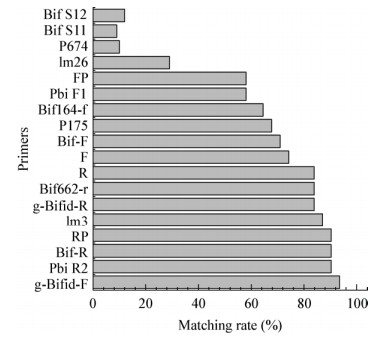
|
| 图 1 引物与双歧杆菌属16S rRNA基因数据库的匹配率 Figure 1 Matching rate of primers with Bifidobacterium 16S rRNA gene database |
|
|
8对匹配率高的引物起始位点如图 2所示。引物相对位置的确定为后续扩增片段长度筛选工作奠定基础。
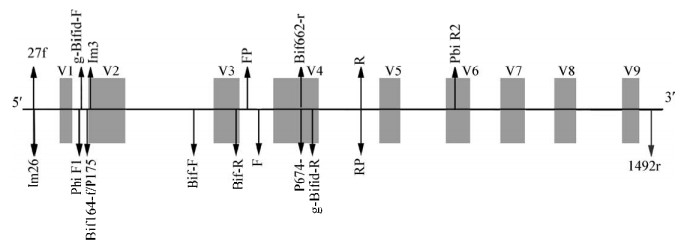
|
| 图 2 引物相对B. longum subsp. infantis 16S rRNA基因序列的位置(NCBI登录号为ATCC 15697) Figure 2 Relative position of various primers on B. longum subsp. infantis 16S rRNA gene sequence (NCBI accession number ATCC 15697) |
|
|
根据上述特征,将高匹配率引物重组,得到以下两者均较优且可扩增大片段(大于800 bp)的组合:Pbi F1/Pbi R2、lm26/R、Bif164-f/lm26和Pbi R2/Bif-F。
2.2.2 引物的序列库比对结果通过MEGA将上述组合与双歧杆菌属和乳酸菌7个属序列进行BLAST比对,结果如图 3所示。综合比对结果发现Bif-F可匹配乳酸菌5个属的序列,说明其特异性差,因此舍弃。Im26双歧杆菌匹配率最低,因此舍弃。其他引物对双歧杆菌特异性均良好。

|
| 图 3 引物在双歧杆菌属与乳酸菌属数据库中的序列匹配数 Figure 3 Matched sequences amount of primers with Bifidobacterium and Lactobacillus 16S rRNA gene database |
|
|
将特异性好的引物再次组合,使其扩增片段长度约600−1 000 bp,以发挥三代测序读长优势,并提高物种预测的准确度,最终组合结果如表 3所示。
| 组合 Combination |
引物名称 Primers name |
序列 Primers sequence (5′→3′) |
扩增片段长度 Amplified fragment length (bp) |
| 1 | Pbi F1 | CCGGAATAGCTCC | 894 |
| Pbi R2 | GACCATGCACCACCTGTGAA | ||
| 2 | Bif164-f | GGGTGGTAATGCCGGATG | 878 |
| Pbi R2 | GACCATGCACCACCTGTGAA |
温度实验显示Pbi F1/Pbi R2特异性差而放弃后续实验。Bif164-f/Pbi R2特异性良好,因此将其确定为最优引物。同时确定实验最高温度60 ℃为最佳退火温度以增加特异性[29]。
Bif164-f/Pbi R2对2株双歧杆菌、20株相近属菌种和蒸馏水的扩增结果如图 4所示。仅2株双歧杆菌的扩增条带明亮,引物表现出良好特异性。

|
| 图 4 Bif164-f/Pbi R2的特异性 Figure 4 Specificity of Bif164-f/Pbi R2 注:1:高卢双歧杆菌;2:长双歧杆菌;3:驴肠球菌;4:屎肠球菌;5:意大利肠球菌;6:埃希氏大肠杆菌;7:瑞士乳杆菌;8:干酪乳杆菌;9:假肠膜明串珠菌;10:唾液乳杆菌;11:植物乳球菌;12:单核细胞增生李斯特菌;13:乳酸片球菌;14:戊糖片球菌;15:铜绿假单胞菌;16:伤寒沙门氏菌;17:嗜热链球菌;18:福氏志贺氏菌;19:金黄色葡萄球菌;20:贝尼氏魏斯氏菌;21:融合魏斯氏菌;22:坎氏魏斯氏菌;23:蒸馏水. Note: 1: Bifidobacterium gallicum; 2: Bifidobacterium longum; 3: Enterococcus asini; 4: Enterococcus faecium; 5: Enterococcus italicus; 6: Escherichia coli; 7: Lactobacillus helveticus; 8: Lactobacillus casei; 9: Leuconostoc pseudomesenteroides; 10: Lactobacillus salivarius; 11: Lactococcus plantarum; 12: Listeria monocytogenes; 13: Pediococcus acidilactici; 14: Pediococcus pentosaceus; 15: Pseudomonas aeruginosa; 16: Salmonella typhi; 17: Streptococcus thermophilus; 18: Shigella flexneri; 19: Staphylococcus aureus; 20: Weissella beninensis; 21: Weissella confusa; 22: Weissella kandleri; 23: Distilled water. |
|
|
引物27f/1492r与Bif164-f/Pbi R2对样品A1、A2和A3的扩增序列随机选取相同序列数,依次分别为1 293、8 173和9 062条。
2.4.1 引物在属水平上扩增结果比较引物对粪便样品中细菌DNA的扩增结果如图 5所示。A1、A2和A3样品中,27f/1492r扩增结果中相对含量大于5%的优势菌属依次分别为Veillonella (57.72%)、Escherichia (16.53%)、Bacteroides (12.89%)和Streptococcus (25.67%)、Haemophilus (10.60%)、Collinsella (8.56%)、Ruminococcus (6.51%)及Eubacterium (20.29%)、Bacteroides (8.53%)、Collinsella (7.83%)、Clostridium (7.27%),而Bifidobacterium相对含量分别为0.07%、0.56%和0.40%。Bif164-f/Pbi R2扩增结果表明3份样品优势菌属为Bifidobacterium,其相对含量分别为96.15%、100%和100%。意味着与细菌通用引物相比,Bif164-f/Pbi R2将样品中双歧杆菌检出的平均相对含量由0.34%提高至98.72%,对粪便中双歧杆菌具有良好的特异性,可用于揭示粪便样品中低丰度双歧杆菌的多样性。

