扩展功能
文章信息
- 田琇, 张利, 刘马峰
- TIAN Xiu, ZHANG Li, LIU Ma-Feng
- 细菌自然转化的分子机制及生物学功能研究进展
- Molecular mechanisms and biological functions of bacteria natural transformation
- 微生物学通报, 2019, 46(7): 1723-1730
- Microbiology China, 2019, 46(7): 1723-1730
- DOI: 10.13344/j.microbiol.china.180952
-
文章历史
- 收稿日期: 2018-11-25
- 接受日期: 2019-03-22
- 网络首发日期: 2019-04-24
2. 四川农业大学动物医学院预防兽医研究所 四川 成都 611130;
3. 动物疫病与人类健康四川省重点实验室 四川 成都 611130
2. Institute of Preventive Veterinary Medicine, College of Veterinary Medicine, Sichuan Agricultural University, Chengdu, Sichuan 611130, China;
3. Key Laboratory of Animal Disease and Human Health of Sichuan Province, Chengdu, Sichuan 611130, China
基因水平转移(Horizontal gene transfer,HGT)是指遗传物质在细菌或其它微生物之间进行互相转移而并非传递给子代的过程。主要存在3种机制:转导(Transduction)、接合转移(Conjugation)和自然转化(Natural transformation)[1]。转导是指由噬菌体介导将细菌DNA或RNA转移到另一个细菌的过程[2];接合转移是指供体菌与受体菌通过细胞间的直接接触来传递大分子DNA (主要是质粒DNA)的过程,此过程主要由Pili所介导[3];而自然转化是指细菌能够自发从胞外环境中摄取游离DNA分子并整合到自身基因组上的过程,该过程仅由受体菌的基因编码与调控,与外源供体DNA无关[4]。该现象最先由Griffith (1928)在肺炎链球菌中发现[2],目前已报道至少83种细菌能够发生自然转化[3, 5],细菌的进化分析结果表明,自然转化现象广泛存在于细菌界[6],因此可能有更多的细菌具有发生自然转化的潜力。截至目前对自然转化机制研究较清楚的主要有肺炎链球菌、枯草芽孢杆菌(Bacillus subtilis,B. subtilis)、奈瑟氏菌、流感嗜血杆菌(Haemophilus influenzae,H. influenzae)以及幽门螺杆菌(Helicobacter pylori,H. pylori)等[6-7]。
作为细菌获取DNA分子的一种潜在机制[8],研究者认为自然转化对细菌适应环境、进化以及新物种形成具有重要的作用[4]。除可以使细菌获得新的性状如耐药性[9-10]、毒力[11]、代谢功能[8]外,还可以为细菌的生长提供食物来源。此外,也有研究报道自然转化可以修复细菌自身DNA损伤[12]。然而最近有观点指出以上所提出的自然转化功能不存在普遍性且证据不足[13]。本文主要就细菌自然转化的作用机制、生物学功能争论的焦点以及未来研究方向等内容做一综述。
1 自然感受态的形成自然界中某些细菌,如奈瑟氏菌[14]、鲍曼不动杆菌[15](Acinetobacter baumannii,A. baumannii)和鸭疫里默氏杆菌(Riemerella anatipestifer,R. anatipestifer)[6]等少数菌在任何生长状态下均具有自然转化的能力,而大部分被发现具有自然转化现象的细菌,如流感嗜血杆菌、霍乱弧菌(Vibrio cholerae,V. cholerae)和肺炎链球菌等只有在某种特定的生理状态下才具有自然转化的能力。如肺炎链球菌自然感受态发生在对数生长早期[16];枯草芽孢杆菌的自然感受态在稳定期形成[17];鸭疫里默氏杆菌[5]以及乙酸钙不动杆菌[18](Acinetobacter acetate,A. calcoaceticus)在对数生长期自然转化频率最高。较为特殊的是,幽门螺杆菌自然感受态在不同生长阶段多次被诱导。Baltrus等发现,幽门螺杆菌26695菌株自然感受态发生于对数生长期;G27菌株自然感受态在对数期以及平台期分别被诱导;而J99菌株在任何生长阶段均能发生自然转化,并且其自然转化频率在生长初期、对数生长期和平台期各出现一次峰值[19]。诱导细菌感受态发生的因素有很多,如群体感应、营养信号或压力刺激等,其具体调控机制尚不清楚。
1.1 诱导自然感受态形成的因素在自然环境中,细菌自然感受态的形成与营养信号有关。营养匮乏是诱导一些细菌自然感受态发生的原因之一[20]。如流感嗜血杆菌,当其生长到达稳定期或将其从营养丰富环境转移到营养缺乏环境中时,会诱导自然感受态的发生[21]。相反地,丰富的几丁质能够诱导霍乱弧菌、创伤弧菌(Vibrio vulnificus)、副溶血性弧菌(Vibrio parahemolyticus)等弧菌属细菌自然感受态形成[22];乙酸钙不动杆菌自然感受态也发生在营养丰富条件下[18]。乙酸钙不动杆菌自然转化主要发生于对数生长期,随后自然转化频率逐渐降低,平台期细胞几乎丧失自然转化能力。研究发现该现象是由于平台期营养匮乏影响了细胞活性造成的[23-24]。
群体感应(Quorum-Sensing,QS)也能够诱导细菌自然感受态的发生。群体感应指的是细菌在生长过程中,由于群体密度的增加,细菌细胞之间会分泌一些小信号分子来调控细菌某些基因的表达,从而适应环境的变化。在霍乱弧菌中首次发现自然转化与群体感应相关[25]。枯草芽孢杆菌自然感受态也发生在高细胞密度时期[17]。
此外,研究发现细菌自然感受态的形成还与一些压力刺激有关,如抗生素可以诱导肺炎链球菌、幽门螺杆菌、鲍曼不动杆菌自然感受态的形成[15, 26-28]。
