扩展功能
文章信息
- 金磊, 王立志, 王之盛, 薛白, 彭全辉
- JIN Lei, WANG Li-Zhi, WANG Zhi-Sheng, XUE Bai, PENG Quan-Hui
- 基于高通量测序技术对山羊盲肠细菌多样性的分析
- Analysis of cecum bacterial diversity of goat based on Illumina MiSeq sequencing
- 微生物学通报, 2019, 46(6): 1423-1433
- Microbiology China, 2019, 46(6): 1423-1433
- DOI: 10.13344/j.microbiol.china.180599
-
文章历史
- 收稿日期: 2018-07-30
- 接受日期: 2018-10-15
- 网络首发日期: 2018-11-13
反刍动物的胃肠道拥有丰富多样的微生物菌群,其不同胃肠道区域的微生物功能偏重点不同,例如,瘤胃微生物的功能主要是消化降解营养物质,尤其是纤维物质,而肠道微生物除了对瘤胃未完全消化的营养物质进行补偿消化外,在保持宿主肠道健康也发挥重要作用[1-5]。对于反刍动物而言,科研工作者以往多数是以瘤胃或粪便样品的微生物为代表,来反映整个胃肠道微生物的结构与组成[6-11]。然而,研究发现,动物胃肠道不同区域的微生物多样性及结构存在显著差异[12-15]。因此,反刍动物瘤胃和粪便微生物并不能真实的反映胃肠道其他部分的微生物结构与组成。研究表明,动物盲肠微生物在宿主营养物质消化和肠道健康方面均扮演着重要角色,例如:盲肠微生物能降解纤维素、半纤维素和不溶性蛋白质等为宿主提供能量,其还是末端免疫器官发育和成熟所必需的[16-18]。目前为止,学者们在对猪[19]、肉鸡[20]和兔[21]等动物的盲肠微生物多样性的研究中均有重要发现,但关于山羊盲肠微生物菌群多样性的研究尚未见报道。针对这一问题,本研究采用高通量测序技术分析山羊盲肠细菌多样性,为深入研究山羊盲肠微生物的组成与营养物质的消化吸收和肠道健康之间的关系奠定基础。
1 材料与方法 1.1 实验动物与样品采集实验动物为12只年龄为10月龄、平均体重为20.70±1.60 kg的雌性努比亚黑山羊。饲养在相同环境下,采用自由采食、饮水的管理方式。参照我国《肉羊饲养标准》(NY/T 816-2004),以每天每头增重0.1 kg为标准,12只山羊自由采食配合饲料(玉米38.47%,羊草35.00%,苜蓿草粉20.00%,豆粕4.50%,食盐0.45%,小苏打0.45%,碳酸钙0.08%,磷酸氢钙0.60%,维生素、矿物质添加剂0.45%)。预饲20 d,实验的第21天屠宰所有山羊,分割出盲肠并进行结扎,以防止相邻肠断的内容物流入盲肠,迅速采集盲肠内容物样品于20 mL的冻存管中,-80 ℃保存备用。
1.2 DNA提取和PCR扩增DNA的提取和纯化参照Guo等[22]描述的方法进行。用琼脂糖凝胶电泳检测提取DNA的纯度和浓度。选择16S rRNA基因的V4区作为扩增和测序的目的片段,引物序列为:515F (5′-GTGCCAGC MGCCGCGGTAA-3′)和806R (5′-GGACTACVSGG GTATCTAAT-3′),然后对目的基因片段进行扩增。PCR反应体系(50 μL):50 ng/μL模板DNA 1.0 μL,10×Buffer 5.0 μL,10 μmol/L引物各1.25 μL,10 mmol/L dNTPs 1.0 μL,5 U/μL Taq酶0.25 μL,补ddH2O至50 μL。PCR反应条件:95 ℃ 2 min;95 ℃ 30 s,55 ℃ 30 s,72 ℃ 30 s,30个循环;72 ℃ 5 min。PCR产物用2%琼脂糖凝胶电泳鉴定并用PCR纯化试剂盒纯化和回收。回收的PCR产物用QuantiFluor™-ST fluorometer进行定量,采用Illumina MiSeq测序平台进行测序。
1.3 生物信息学分析高通量测序的原始数据为Fastq文件格式,先用QIIME 1.8.0软件[23]对原始数据进行初次质控,去除序列平均碱基质量值< Q20、未知碱基数 > 6及同聚物太大的序列,保留240-500 bp长度的序列。去除Barcode和引物序列,然后用Mothur[24]拼接质控后的序列,将相似度大于97%的序列使用Uclust[25]聚类为一个分类操作单元(Operational taxonomic units,OTU)。挑选相对丰度最高的OTU代表序列与基因数据库(Green genes)中的参考序列进行比对[26],使用RDF分类器[27]对代表序列进行物种注释。基于OTU table和rep_set. tree文件及抽样的最大深度绘制稀释曲线并统计各样品α多样性指数;结合各个样品的OTU种类及其丰度进行计算,获得样品间Unweighted UniFrac距离矩阵,利用加权组平均法(UPGMA)进行聚类分析,绘制PCoA聚类图[28-29]。用OriginPro 9.0和Rx 64 3.0.2软件绘制优势菌门柱形图和共享属聚类热图。基于16S rRNA基因扩增子测序结果,对盲肠微生物的基因进行PICRUSt功能预测[30-31]。