|
| 图 5 基于不同引物比较样品中细菌菌群结构 Figure 5 Comparison of bacterial community structure in samples based on different primers 注:27f/1492r (A)与Bif164-f/Pbi R2 (B)检测粪便样品菌群(A1)的属水平多样性;27f/1492r (C)与Bif164-f/Pbi R2 (D)检测粪便样品菌群(A2)的属水平多样性;27f/1492r (E)与Bif164-f/Pbi R2 (F)检测粪便样品菌群(A3)的属水平多样性. Note: Bacterial diversity of fecal sample (A1) identified using 27f/1492r (A) and Bif164-f/Pbi R2 (B) at the genus level; Bacterial diversity of fecal sample (A2) identified using 27f/1492r (C) and Bif164-f/Pbi R2 (D) at the genus level; Bacterial diversity of fecal sample (A3) identified using 27f/1492r (E) and Bif164-f/Pbi R2 (F) at the genus level. |
|
|
2种引物对3份样品扩增序列的种水平双歧杆菌多样性分析见图 6B。27f/1492r对A1、A2与A3扩增序列的生物学注释显示,3份样品分别含B. longum subsp. longum和B. adolescentis、B. breve、B. longum subsp. longum及B. animalis subsp. lactis、B. breve、B. pseudocatenulatum、B. bifidum;而Bif164-f/Pbi R2的3份扩增子生物学注释信息则显示,3份样品分别含B. breve、B. longum和B. longum subsp. longum、B. catenulatum、B. animalis subsp. lactis、B. adolescentis、B. dentium、B. kashiwanohense及B. stercoris、B. breve、B. catenulatum、B. bifidum、B. longum subsp. longum、B. thermophilum、B. animalis subsp. lactis、B. longum subsp. suis。表明Bif164-f/Pbi R2对样品中双歧杆菌的扩增效果明显优于27f/1492r。

|
| 图 6 不同引物的双歧杆菌特异性及多样性检测 Figure 6 Bifidobacterial specificity and diversity detection of different primers 注:A:不同引物的粪便样品细菌DNA扩增子中非双歧杆菌属检出率;B:不同引物检测粪便样品中双歧杆菌的种水平多样性;M1、M2、M3、M4为母亲粪便样品;I1、I2、I3、I4为婴儿粪便样品[36];ITS表示引物Probio-bif_Uni/Probio-bif_Rev;27f表示引物27f/1492r;Bif164表示引物Bif164-f/Pbi R2;黑色代表有;白色代表无. Note: A: Detection rates of non-Bifidobacterium in the DNA amplicons of fecal bacterial micyobiota in using various primers; B: Bifidobacterial diversity of fecal samples identified using various primers at the species level; M1, M2, M3 and M4 were fecal samples from mothers, and I1, I2, I3 and I4 were fecal samples from infants[36]; ITS denotes the primer Probio-bif_Uni/Probio-bif_Rev, 27f denotes the primer 27f/1492r, and Bif164 denotes the primer Bif164-f/Pbi R2; Black mains the presence, and white mains the absence of Bifidobacteria. |
|
|
本文纳入Milani等[36]设计的双歧杆菌引物Probio-bif_Uni/Probio-bif_Rev进行比较分析。引物Bif164-f/Pbi R2、27f/1492r与Probio-bif_Uni/ Probio-bif_Rev对11份样品的扩增注释结果如图 6A所示,其检出非双歧杆菌物种的最高比例依次分别为3.85%、99.60%和75.36%。引物Bif164-f/Pbi R2与Probio-bif_Uni/Probio-bif_Rev的种水平双歧杆菌多样性如图 6B所示,Bif164-f/Pbi R2对样品扩增序列的生物学注释信息显示,婴儿粪便样品以B. breve和B. longum为主,母亲粪便样品中包含B. adolescentis、B. longum、B. animalis、B. dentium和B. stercoris,与Milani等[36]的研究结果一致。在此基础上,引物Bif164-f/Pbi R2的2份母亲粪便样品细菌DNA的扩增子还检测到了B. breve、B. bifidum、B. catenulatum、B. kashiwanohense和B. thermophilum,而引物Probio-bif_Uni/Probio-bif_Rev的4份粪便样品细菌DNA的扩增子中检测到B. angulatum和B. pseudolongum。