1.2 自然感受态的调控因子和通路大多数细菌自然感受态的形成依赖于CRP和Sxy调控子[29]。通常情况下,自然感受态诱导信号促使胞内第二信使cAMP水平升高,后者与其受体蛋白CRP形成有活性的cAMP-CRP复合物,并结合在sxy基因启动子区域的相应位点进一步激活sxy的转录。Sxy是参与自然转化相关基因的激活剂,属于转录辅助调控因子[21]。自然转化相关25个基因的表达均受Sxy的直接调控[30]。Sxy可以帮助cAMP-CRP复合物结合至自然转化相关基因启动子区域激活自然转化。如胞内嘌呤碱基匮乏诱导流感嗜血杆菌自然感受态发生,该过程中cAMP-CRP复合物的结合位点为sxy启动子区域的CRP-S位点,以及自然转化相关基因启动子区域的CRE位点[29, 31]。而几丁质丰富条件下,几丁质低聚物-几丁质二糖诱导霍乱弧菌自然感受态的形成,该过程中cAMP-CRP复合物除了参与调控霍乱弧菌tfoX (sxy同系物)的转录外[32],还上调自然转化相关基因的表达水平,参与霍乱弧菌在几丁质表面定殖以及几丁质的分解利用等[33]。此外,tfoX转录水平还受sRNA tfoR调控,转录调控因子TfoS参与该过程[34]。
而在群体感应诱导霍乱弧菌自然感受态的过程中,除TfoX外,还依赖于QS调节蛋白HapR的参与。研究表明群体感应促进霍乱弧菌局部自诱导物CAI-1和CAI-2的转录,当细胞密度较低时,自诱导物的缺乏致使hapR mRNA降解,QS调节蛋白HapR不表达,霍乱弧菌丧失自然转化能力;当细胞密度较高时自诱导物激活HapR[35],后者抑制细胞外核酸酶Dns的表达,从而保护细胞外DNA不被降解[36-37]。同时,HapR上调多个自然转化相关基因,如comEA、comEC、pilA等的表达[22]。Lo-Scrudato等发现以上调控系统是由QS和TfoX依赖性的转录调节子QstR调控的。TfoX和HapR协同激活QstR调节子,后者结合至HapR所调节基因的上游区域,实现对自然转化相关基因表达的调控[38-39]。
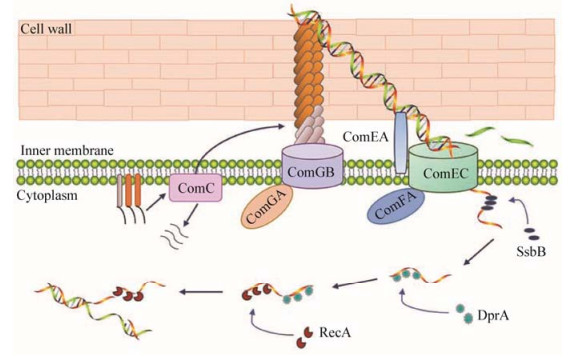
2 自然转化发生过程 2.1 革兰氏阳性菌的DNA转运过程 2.1.1 DNA分子的结合在革兰氏阳性菌中,由comEA及comGB操纵子编码的蛋白质是自然转化过程的必需蛋白。以枯草芽孢杆菌为例(图 1),ComEA已被鉴定为枯草芽孢杆菌的DNA受体,其C-端区域位于细胞膜外侧,具有结合DNA的活性[40]。多胞型ComGB蛋白能够修饰细胞壁形成一个通道,使得外源DNA能够透过细胞壁与膜结合受体ComEA结合[41]。
DNA转运机制由comC和comG操纵子编码。comG操纵子仅在革兰氏阳性菌中发现且高度保守[42]。由comG编码的蛋白质主要有:转运NTP酶ComGA[43],多胞型膜蛋白ComGB[41],以及由肽酶ComC加工成伪菌毛的4个伪菌毛蛋白(ComGC、ComGD、ComGE和ComGG)[43]。当穿过细胞壁的外源DNA分子与DNA结合蛋白ComEA结合后,由ComEA将其呈递给位于胞质膜上的跨膜蛋白ComEC (图 1)[5, 44],ATP酶ComFA为该过程提供能量支撑[45]。随后核酸内切酶NucA将位于ComEC上的双链DNA分子剪切成单链,随后一条链经ComEC进入到胞质,另一条链被降解成核苷酸或其它酸溶性产物,释放到细胞外环境或周质中[46]。
2.2 革兰氏阴性菌的DNA转运过程 2.2.1 DNA分子的摄取大多数感受态细菌能够结合任何来源的DNA,但对于某些革兰氏阴性菌如奈瑟氏菌来说,仅含有特定DNA摄取序列(DNA uptake sequence,DUS)的DNA才能被识别并转运[47]。DUS是广泛存在于细菌基因组中的一段短的碱基序列,如淋病奈瑟氏菌(Neisseria gonorrhoeae,N. gonorrhoeae)的DUS为5′-GCCGTCTGAA-3′[48]。含有DUS的DNA被DUS受体(DUS-R)识别后传递至分泌素PilQ。PilQ (图 2)是一种位于细胞外膜上的跨膜蛋白,中央有一个直径约6 nm的中央腔,允许双链DNA分子(约2.4 nm)跨过外膜及细胞壁,从而进入胞内被ComE蛋白识别[49]。
研究表明大部分革兰氏阴性菌利用IV型Pili (Tfp)来转运DNA分子[7]。Tfp由结构亚单位菌毛蛋白聚合而成,能够凸出于细胞内膜表面。以淋病奈瑟氏菌为例(图 2),菌毛蛋白前体肽酶PilD将菌毛蛋白前体加工组装成菌毛蛋白,然后通过多胞型膜蛋白PilG进入到周质。PilF和PilT为ATP酶,为菌毛蛋白的组装与拆卸提供所需能量[7]。DNA分子在先导蛋白PilP的协助下通过PilQ跨过外膜和细胞壁进入到周质,带有DUS的DNA分子被类菌毛蛋白ComP识别,随后由DNA结合蛋白ComE将DNA分子传递到膜通道蛋白ComA,随后双链DNA分子被解链成单链,一条链进入到胞质,另一条被降解成核苷酸或其它酸溶性产物,释放到细胞外环境或周质中[7, 50]。通过对鸭疫里默氏杆菌基因组序列分析发现其存在comE、comM、dprA、ssb等同源基因,但并未发现编码Pili结构的基因,因此可能存在新型DNA转运机制[5]。
2.3 非转化ssDNA分子的降解在革兰氏阳性菌如枯草芽孢杆菌中,当DNA分子与细菌细胞表面DNA受体结合后,具有核酸内切酶活性的膜内蛋白质NucA能够将结合在细菌细胞表面的DNA分子剪切成单链。NucA具有提高DNA环化率的作用,使得只有一条单链DNA分子进入细胞质,而另一条链被降解为核苷酸或其它酸溶性产物,然后释放到外界环境中[6]。对于肺炎链球菌而言,位于细胞膜表面的核酸酶EndA在自然转化的发生中起重要作用。研究发现该基因缺失后DNA仍能发生降解,表明还存在其它降解DNA的蛋白质[52]。对革兰氏阴性菌来说,如淋病奈瑟氏菌[6],双链DNA分子跨过外膜到达位于内膜上的ComA时发生解链,一条链转运进入到胞质,而另一条链降解成核苷酸释放到周质,随后由特定核苷酸转运蛋白转运到胞质,为自然转化过程提供碳源或能量[2]。