2 结果与分析 2.1 样品序列及OTU统计本次实验测序共获得813 496条有效序列,平均每个样品含有76 753±6 574条序列。基于相似性大于97%的原则,将获得的有效序列进行聚类,共获得6 883个OTU,平均每个样品含有3 583±456个OTU。每个样品有效序列和OTU数详见表 1。由表 1可知,所有样品的Coverage (测序深度指数)数值均为0.99,此外,由图 1也可以看出,所有样品的稀释曲线最终均趋于平缓,说明此次测序深度全面地覆盖了山羊的盲肠微生物,测序数据量足够大,可以反映样本中绝大多数的细菌信息。在相同测序深度下,OTU数由高到低的样品依次为C10、C5、C4、C8、C2、C9、C11、C11、C3、C7、C6和C12。
| Sample ID | Reads | OTUs | Chao1 | Shannon | Simpson | Coverage |
| C1 | 66 846 | 3 205 | 2 199.36 | 8.64 | 0.99 | 0.99 |
| C2 | 69 303 | 3 160 | 2 187.50 | 8.05 | 0.98 | 0.99 |
| C3 | 82 777 | 3 953 | 2 399.76 | 9.05 | 0.99 | 0.99 |
| C4 | 71 549 | 3 517 | 2 123.16 | 8.87 | 0.99 | 0.99 |
| C5 | 67 105 | 4 260 | 2 111.81 | 8.53 | 0.99 | 0.99 |
| C6 | 78 140 | 4 260 | 2 239.10 | 8.92 | 0.99 | 0.99 |
| C7 | 75 284 | 3 288 | 2 131.86 | 8.89 | 0.99 | 0.99 |
| C8 | 80 747 | 2 918 | 2 196.74 | 7.75 | 0.97 | 0.99 |
| C9 | 78 573 | 4 112 | 2 050.11 | 8.57 | 0.99 | 0.99 |
| C10 | 83 782 | 3 583 | 2 402.96 | 8.69 | 0.99 | 0.99 |
| C11 | 82 340 | 3 493 | 2 390.23 | 8.79 | 0.99 | 0.99 |
| C12 | 84 590 | 3 271 | 2 179.22 | 8.01 | 0.98 | 0.99 |
| Mean | 76 753 | 3 583 | 2 217.65 | 8.56 | 0.98 | 0.99 |
| Standard deviation | 6 574 | 456 | 119.26 | 0.41 | 0.01 | 0.00 |

|
| 图 1 12个样品的稀释曲线 Figure 1 Rarefaction curves of 12 samples |
|
|
α多样性指数是反映样品中物种丰富度(Chao1)和均匀度(Shannon和Simpson)的指标。由表 2可知,12个样品的Shannon和Simpson多样性指数的标准差较小,表明所有样品的微生物菌落的均匀度基本一致,而所有样品的Chao1指数标准差较大,表明12个样品的物种丰富度存在个体差异。β多样性分析主要用于评价样品间微生物的相似性。结合样本的OTU的种类及其相对丰度,计算样本间加权遗传距离矩阵,利用距离矩阵绘制PCoA聚类图,分析所有样品间的相似性。由图 2可知,第1主成分的贡献率为16.44%,第2主成分的贡献率为12.21%。12个盲肠内容物样品中,其中3个样品并未依据所处的组别与其余9个样品相互聚类到一起,表明3个样品与其余的样品差异较大。整体来看,12个样品的相似性大于其差异性。
| Taxa | Ruminococcaceae | Clostridiales | Lachnospiraceae | Clostridium | Bacteroidales | Bacteroidaceae | Clostridiaceae | Ruminococcus |
| C1 | 22.25 | 20.25 | 5.77 | 5.02 | 4.98 | 3.03 | 2.73 | 2.10 |
| C2 | 22.99 | 22.65 | 6.11 | 3.93 | 2.51 | 1.68 | 3.11 | 4.88 |
| C3 | 32.01 | 12.45 | 5.19 | 6.89 | 6.30 | 4.11 | 1.04 | 2.45 |