在测序量有限的情况下,本研究筛选出的引物Bif164-f/Pbi R2扩增子的种水平多样性更高,表明其在反映粪便样品中双歧杆菌种水平多样性方面表现优异,具有实用价值。需要说明的是Bif164-f/Pbi R2与Probio-bif_Uni/ Probio-bif_Rev分别是基于双歧杆菌16S rRNA基因与ITS序列设计的引物,二者应用的测序平台分别为三代测序与二代测序,其扩增片段的长度范围分别为500−900 bp与350−600 bp。样品A1、A2、A3、I1、I2、I3、I4、M1、M2、M3和M4测序量分别为1 293、8 173、9 062、310 287、141 904、132 208、198 178、75 644、68 988、97 733和334 143条。
3 讨论与结论双歧杆菌是公认的有益菌,具有缓解便秘、改善乳糖不耐症和抑制有害菌等益生功能[5-8, 37]。越来越多的研究认为益生菌可能是通过维持肠道健康的稳态达到益生作用[38-39]。然而肠道中菌属变化是微量的,并且双歧杆菌属于肠道菌群中的低丰度物种,常规而普遍的测序手段很难捕获其大量的有效数据,这极大地阻碍了其多样性及变化规律研究的深入。特异性引物可以辅助放大变化趋势,因此适于低丰度双歧杆菌的特异性引物至关重要。本文通过理论与实验相结合的方法筛选出一对适用于粪便样品中双歧杆菌测序的特异性引物,并通过PacBio SMRT对其进行验证,同时为引物筛选提供了一种新思路。
本实验筛选优化获得了一对双歧杆菌引物Bif164-f/Pbi R2,其扩增子分析结果显示婴儿粪便样品以B. breve和B. longum为主,母亲粪便样品以B. adolescentis、B. catenulatum、B. longum和B. animalis为主,与相关报道[36, 40-42]指出的肠道双歧杆菌优势种一致。婴儿粪便样品的双歧杆菌多样性较健康成人粪便样品低[36, 43],符合婴儿时期双歧杆菌物种的数量偏低且随着年龄的增长而减少的规律[44],说明引物Bif164-f/Pbi R2是可靠的。此外,Bif164-f/Pbi R2在扩增多样性方面明显优于27f/1492r与Probio-bif_Uni/Probio- bif_Rev,有限测序量下,引物Bif164-f/Pbi R2较引物Probio-bif_Uni/Probio-bif_Rev和27f/1492r从母亲粪便样品中在种水平检测到了更多的双歧杆菌种,而引物Bif164-f/Pbi R2扩增的目的片段大于800 bp,可以发挥PacBio SMRT测序平台“长读长”的优势并提高物种预测的准确度。同时,Bif164-f/Pbi R2在特异性方面也表现良好。测序量相同时,引物Bif164-f/Pbi R2对粪便样品中的双歧杆菌检出相对含量提高了98.38%,较细菌通用引物27f/1492r能更有效地识别粪便样品中的双歧杆菌属,有助于观察不同样品间或相同样品不同阶段的双歧杆菌种变化规律。引物Bif164-f/Pbi R2的筛选与优化充分整合了现有资源,较一些仅通过实验比较或者仅在NCBI上比对进行引物设计、筛选的方案节省了人力物力[45-46]。需要说明的是本研究筛选的双歧杆菌特异性测序引物是适用于所有三代测序技术的。
综上所述,本研究筛选出一对适用于粪便样品中双歧杆菌测序的特异性引物,同时为引物筛选优化提供了新思路,为种水平深入解析样品中低丰度双歧杆菌多样性奠定了基础。
| [1] |
Biavati B, Vescovo M, Torriani S, et al. Bifidobacteria: history, ecology, physiology and applications[J]. Annals of Microbiology, 2000, 50: 117-131. |
| [2] |
Schell MA, Karmirantzou M, Snel B, et al. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract[J]. Proceedings of the National Academy of Sciences of the United States of America, 2002, 99(22): 14422-14427. DOI:10.1073/pnas.212527599 |
| [3] |
Tissier H. Recherches sur la flore intestinale des nourrissons (état normal et pathologique)[D]. Paris: Master's Thesis of G. Carre and C. Naud, 1900
|
| [4] |
Buchanan RE, Gibbons NE. Berger Handbook of Bacteria Identification[M]. 8th ed. Beijing: Science Press, 1984: 926. (in Chinese) 布坎南 RE, 吉本斯 NE. 伯杰细菌鉴定手册[M]. 8版. 北京: 科学出版社, 1984: 926. |
| [5] |
Arunachalam K, Gill HS, Chandra RK. Enhancement of natural immune function by dietary consumption of Bifidobacterium lactis (HN019)[J]. European Journal of Clinical Nutrition, 2000, 54(3): 263-267. DOI:10.1038/sj.ejcn.1600938 |
| [6] |
Tanaka K, Tsukahara T, Yanagi T, et al. Bifidobacterium bifidum OLB6378 simultaneously enhances systemic and mucosal humoral immunity in low birth weight infants: A non-randomized study[J]. Nutrients, 2017, 9(3): 195. DOI:10.3390/nu9030195 |