2.4 DNA重组加工过程无论是革兰氏阳性菌还是革兰氏阴性菌,当外源DNA分子转运至细菌细胞质后,主要有SsbB、DprA和RecA三种蛋白参与外源DNA与宿主基因组DNA的重组过程。其中DprA和RecA的同源物存在于几乎所有被测序的细菌当中且具有结构和功能的保守性[3]。SsbB能够进一步保护ssDNA免受内源性核酸酶的降解[53]。它是一种ssDNA结合蛋白,能够特异性地结合转运至胞质中的单链DNA分子并呈递给DprA。DprA为DNA加工蛋白,最先在流感嗜血杆菌中被鉴定,具有高度保守性,能够特异地招募RecA[54]。研究发现多种具有自然转化能力的细菌在敲除dprA后转化效率显著降低[55-57]。相似地,鸭疫里默氏杆菌dprA缺失后自然转化不再发生(未发表)。RecA是大分子DNA (DNA序列长度大于250 bp)发生同源重组的必需蛋白,能够招募com调节基因促进ssDNA分子与基因组DNA整合,大多数细菌缺失recA后,自然转化不再发生[58],包括鸭疫里默氏杆菌(未发表)。Overballe-Petersen等敲除鲍曼不动杆菌recA基因后发现,极短DNA片段的同源重组仍可发生,表明极短片段DNA (DNA序列长度小于100 bp)的同源重组不需要RecA的参与[59]。
3 自然转化的生物学功能 3.1 作为食物来源自然转化摄取的DNA能够作为细菌微生物的食物来源,尤其当所摄取的DNA与细菌基因组不具有同源性而不发生重组时。研究表明细菌所摄取的DNA分子能够提供自然转化所需的碳源、磷酸盐来源[37]以及能量来源[60-61]。一些细菌自身基因组复制过程中,在细胞密度过高或者营养缺乏的情况下会摄取现成的碱基和核苷酸满足自身需求,而环境中DNA分子是细菌获得核苷酸的最有效来源。与此观点相一致的是流血嗜血杆菌在营养限制性环境下激发自然感受态的形成[21]。然而,此种观点并不具备普遍性且缺乏有力的证据。如:链球菌和乙酸钙不动杆菌的自然感受态在营养丰富时发生[18, 62];奈瑟氏菌和流感嗜血杆菌等多种细菌只识别并转运与其同源的核苷酸序列[63];而在霍乱弧菌中外源的胞嘧啶核苷反而会抑制自然感受态的发生[15]。
3.2 修复DNA损伤自然转化的另外一种生物学功能被认为是用于基因组的修复。如在链球菌、幽门螺杆菌等缺乏DNA损伤诱导(SOS)反应的细菌,DNA损伤试剂可以诱导其感受态的发生[26-27]。然而此证据也缺乏普遍性,如同样缺乏SOS反应的弯曲杆菌(Campylobacter)的自然感受态并不受抗生素和DNA损伤试剂的诱导[64],而且在鲍曼不动杆菌中不损伤DNA的抗生素可以诱导自然感受态的发生[28]。有证据表明,自然感受态本身有助于肺炎链球菌在抗生素压力下的存活,与重组并无直接关联[65]。
3.3 产生新的基因型自然转化对于生物进化具有重要作用,可以促成新物种的形成以及新耐药性状的产生。研究发现,当同源重组发生在基因组等位基因中编码有害性状的基因上时,极大地促进了细菌清除这些有害突变的几率[66]。此外,自然转化在将无害细菌转化为人类主要病原体的过程中起着重要作用,Rasko等发现自然转化致使两种不同的大肠杆菌病原体重组为高致病性的共生大肠杆菌[67]。自然转化也可能参与耐药基因簇以及毒力岛的转移。如Perron等发现鲍曼不动杆菌中耐药基因的高频率重组致使多重耐药菌株(MDR)的产生[68]。尽管有证据表明自然转化促进了细菌的进化,但单个细胞中DNA摄取与重组的影响尚属未知。
4 展望尽管自然转化现象发现至今已有近百年的历史,但自然转化的机制以及对于细菌的生物学功能仍有许多未解之谜。如我们近期发现鸭疫里默氏杆菌具有自然转化现象[5],但通过基因组序列分析并未发现编码Pili结构的基因,说明存在一种新型的转运机制。在自然转化的调控方面,几乎每种细菌的调控方式都有所不同,自然转化调控潜在的生物学意义仍有待进一步研究。除此之外,自然转化对细菌生理的作用仍未有定论,尽管提出了3种功能假设,但都有缺陷和不足之处。可以推测自然转化的发生对细菌的影响很可能是多元性的,细菌为了满足自身不同的需求摄取外源具有同源性的DNA序列。比如流感嗜血杆菌在营养缺乏时摄取外源DNA作为营养来源。但是不可否认的是,自然转化作为水平基因转移方式之一,在细菌适应环境、新物种形成以及进化方面具有重要的作用。因此,研究自然转化的发生机制意义重大,未来研究中应进一步探究自然转化的作用和调控机制以及在特定细菌中的生物学功能,如自然转化的发生时期、DNA内化机制、编码DNA转运系统基因的功能,以及通过蛋白质-蛋白质(DNA)相互作用分析技术进一步研究DNA转运系统各种复合物的组装和转运过程等。
| [1] |
Claverys JP, Martin B, Polard P. The genetic transformation machinery: composition, localization, and mechanism[J]. FEMS Microbiology Reviews, 2009, 33(3): 643-656. DOI:10.1111/j.1574-6976.2009.00164.x |
| [2] |
Seitz P, Blokesch M. Cues and regulatory pathways involved in natural competence and transformation in pathogenic and environmental Gram-negative bacteria[J]. FEMS Microbiology Reviews, 2013, 37(3): 336-363. DOI:10.1111/j.1574-6976.2012.00353.x |
| [3] |
Johnston C, Martin B, Fichant G, et al. Bacterial transformation: distribution, shared mechanisms and divergent control[J]. Nature Reviews Microbiology, 2014, 12(3): 181-196. DOI:10.1038/nrmicro3199 |
| [4] |
Johnsborg O, Eldholm V, Håvarstein L. Natural genetic transformation: prevalence, mechanisms and function[J]. Research in Microbiology, 2007, 158(10): 767-778. DOI:10.1016/j.resmic.2007.09.004 |
| [5] |