| C4 | 28.54 | 12.57 | 6.31 | 8.43 | 6.83 | 3.85 | 1.96 | 2.03 |
| C5 | 27.56 | 11.73 | 6.53 | 10.95 | 10.61 | 3.96 | 1.92 | 2.15 |
| C6 | 24.61 | 13.01 | 7.19 | 7.82 | 4.74 | 4.02 | 1.24 | 2.59 |
| C7 | 25.81 | 11.31 | 7.43 | 7.58 | 6.86 | 5.46 | 1.39 | 1.86 |
| C8 | 17.37 | 9.93 | 5.77 | 7.73 | 6.30 | 4.42 | 1.87 | 2.33 |
| C9 | 25.47 | 11.17 | 7.43 | 8.28 | 11.98 | 4.58 | 1.03 | 2.44 |
| C10 | 23.53 | 10.62 | 7.84 | 5.82 | 8.94 | 5.26 | 1.83 | 1.69 |
| C11 | 24.43 | 10.91 | 7.98 | 8.01 | 7.70 | 5.37 | 1.96 | 2.34 |
| C12 | 21.71 | 10.15 | 8.02 | 10.89 | 14.99 | 4.78 | 1.80 | 2.26 |
| Mean | 24.69 | 13.06 | 6.79 | 7.61 | 7.72 | 4.21 | 1.82 | 2.42 |
| Standard deviation | 3.71 | 4.06 | 0.97 | 2.07 | 3.43 | 1.06 | 0.63 | 0.81 |

|
| 图 2 PCoA聚类分析 Figure 2 Cluster analysis by Principal Coordinate Analysis |
|
|
本研究中,共注释得到12个门。相对丰度排名前10的门均依次分别为厚壁菌门(Firmicutes)、拟杆菌门(Bacteroidetes)、疣微菌门(Verrucomicrobia)、软壁菌门(Tenericutes)、变形菌门(Proteobacteria)、广古菌门(Euryarchaeota)、黏胶球形菌门(Lentisphaerae)、螺旋体门(Spirochaetes)、纤维杆菌门(Fibrobacteres)、放线菌门(Actinobacteria)和浮霉状菌门(Planctomycetes),其物种组成如图 3所示。其中所有样品的最优菌门均为Firmicutes,其次为Bacteroidetes。各样品Firmicutes和Bacteroidetes的相对丰度见表 2,12个样品Firmicutes和Bacteroidetes的平均相对丰度为66.05%±5.79%、20.26%±4.05% (Mean±Std)。
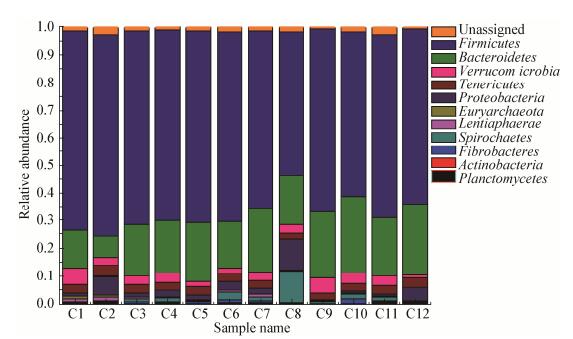
|
| 图 3 门水平上的菌群组成 Figure 3 Composition of microbial at the phylum level |
|
|
在属水平,所有样品共检测到112个属。限于属种类多且部分属的相对丰度较低,几乎没有研究价值或培养技术等的限制无法对其功能进行研究,因而仅对相对丰度大于0.1%的47个属进行了关注。各样品相对丰度大于0.1%的微生物共有47个,其中包括未分类至属的微生物16个。针对相对丰度前30的共享菌群绘制聚类热图(图 4),这30个共享微生物占总菌群相对丰度的78.28%-89.41%。由图 4可知,C9和C12、C4和C11、C3和C7、C11和C2分别聚为一簇,表明它们的菌群相似性相对高于同组内其他样品。一般将相对丰度大于1%的微生物称为优势菌群,优势菌群共有8个,未分类至属水平的共6个物种,分别为瘤胃球菌科(f_Ruminococcaceae)、梭菌目(o_Clostridiales)、毛螺旋菌科(f_Lachnospiraceae)、拟杆菌目(o_Bacteroidales)、拟杆菌科(f_Bacteroidaceae)和梭菌科(f_Clostridiaceae)。优势菌属为瘤胃球菌属(g_Ruminococcus)和梭菌属(g_Clostridium),其各样品优势菌群的相对丰度详见表 2。各样品相对丰度最高的微生物为f_Ruminococcaceae,其各样品相对丰度的变化范围为17.37%-32.01%,12个样品平均相对丰度为24.69%±3.71%。各样品相对丰度最高的属为g_Clostridium,其各样品相对丰度的变化范围为5.02%-10.89%,12个样品平均相对丰度为7.61%±2.07%。

|