| [7] |
Qiao RJ. Efficacy of Bifidobacterium triple viable in treatment of secondary lactose intolerance in infants with rotavirus enteritis[J]. Chinese Journal of Modern Drug Application, 2013, 7(19): 129-130. (in Chinese) 乔瑞君. 双歧杆菌三联活菌治疗婴幼儿轮状病毒肠炎继发性乳糖不耐受的疗效观察[J]. 中国现代药物应用, 2013, 7(19): 129-130. DOI:10.3969/j.issn.1673-9523.2013.19.107 |
| [8] |
Liu J, Huang XE. Efficacy of Bifidobacterium tetragenous viable bacteria tablets for cancer patients with functional constipation[J]. Asian Pacific Journal of Cancer Prevention, 2015, 15(23): 10241-10244. DOI:10.7314/APJCP.2014.15.23.10241 |
| [9] |
Wang TT. The interactions between structural shifts of gut microbiota and development of colorectal cancer[D]. Shanghai: Doctoral Dissertation of Shanghai Jiaotong University, 2012 (in Chinese) 王婷婷.肠道菌群结构变化与结直肠癌发生发展关系的研究[D].上海: 上海交通大学博士学位论文, 2012 http://cdmd.cnki.com.cn/Article/CDMD-10248-1012016757.htm |
| [10] |
Mizuno S, Masaoka T, Naganuma M, et al. Bifidobacterium-rich fecal donor may be a positive predictor for successful fecal microbiota transplantation in patients with irritable bowel syndrome[J]. Digestion, 2017, 96(1): 29-38. DOI:10.1159/000471919 |
| [11] |
Jia ZT, Li YX. Research progress in the function and application of Bifidobacterium[J]. Modern Animal Husbandry, 2014(10): 6-8. (in Chinese) 贾子堂, 李艳霞. 双歧杆菌功能与应用的研究进展[J]. 当代畜禽养殖业, 2014(10): 6-8. DOI:10.3969/j.issn.1005-5959.2014.10.003 |
| [12] |
Xin RZ, Ding M. A study on the independent counting method of every probiotics and the mixed fermentation technology of functional acidophilus milk[J]. Food and Fermentation Industries, 2006, 32(7): 151-156. (in Chinese) 辛若竹, 丁梅. 功能性酸乳中每种益生菌的单独计数方法及其混合发酵工艺的研究[J]. 食品与发酵工业, 2006, 32(7): 151-156. DOI:10.3321/j.issn:0253-990X.2006.07.039 |
| [13] |
Huo DX, Zhang JC, Bai N, et al. Selective medium to isolate human Bifidobacterium[J]. Acta Microbiologica Sinica, 2014, 54(4): 433-441. (in Chinese) 霍冬雪, 张家超, 白娜, 等. 适用于分离人肠道中双歧杆菌的选择性培养基[J]. 微生物学报, 2014, 54(4): 433-441. |
| [14] |
Aires J, Anglade P, Baraige F, et al. Proteomic comparison of the cytosolic proteins of three Bifidobacterium longum human isolates and B. longum NCC2705[J]. BMC Microbiology, 2010, 10(1): 29. DOI:10.1186/1471-2180-10-29 |
| [15] |
Suau A, Bonnet R, Sutren M, et al. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut[J]. Applied and Environmental Microbiology, 1999, 65(11): 4799-4807. |
| [16] |
Kheadr E, Dabour N, Von Ah U, et al. Genetic and phenotypic diversity of Bifidobacterium thermacidophilum fecal isolates from newborns[J]. Canadian Journal of Microbiology, 2007, 53(12): 1348-1359. DOI:10.1139/W07-101 |