Liu MF, Zhang L, Huang L, et al. Use of natural transformation to establish an easy knockout method in Riemerella anatipestifer[J]. Applied and Environmental Microbiology, 2017, 83(9): e00127-17. |
| [6] |
Chen I, Dubnau D. DNA uptake during bacterial transformation[J]. Nature Reviews Microbiology, 2004, 2(3): 241-249. DOI:10.1038/nrmicro844 |
| [7] |
Averhoff B. DNA transport and natural transformation in mesophilic and thermophilic bacteria[J]. Journal of Bioenergetics and Biomembranes, 2004, 36(1): 25-33. |
| [8] |
Mao JW, Lu T. Population-dynamic modeling of bacterial horizontal gene transfer by natural transformation[J]. Biophysical Journal, 2016, 110(1): 258-268. |
| [9] |
Hamilton HL, Domínguez NM, Schwartz KJ, et al. Neisseria gonorrhoeae secretes chromosomal DNA via a novel type Ⅳ secretion system[J]. Molecular Microbiology, 2010, 55(6): 1704-1721. |
| [10] |
Domingues S, Harms K, Fricke WF, et al. Natural transformation facilitates transfer of transposons, integrons and gene cassettes between bacterial species[J]. PLoS Pathogens, 2012, 8(8): e1002837. DOI:10.1371/journal.ppat.1002837 |
| [11] |
Coupat-Goutaland B, Bernillon D, Guidot A, et al. Ralstonia solanacearum virulence increased following large interstrain gene transfers by natural transformation[J]. Molecular Plant-Microbe Interactions, 2011, 24(4): 497-505. DOI:10.1094/MPMI-09-10-0197 |
| [12] |
Brown CT, Fishwick LK, Chokshi BM, et al. Whole-genome sequencing and phenotypic analysis of Bacillus subtilis mutants following evolution under conditions of relaxed selection for sporulation[J]. Applied and Environmental Microbiology, 2011, 77(19): 6867-6877. DOI:10.1128/AEM.05272-11 |
| [13] |
Ambur OH, Engelstädter J, Johnsen PJ, et al. Steady at the wheel: conservative sex and the benefits of bacterial transformation[J]. Philosophical Transactions of the Royal Society B: Biological Sciences, 2016, 371(1706): 20150528. DOI:10.1098/rstb.2015.0528 |
| [14] |
Biswas GD, Thompson SA, Sparling PF. Gene transfer in Neisseria gonorrhoeae[J]. Clinical Microbiology Reviews, 1989, 2(2 Suppl): S24-S28. |
| [15] |
Blokesch M. Natural competence for transformation[J]. Current Biology, 2016, 26(21): R1126-R1130. DOI:10.1016/j.cub.2016.08.058 |
| [16] |
Marks LR, Reddinger RM, Hakansson AP. High levels of genetic recombination during nasopharyngeal carriage and biofilm formation in Streptococcus pneumoniae[J]. mBio, 2012, 3(5): e00200-12. |
| [17] |
Spizizen J. Transformation of biochemically deficient strains of Bacillus subtilis by deoxyribonucleate[J]. Proceedings of the National Academy of Sciences of the United States of America, 1958, 44(10): 1072-1078. DOI:10.1073/pnas.44.10.1072 |
| [18] |