| 图 4 共享属聚类热图 Figure 4 Shared genus cluster heatmap 注:越偏蓝色表示相对丰度越高,越偏绿色表示丰度越低.对于不能分类到属水平,但仍然出现在各样品中的共享序列,用其最高分类水平表示. Note: The closer to blue, the higher of their relative abundance, while the closer to green, the lower of their relative abundance. The shared sequences, which could not be classified to any genus but still existed in samples, were expressed using the highest classification level that can be assigned to them. |
|
|
PICRUSt对盲肠微生物的基因进行功能预测,图 5展示了34种功能基因。相对丰度从高到低的基因其功能依次为膜转运、碳水化合物代谢、氨基酸代谢、能量代谢、辅助因子和维生素代谢、脂质代谢等。通过对34种功能基因进行归类,由图 6可知,盲肠微生物最主要的基因功能为代谢功能类,其各样品的相对丰度分别为C1为46.26%,C2为46.01%,C3为46.59%,C4为46.45%,C5为46.49%,C6为46.24%,C7为46.63%,C8为45.82%,C9为46.98%,C10为47.14%,C11为46.76%,C12为46.71%,平均相对丰度为46.51%±0.38%。
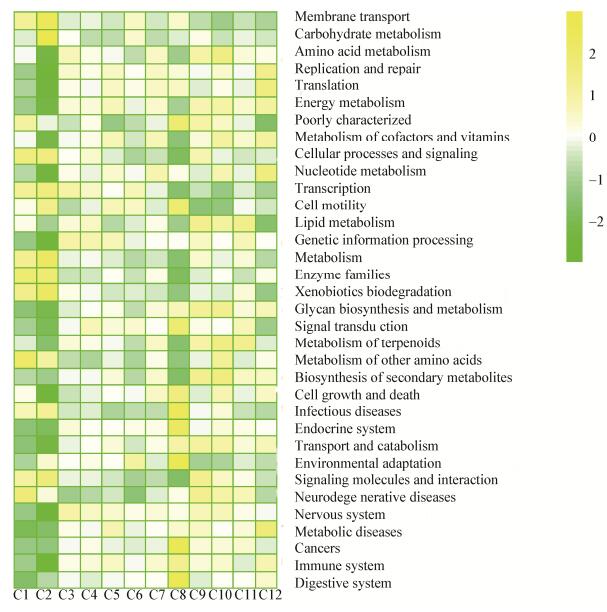
|
| 图 5 功能基因的相对丰度组成 Figure 5 The composition of the relative abundance contributed by the 34 genes 注:越偏黄色表示相对丰度越高,越偏绿色表示丰度越低. Note: The closer to yellow, the higher of their relative abundance, while the closer to green, the lower of their relative abundance. |
|
|

|
| 图 6 功能基因的分类 Figure 6 Classification of functional genes |
|
|
反刍动物肠道微生物可帮助宿主从饲料中获取养分,并能维持肠道健康。目前为止,有关反刍动物肠道微生物组成和结构的研究较少[32-33],就盲肠微生物多样性的研究还未见报道。因此,本试验采用二代测序技术研究了山羊盲肠微生物的物种组成及其丰度。通过稀释曲线和Coverage指数分析表明,此次测序数据比较全面的覆盖了12个研究对象的盲肠微生物,能够真实全面的反映其组成情况。综合本研究PCoA图以及门和属水平上各样品中优势菌群的相对丰度可知,不同山羊个体之间盲肠微生物的多样性及相对丰度存在差异,例如:各样品厚壁菌门与瘤胃球菌科相对丰度变化范围分别为52.02%-72.57%和17.37%- 32.01%。研究表明,品种、性别、饮食等因素均一致的实验个体之间,其菌群个体差异是普遍存在的,通过对优势菌群的分析,可以规避个体差异得到科学的实验结果[34-35]。
动物胃肠道中厚壁菌门最主要的功能是降解纤维类物质,而拟杆菌门主要降解非纤维物质[36-37]。研究发现,厚壁菌门与拟杆菌门的相对比例能够反映宿主对能量的吸收和储藏的能力[38-39]。已有研究发现,肠道中拟杆菌门的相对丰度与宿主体脂含量呈显著负相关[40-41],其可以通过增加脂肪分解和脂肪酸氧化,增加脂肪的降解,从而降低了宿主体脂含量[42]。本研究中,山羊盲肠微生物相对丰度最高的是厚壁菌门(Firmicutes),其次为拟杆菌门(Bacteroidetes)。这与以往在鸡[12]和猪[13]盲肠微生物的研究结果一致,说明反刍动物与单胃动物盲肠微生物的组成存在共性。王继文等[11]研究发现,山羊瘤胃中相对丰度最高的是拟杆菌门,其次为厚壁菌门。de Jesús-Laboy等[43]报道,山羊粪便相对丰度最高的门为厚壁菌门,其次为拟杆菌门。以上研究结果表明,反刍动物盲肠与瘤胃微生物的组成存在差异,而与粪便微生物的组成较相似,推测动物胃肠道生理部位越接近其微生物的相似度可能越高。已有研究发现,同一品种的动物,各肠段菌群组成存在特异性[44-45],这些现象可能是由不同胃肠道部位的pH值、食糜蠕动、分泌物和氧化还原电位等因素的差异造成的。