| [17] |
Duranti S, Turroni F, Milani C, et al. Exploration of the genomic diversity and core genome of the Bifidobacterium adolescentis phylogenetic group by means of a polyphasic approach[J]. Applied and Environmental Microbiology, 2013, 79(1): 336-346. DOI:10.1128/AEM.02467-12 |
| [18] |
Endo A, Futagawa-Endo Y, Dicks LMT. Lactobacillus and Bifidobacterium diversity in horse feces, revealed by PCR-DGGE[J]. Current Microbiology, 2009, 59(6): 651-655. DOI:10.1007/s00284-009-9498-4 |
| [19] |
Mosher JJ, Bowman B, Bernberg EL, et al. Improved performance of the PacBio SMRT technology for 16S rDNA sequencing[J]. Journal of Microbiological Methods, 2014, 104: 59-60. DOI:10.1016/j.mimet.2014.06.012 |
| [20] |
Li QS, Li Y, Song JY, et al. High-accuracy de novo assembly and SNP detection of chloroplast genomes using a SMRT circular consensus sequencing strategy[J]. New Phytologist, 2014, 204(4): 1041-1049. DOI:10.1111/nph.12966 |
| [21] |
Lemos LN, Fulthorpe RR, Triplett EW, et al. Rethinking microbial diversity analysis in the high throughput sequencing era[J]. Journal of Microbiological Methods, 2011, 86(1): 42-51. DOI:10.1016/j.mimet.2011.03.014 |
| [22] |
Grmanová M, Vlková E, Rada V, et al. Survival of bifidobacteria in adult intestinal tract[J]. Folia Microbiologica, 2010, 55(3): 281-285. DOI:10.1007/s12223-010-0042-5 |
| [23] |
Jiang M, Yao P, Yang T, et al. Real-time fluorescence quantitative-PCR analysis of the variation of Bifidobacterium spp., Faecalibacterium prausnitzii and Bacteroides in patients with ulcerative colitis[J]. Chinese Journal of Microecology, 2013, 25(11): 1245-1249, 1254. (in Chinese) 蒋曼, 姚萍, 杨涛, 等. 实时荧光定量PCR法研究溃疡性结肠炎患者肠道双歧杆菌属、柔嫩梭菌属及拟杆菌属量的变化[J]. 中国微生态学杂志, 2013, 25(11): 1245-1249, 1254. |
| [24] |
Bonjoch X, Ballesté E, Blanch AR. Multiplex PCR with 16S rRNA gene-targeted primers of Bifidobacterium spp. to identify sources of fecal pollution[J]. Applied and Environmental Microbiology, 2004, 70(5): 3171-3175. DOI:10.1128/AEM.70.5.3171-3175.2004 |
| [25] |
Xi XX. Correlations between the gut microbiota of newborns and microbial communities of multiple body habitats in mothers[D]. Inner Mongolia: Doctoral Dissertation of Inner Mongolia Agricultural University, 2017 (in Chinese) 席晓霞.初生婴儿肠道菌群与母体各部位菌群相关性研究[D].内蒙古: 内蒙古农业大学博士学位论文, 2017 http://cdmd.cnki.com.cn/Article/CDMD-10129-1017212377.htm |
| [26] |
Rinttilä T, Kassinen A, Malinen E, et al. Development of an extensive set of 16S rDNA-targeted primers for quantification of pathogenic and indigenous bacteria in faecal samples by real-time PCR[J]. Journal of Applied Microbiology, 2004, 97(6): 1166-1177. DOI:10.1111/jam.2004.97.issue-6 |
| [27] |