Palmen R, Vosman B, Buijsman P, et al. Physiological characterization of natural transformation in Acinetobacter calcoaceticus[J]. Journal of General Microbiology, 1993, 139(2): 295-305. DOI:10.1099/00221287-139-2-295 |
| [19] |
Baltrus DA, Guillemin K. Multiple phases of competence occur during the Helicobacter pylori growth cycle[J]. FEMS Microbiology Letters, 2010, 255(1): 148-155. |
| [20] |
Claverys JP, Prudhomme M, Martin B. Induction of competence regulons as a general response to stress in Gram-positive bacteria[J]. Annual Review of Microbiology, 2006, 60(1): 451-475. DOI:10.1146/annurev.micro.60.080805.142139 |
| [21] |
Sinha S, Mell J, Redfield R. The availability of purine nucleotides regulates natural competence by controlling translation of the competence activator Sxy[J]. Molecular Microbiology, 2013, 88(6): 1106-1119. DOI:10.1111/mmi.2013.88.issue-6 |
| [22] |
Metzger LC, Blokesch M. Regulation of competence-mediated horizontal gene transfer in the natural habitat of Vibrio cholerae[J]. Current Opinion in Microbiology, 2016, 30: 1-7. DOI:10.1016/j.mib.2015.10.007 |
| [23] |
Herzberg C, Friedrich A, Averhoff B. comB, a novel competence gene required for natural transformation of Acinetobacter sp. BD413: identification, characterization, and analysis of growth-phase-dependent regulation[J]. Archives of Microbiology, 2000, 173(3): 220-228. DOI:10.1007/s002039900134 |
| [24] |
Porstendörfer D, Gohl O, Mayer F, et al. ComP, a pilin-like protein essential for natural competence in Acinetobacter sp. Strain BD413: regulation, modification, and cellular localization[J]. Journal of Bacteriology, 2000, 182(13): 3673-3680. DOI:10.1128/JB.182.13.3673-3680.2000 |
| [25] |
Suckow G, Seitz P, Blokesch M. Quorum sensing contributes to natural transformation of Vibrio cholerae in a species-specific manner[J]. Journal of Bacteriology, 2011, 193(18): 4914-4924. DOI:10.1128/JB.05396-11 |
| [26] |
Dorer MS, Fero J, Salama NR. DNA damage triggers genetic exchange in Helicobacter pylori[J]. PLoS Pathogens, 2010, 6(7): e1001026. DOI:10.1371/journal.ppat.1001026 |
| [27] |
Prudhomme M, Attaiech L, Sanchez G, et al. Antibiotic stress induces genetic transformability in the human pathogen Streptococcus pneumoniae[J]. Science, 2006, 313(5783): 89-92. DOI:10.1126/science.1127912 |
| [28] |
Quinn B, Martinez J, Liu C, et al. The effect of sub-inhibitory concentrations of antibiotics on natural transformation in Acinetobacter baumannii[J]. International Journal of Antimicrobial Agents, 2018, 51(5): 809-810. DOI:10.1016/j.ijantimicag.2018.01.026 |
| [29] |
Cameron ADS, Redfield RJ. CRP binding and transcription activation at CRP-S sites[J]. Journal of Molecular Biology, 2008, 383(2): 313-323. DOI:10.1016/j.jmb.2008.08.027 |
| [30] |