研究发现,瘤胃球菌科中有些菌属能够产生纤维素酶、淀粉酶等碳水化合物降解酶[46]。此外,隶属于瘤胃球菌科的瘤胃球菌属所含的白色瘤胃球菌和黄色瘤胃球菌能分泌大量的纤维素酶和半纤维素酶[47],说明瘤胃球菌科微生物具有广泛的碳水化合物降解活性。本试验得出,山羊盲肠属水平的微生物相对丰度最高的为未分类的瘤胃球菌科(f_Ruminococcaceae),且瘤胃球菌属(g_Ruminococcus)在各样品中的相对丰度也较高。Gu等[48]研究发现,瘤胃球菌科是小鼠大肠和粪便中的优势菌群,与本试验的结果相似。本试验结果表明,山羊后肠可能存在活跃的碳水化合物发酵。以往的研究发现,反刍动物瘤胃的优势菌属普遍为普雷沃氏菌属,例如:山羊[11]、奶牛[49]、肉牛[50]和绵羊[51]等。值得注意的是,本研究中,虽然山羊盲肠相对丰度最高的为未分类的瘤胃球菌科,但普雷沃氏菌属在各样品其相对丰度均较高。而普雷沃氏菌属几乎可以单独完成蛋白质、糖类和碳水化合物等营养物质的降解任务,它在胃肠道不同部位的分布与其高度遗传变异有关[52-53]。从本试验结果中可知,以往采用瘤胃微生物并不能真实的反映肠道微生物的分布情况。Frey等[54]研究报道,食糜从胃肠道的一个部位到另一个部位时,其内的微生物结构与组成会发生显著变化。因此,胃肠道不同部位间微生物差异有可能是因为微生物随着食糜流向肠道时,肠道内酸性环境会溶解微生物细胞,部分细菌会降解甚至死亡,到达后肠的瘤胃微生物减少,导致胃肠道微生物的组成会发生改变。
本研究中,未分类的拟杆菌科(f_Bacteroidaceae)、未分类的毛螺旋菌科(f_Lachnospiraceae)和梭菌属(g_Clostridium)均为盲肠优势菌群。研究表明,隶属于拟杆菌目的脆弱拟杆菌能促进肠道免疫系统的发育[55]。研究发现,毛螺旋菌科有抗肠炎的功能[56],因为毛螺旋菌科(Lachnospiraceae)中的多个菌属为丁酸生成菌属,可以发酵纤维物质产生丁酸,促进盲肠防御素基因的表达,从而有效地抑制病源微生物侵袭[57-58]。梭菌属包括一种普遍存在于哺乳动物肠道内的益生菌-丁酸梭菌,它能通过发酵纤维物质,代谢产生丁酸刺激粘膜免疫反应,从而抑制肠道有害菌的生长[59]。这些结果暗示了盲肠微生物在维持肠道健康方面发挥着重要作用。同时,以上的3种微生物均能分泌纤维素酶,帮助宿主消化纤维素[60-63]。本研究结果再次提示我们,山羊盲肠可能存在活跃的碳水化合物发酵,并且已有研究表明,反刍动物的后肠道是可以补偿消化过瘤胃的纤维物质[64-65]。同时,说明盲肠微生物可能主要是通过影响消化代谢产物来调节肠道免疫功能。
本研究的PICRUSt基因预测也暗示了盲肠菌群在营养物质的消化代谢方面发挥重要作用,因为盲肠微生物最主要的功能类别为代谢功能类,包括能量代谢、糖酵解途径、碳水化合物代谢、脂肪代谢和氨基酸代谢等。说明盲肠微生物能够影响山羊的代谢从而影响其生长性能。此外,很多菌群的功能仍不清楚,这将是今后研究的方向。本试验中,优势菌属相当大一部分为尚未命名的菌属,说明有关山羊盲肠微生物在属水平上的研究还不够深入。
4 结论山羊盲肠微生物与瘤胃微生物组成存在差异,与粪便微生物组成相似。与单胃动物盲肠微生物相比,存在一定的共性。山羊盲肠中相对丰度最高的门为是厚壁菌门(Firmicutes),其次为拟杆菌门(Bacteroidetes)。盲肠微生物的优势菌群主要为纤维降解菌类和与肠道健康相关的微生物,包括瘤胃球菌属(Ruminococcus)、梭菌属(Clostridium)和毛螺旋菌科(Lachnospiraceae)等。
| [1] |
Dodd D, Spitzer MH, van Treuren W, et al. A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites[J]. Nature, 2017, 551(7682): 648-652. DOI:10.1038/nature24661 |
| [2] |
Kadoki M, Patil A, Thaiss CC, et al. Organism-level analysis of vaccination reveals networks of protection across tissues[J]. Cell, 2017, 171(2): 398-413. DOI:10.1016/j.cell.2017.08.024 |
| [3] |
Cervantes-Barragan L, Chai JN, Tianero MD, et al. Lactobacillus reuteri induces gut intraepithelial CD4+CD8αα+ T cells[J]. Science, 2017, 357(6353): 806-810. DOI:10.1126/science.aah5825 |
| [4] |
Koppel N, Maini RV, Balskus EP. Chemical transformation of xenobiotics by the human gut microbiota[J]. Science, 2017, 356(6344): 1246-1257. |