Wen YP, Hou QC, Zhang HP. Effect of natural fermented Koumiss on human gut microbiota — based on PacBio sequencing technology[J]. Chinese Dairy Industry, 2017, 45(2): 4-7. (in Chinese) 温永平, 侯强川, 张和平. 自然发酵酸马奶对人体肠道菌群的影响——基于PacBio SMRT测序技术[J]. 中国乳品工业, 2017, 45(2): 4-7. DOI:10.3969/j.issn.1001-2230.2017.02.001 |
| [28] |
Matsuki T, Watanabe K, Fujimoto J, et al. Development of 16S rRNA-gene-targeted group-specific primers for the detection and identification of predominant bacteria in human feces[J]. Applied and Environmental Microbiology, 2002, 68(11): 5445-5451. DOI:10.1128/AEM.68.11.5445-5451.2002 |
| [29] |
Kok RG, de Waal A, Schut F, et al. Specific detection and analysis of a probiotic Bifidobacterium strain in infant feces[J]. Applied and Environmental Microbiology, 1996, 62(10): 3668-3672. |
| [30] |
Kaufmann P, Pfefferkorn A, Teuber M, et al. Identification and quantification of Bifidobacterium species isolated from food with genus-specific 16S rRNA-targeted probes by colony hybridization and PCR[J]. Applied and Environmental Microbiology, 1997, 63(4): 1268-1273. |
| [31] |
Roy D, Sirois S. Molecular differentiation of Bifidobacterium species with amplified ribosomal DNA restriction analysis and alignment of short regions of the ldh gene[J]. FEMS Microbiology Letters, 2000, 191(1): 17-24. DOI:10.1111/fml.2000.191.issue-1 |
| [32] |
Gómez-Doñate M, Ballesté E, Muniesa M, et al. New molecular quantitative PCR assay for detection of host-specific Bifidobacteriaceae suitable for microbial source tracking[J]. Applied and Environmental Microbiology, 2012, 78(16): 5788-5795. DOI:10.1128/AEM.00895-12 |
| [33] |
Huang YC, Cheng AC, Wang MS, et al. The establishment of 16S rDNA group-specific PCR method for the detection of Bifidobacterium and application in duck intestinal tract[A]//The third session of Eighth National Symposium on the Development of animal micro-ecological enterprises[C]. Guangzhou: China Society of animal science and veterinary Microecology, 2006: 382-386 (in Chinese) 黄永成, 程安春, 汪铭书, 等.双歧杆菌16S rDNA群特异性PCR方法的建立及对鸭肠道双歧杆菌的检测[A]//第三届第八次全国学术研讨会暨动物微生态企业发展战略论坛[C].广州: 中国畜牧兽医学会动物微生态学分会, 2006: 382-386 |
| [34] |
Dong MS, Jiang HH, Zhang L, et al. Identification of Bifidobactria by PCR and RAPD fingerprinting[J]. Chinese Animal Products and Food, 2000, 7(5): 201-203. (in Chinese) 董明盛, 江汉湖, 张磊, 等. PCR技术结合RAPD图谱鉴定食品中双歧杆菌的研究[J]. 中国畜产与食品, 2000, 7(5): 201-203. |
| [35] |
Wu YJ, Wang JF. 16S rDNA-based identification of Bifidobacterium in foods[J]. Food Science, 2009, 30(18): 359-361. (in Chinese) 吴拥军, 王嘉福. 应用16S rDNA鉴定食品中的双歧杆菌[J]. 食品科学, 2009, 30(18): 359-361. DOI:10.3321/j.issn:1002-6630.2009.18.084 |
| [36] |