Redfield RJ, Cameron AD, Qian Q, et al. A novel CRP-dependent regulon controls expression of competence genes in Haemophilus influenzae[J]. Journal of Molecular Biology, 2005, 347(4): 735-747. DOI:10.1016/j.jmb.2005.01.012 |
| [31] |
Sinha S, Mell JC, Redfield RJ. Seventeen Sxy-dependent cyclic AMP receptor protein site-regulated genes are needed for natural transformation in Haemophilus influenzae[J]. Journal of Bacteriology, 2012, 194(19): 5245-5254. DOI:10.1128/JB.00671-12 |
| [32] |
Wu R, Zhao M, Li J, et al. Direct regulation of the natural competence regulator gene tfoX by cyclic AMP (cAMP) and cAMP receptor protein (CRP) in Vibrios[J]. Scientific Reports, 2015, 5: 14921. DOI:10.1038/srep14921 |
| [33] |
Blokesch M. Chitin colonization, chitin degradation and chitin-induced natural competence of Vibrio cholerae are subject to catabolite repression[J]. Environmental Microbiology, 2012, 14(8): 1898-1912. DOI:10.1111/j.1462-2920.2011.02689.x |
| [34] |
Yamamoto S, Izumiya H, Mitobe J, et al. Identification of a chitin-induced small RNA that regulates translation of the tfoX gene, encoding a positive regulator of natural competence in Vibrio cholerae[J]. Journal of Bacteriology, 2011, 193(8): 1953-1965. DOI:10.1128/JB.01340-10 |
| [35] |
Lenz DH, Mok KC, Lilley BN, et al. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae[J]. Cell, 2004, 118(1): 69-82. DOI:10.1016/j.cell.2004.06.009 |
| [36] |
Blokesch M, Schoolnik GK. The extracellular nuclease Dns and its role in natural transformation of Vibrio cholerae[J]. Journal of Bacteriology, 2008, 190(21): 7232-7240. DOI:10.1128/JB.00959-08 |
| [37] |
Seper A, Fengler VHI, Roier S, et al. Extracellular nucleases and extracellular DNA play important roles in Vibrio cholerae biofilm formation[J]. Molecular Microbiology, 2011, 82(4): 1015-1037. DOI:10.1111/j.1365-2958.2011.07867.x |
| [38] |
Lo Scrudato M, Blokesch M. A transcriptional regulator linking quorum sensing and chitin induction to render Vibrio cholerae naturally transformable[J]. Nucleic Acids Research, 2013, 41(6): 3644-3658. DOI:10.1093/nar/gkt041 |
| [39] |
Jaskólska M, Stutzmann S, Stoudmann C, et al. QstR-dependent regulation of natural competence and type Ⅵ secretion in Vibrio cholerae[J]. Nucleic Acids Research, 2018, 46(20): 10619-10634. |
| [40] |
Provvedi R, Dubnau D. ComEA is a DNA receptor for transformation of competent Bacillus subtilis[J]. Molecular Microbiology, 2010, 31(1): 271-280. |
| [41] |
Dubnau D, Provvedi R. Internalizing DNA[J]. Research in Microbiology, 2000, 151(6): 475-480. DOI:10.1016/S0923-2508(00)00166-2 |
| [42] |
Laurenceau R, Pehau-Arnaudet G, Baconnais S, et al. A type Ⅳ pilus mediates DNA binding during natural transformation in Streptococcus pneumoniae[J]. PLoS Pathogens, 2013, 9(6): e1003473. DOI:10.1371/journal.ppat.1003473 |