| [5] |
Bauer PV, Duca FA, Waise T, et al. Metformin alters upper small intestinal microbiota that impact a glucose-sglt1-sensing glucoregulatory pathway[J]. Cell Metabolism, 2018, 27(1): 101-117. DOI:10.1016/j.cmet.2017.09.019 |
| [6] |
Lettat A, Nozière P, Silberberg M, et al. Rumen microbial and fermentation characteristics are affected differently by bacterial probiotic supplementation during induced lactic and subacute acidosis in sheep[J]. BMC Microbiology, 2012, 12(1): 142. |
| [7] |
Riyanti L, Suryahadi, Evvyernie D. In vitro fermentation characteristics and rumen microbial population of diet supplemented with saccharomyces cerevisiae and rumen microbe probiotics[J]. Media Peternakan, 2016, 39(1): 40-45. |
| [8] |
Ramírez-Restrepo CA, Tan C, O'Neill CJ, et al. Methane production, fermentation characteristics, and microbial profiles in the rumen of tropical cattle fed tea seed saponin supplementation[J]. Animal Feed Science and Technology, 2016, 216: 58-67. DOI:10.1016/j.anifeedsci.2016.03.005 |
| [9] |
Zhang XJ, Wang LZ. Effects of dietary neutral detergent fibre level on structure and composition of rumen bacteria in goats[J]. Chinese Journal of Animal Nutrition, 2018, 30(4): 1377-1386. (in Chinese) 张雪娇, 王立志. 饲粮中性洗涤纤维水平对山羊瘤胃细菌结构及组成的影响[J]. 动物营养学报, 2018, 30(4): 1377-1386. DOI:10.3969/j.issn.1006-267x.2018.04.020 |
| [10] |
Jin L, Zhou ML, Wang LL, et al. Differences of microbial structure and composition in goat rumens with different energy metabolism efficiency[J]. Microbiology China, 2018, 45(1): 91-101. (in Chinese) 金磊, 周美丽, 王禄禄, 等. 不同能量代谢率的山羊瘤胃微生物结构与组成的差异性[J]. 微生物学通报, 2018, 45(1): 91-101. |
| [11] |
Wang JW, Wang LZ, Yan TH, et al. Diversity of ruminal and fecal microbiota of goat[J]. Chinese Journal of Animal Nutrition, 2015, 27(8): 2559-2571. (in Chinese) 王继文, 王立志, 闫天海, 等. 山羊瘤胃与粪便微生物多样性[J]. 动物营养学报, 2015, 27(8): 2559-2571. DOI:10.3969/j.issn.1006-267x.2015.08.030 |
| [12] |
Zhao LL, Wang G, Siegel P, et al. Quantitative genetic background of the host influences gut microbiomes in chickens[J]. Scientific Reports, 2013, 3(5): 1163. |