Milani C, Lugli GA, Turroni F, et al. Evaluation of bifidobacterial community composition in the human gut by means of a targeted amplicon sequencing (ITS) protocol[J]. FEMS Microbiology Ecology, 2014, 90(2): 493-503. |
| [37] |
Ambalam P, Kondepudi KK, Balusupati P, et al. Prebiotic preferences of human lactobacilli strains in co-culture with bifidobacteria and antimicrobial activity against Clostridium difficile[J]. Journal of Applied Microbiology, 2015, 119(6): 1672-1682. DOI:10.1111/jam.12953 |
| [38] |
Yang G, Peng M, Tian XL, et al. Molecular ecological network analysis reveals the effects of probiotics and florfenicol on intestinal microbiota homeostasis: an example of sea cucumber[J]. Scientific Reports, 2017, 7(1): 4778. DOI:10.1038/s41598-017-05312-1 |
| [39] |
Binek M, Kizerwetter-Świda M, Cisek AA, et al. Mechanisms of maintenance of intestinal homeostasis by autochthonic microbiota and probiotics[J]. Medycyna Weterynaryjna, 2016, 72(10): 611-615. DOI:10.21521/mw.5575 |
| [40] |
Arboleya S, Watkins C, Stanton C, et al. Gut bifidobacteria populations in human health and aging[J]. Frontiers in Microbiology, 2016, 7: 1204. |
| [41] |
Wang XH, Yang Y, Xu X, et al. Influence of different feed fashion on intestinal microflora in infants[J]. Chinese Journal of Child Health Care, 2004, 12(1): 40-42. (in Chinese) 王小卉, 杨毅, 徐秀, 等. 不同喂养方式对婴儿肠道菌群的影响[J]. 中国儿童保健杂志, 2004, 12(1): 40-42. DOI:10.3969/j.issn.1008-6579.2004.01.014 |
| [42] |
Reuter G. The Lactobacillus and Bifidobacterium microflora of the human intestine: composition and succession[J]. Current Issues in Intestinal Microbiology, 2001, 2(2): 43-53. |
| [43] |
Zhang LL, Miao JJ, Li M, et al. Colonization and development of intestinal bifidobacteria in early infancy[J]. Chinese Journal of Contemporary Pediatrics, 2015, 17(3): 207-212. (in Chinese) 张玲琳, 苗俊杰, 李鸣, 等. 婴儿早期肠道双歧杆菌构建规律的初探[J]. 中国当代儿科杂志, 2015, 17(3): 207-212. |
| [44] |
Ouwehand AC, Isolauri E, Kirjavainen PV, et al. Adhesion of four Bifidobacterium strains to human intestinal mucus from subjects in different age groups[J]. FEMS Microbiology Letters, 1999, 172(1): 61-64. DOI:10.1111/fml.1999.172.issue-1 |
| [45] |
Bao WB, Chen GH, Shu JT, et al. Screening of peafowl microsatellite primers and analysis of genetic diversity[J]. Hereditas, 2006, 28(10): 1242-1246. (in Chinese) 包文斌, 陈国宏, 束婧婷, 等. 孔雀微卫星引物筛选及其遗传多样性分析[J]. 遗传, 2006, 28(10): 1242-1246. DOI:10.3321/j.issn:0253-9772.2006.10.010 |
| [46] |
Xu PH, Luo WS, He EM, et al. Improvement of specific primer design on 16S rDNA and its application on skin microbes detection of Trachidermus fasciatus[J]. Journal of Fudan University (Natural Science), 2018, 57(1): 59-67. (in Chinese) 徐鹏昊, 罗武松, 何恩明, 等. 16S rDNA特异性引物设计优化及其在淞江鲈体表微生物鉴定中的应用[J]. 复旦学报:自然科学版, 2018, 57(1): 59-67. |
 2019, Vol. 46
2019, Vol. 46