| [43] |
Chung YS, Dubnau D. ComC is required for the processing and translocation of ComGC, a pilin-like competence protein of Bacillus subtilis[J]. Molecular Microbiology, 2010, 15(3): 543-551. |
| [44] |
Hahn J, Inamine G, Kozlov Y, et al. Characterization of comE, a late competence operon of Bacillus subtilis required for the binding and uptake of transforming DNA[J]. Molecular Microbiology, 2010, 10(1): 99-111. |
| [45] |
Londoño-Vallejo JA, Dubnau D. comF, a Bacillus subtilis late competence locus, encodes a protein similar to ATP-dependent RNA/DNA helicases[J]. Molecular Microbiology, 2010, 9(1): 119-131. |
| [46] |
Provvedi R, Chen I, Dubnau D. NucA is required for DNA cleavage during transformation of Bacillus subtilis[J]. Molecular Microbiology, 2010, 40(3): 634-644. |
| [47] |
Maughan H, Wilson LA, Redfield RJ. Bacterial DNA uptake sequences can accumulate by molecular drive alone[J]. Genetics, 2010, 186(2): 613-627. DOI:10.1534/genetics.110.119438 |
| [48] |
Tettelin H, Saunders NJ, Heidelberg J, et al. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58[J]. Science, 2000, 287(5459): 1809-1815. DOI:10.1126/science.287.5459.1809 |
| [49] |
Cehovin A, Simpson PJ, Mcdowell MA, et al. Specific DNA recognition mediated by a type Ⅳ pilin[J]. Proceedings of the National Academy of Sciences of the United States of America, 2013, 110(8): 3065-3070. DOI:10.1073/pnas.1218832110 |
| [50] |
Hepp C, Maier B. Kinetics of DNA uptake during transformation provide evidence for a translocation ratchet mechanism[J]. Proceedings of the National Academy of Sciences of the United States of America, 2016, 113(44): 12467-12472. DOI:10.1073/pnas.1608110113 |
| [51] |
Aas FE, Wolfgang M, Frye S, et al. Competence for natural transformation in Neisseria gonorrhoeae: components of DNA binding and uptake linked to type Ⅳ pilus expression[J]. Molecular Microbiology, 2010, 46(3): 749-760. |
| [52] |
Bergé M, Moscoso M, Prudhomme M, et al. Uptake of transforming DNA in Gram-positive bacteria: a view from Streptococcus pneumoniae[J]. Molecular Microbiology, 2002, 45(2): 411-421. DOI:10.1046/j.1365-2958.2002.03013.x |
| [53] |
Morrison DA, Mortier-Barrière I, Attaiech L, et al. Identification of the major protein component of the pneumococcal eclipse complex[J]. Journal of Bacteriology, 2007, 189(17): 6497-6500. DOI:10.1128/JB.00687-07 |
| [54] |
Dwivedi GR, Srikanth KD, Anand P, et al. Insights into the functional roles of N-terminal and C-terminal domains of Helicobacter pylori DprA[J]. PLoS One, 2015, 10(7): e0131116. DOI:10.1371/journal.pone.0131116 |
| [55] |
Takata T, Ando T, Israel DA, et al. Role of dprA in transformation of Campylobacter jejuni[J]. FEMS Microbiology Letters, 2010, 252(1): 161-168. |
| [56] |
Smeets LC, Bijlsma JJE, Kuipers EJ, et al. The dprA gene is required for natural transformation of Helicobacter pylori[J]. FEMS Immunology & Medical Microbiology, 2000, 27(2): 99-102. |
| [57] |
Yadav T, Carrasco B, Serrano E, et al. Roles of Bacillus subtilis DprA and SsbA in RecA-mediated genetic recombination[J]. Journal of Biological Chemistry, 2014, 289(40): 27640-27652. DOI:10.1074/jbc.M114.577924 |
| [58] |
Sagi D, Tlusty T, Stavans J. High fidelity of RecA-catalyzed recombination: a watchdog of genetic diversity[J]. Nucleic Acids Research, 2010, 34(18): 5021-5031. |
| [59] |
Overballe-Petersen S, Harms K, Orlando LAA, et al. Bacterial natural transformation by highly fragmented and damaged DNA[J]. Proceedings of the National Academy of Sciences of the United States of America, 2013, 110(49): 19860-19865. DOI:10.1073/pnas.1315278110 |
| [60] |
Allemand JF, Maier B, Smith DE. Molecular motors for DNA translocation in prokaryotes[J]. Current Opinion in Biotechnology, 2012, 23(4): 503-509. DOI:10.1016/j.copbio.2011.12.023 |
| [61] |
Burton B, Dubnau D. Membrane-associated DNA transport machines[J]. Cold Spring Harbor Perspectives in Biology, 2010, 2(7): a000406. |
| [62] |
Straume D, Stamsås GA, Håvarstein LS. Natural transformation and genome evolution in Streptococcus pneumoniae[J]. Infection, Genetics and Evolution, 2015, 33: 371-380. DOI:10.1016/j.meegid.2014.10.020 |
| [63] |
Mell JC, Redfield RJ. Natural competence and the evolution of DNA uptake specificity[J]. Journal of Bacteriology, 2014, 196(8): 1471-1483. DOI:10.1128/JB.01293-13 |
| [64] |
Vegge CS, Brondsted L, Ligowska-Marzęta M, et al. Natural transformation of Campylobacter jejuni occurs beyond limits of growth[J]. PLoS One, 2012, 7(9): e45467. DOI:10.1371/journal.pone.0045467 |
| [65] |
Engelmoer DJP, Rozen DE. Competence increases survival during stress in Streptococcus pneumoniae[J]. Evolution, 2011, 65(12): 3475-3485. DOI:10.1111/evo.2011.65.issue-12 |
| [66] |
De Visser JAGM, Elena SF. The evolution of sex: empirical insights into the roles of epistasis and drift[J]. Nature Reviews Genetics, 2007, 8(2): 139-149. DOI:10.1038/nrg1985 |
| [67] |
Rasko DA, Webster DR, Sahl JW, et al. Origins of the E. coli strain causing an outbreak of hemolytic-uremic syndrome in Germany[J]. New England Journal of Medicine, 2016, 187(2): 709-717. |
| [68] |
Perron GG, Lee AEG, Wang Y, et al. Bacterial recombination promotes the evolution of multi-drug-resistance in functionally diverse populations[J]. Proceedings of the Royal Society B: Biological Sciences, 2012, 279(1733): 1477-1484. DOI:10.1098/rspb.2011.1933 |
 2019, Vol. 46
2019, Vol. 46