| [13] |
Liu XF, Fan HL, Ding XB, et al. Analysis of the gut microbiota by high-throughput sequencing of the V5-V6 regions of the 16S rRNA gene in donkey[J]. Current Microbiology, 2014, 68(5): 657-662. DOI:10.1007/s00284-014-0528-5 |
| [14] |
Dougal K, Harris PA, Edwards A, et al. A comparison of the microbiome and the metabolome of different regions of the equine hindgut[J]. FEMS Microbiology Ecology, 2012, 82(3): 642-652. DOI:10.1111/j.1574-6941.2012.01441.x |
| [15] |
Gu SH, Chen DD, Zhang JN, et al. Bacterial community mapping of the mouse gastrointestinal tract[J]. PLoS One, 2013, 8(10): e74957. DOI:10.1371/journal.pone.0074957 |
| [16] |
Wenk C. The role of dietary fibre in the digestive physiology of the pig[J]. Animal Feed Science and Technology, 2001, 90(1/2): 21-33. |
| [17] |
Guarner F, Malagelada JR. Gut flora in health and disease[J]. The Lancet, 2003, 361(9356): 512-519. DOI:10.1016/S0140-6736(03)12489-0 |
| [18] |
Skřivanová E, Molatová Z, Skřivanová V, et al. Inhibitory activity of rabbit milk and medium-chain fatty acids against enteropathogenic Escherichia coli O128[J]. Veterinary Microbiology, 2009, 135(3/4): 358-362. |
| [19] |
Kang RM, Li Y, Lü XB, et al. Analysis of the cecum microbial diversity of different pig breeds by Illumina amplicon sequencing of 16S rDNA tag[J]. China Animal Husbandry & Veterinary Medicine, 2017, 44(11): 3121-3129. (in Chinese) 康润敏, 李瑶, 吕学斌, 等. 利用16S rDNA扩增子测序技术分析不同品种猪盲肠微生物菌落多样性[J]. 中国畜牧兽医, 2017, 44(11): 3121-3129. |
| [20] |
Lin YC, Xu S, Ni XQ, et al. Diversity of the cecal microbiome of broiler chicken based on Illumina MiSeq sequencing platform[J]. Journal of China Agricultural University, 2016, 21(12): 65-73. (in Chinese) 林奕岑, 徐帅, 倪学勤, 等. 利用Illumina MiSeq测序平台分析肉鸡盲肠微生物多样性[J]. 中国农业大学学报, 2016, 21(12): 65-73. |
| [21] |
Xu YJ, Zhang YK, Shen XM, et al. Caecal microbiota diversity analysis of weaning rabbits using Illumina MiSeq sequencing technology[J]. Chinese Journal of Animal Nutrition, 2015, 27(9): 2793-2802. (in Chinese) 许宇静, 张煜坤, 沈雪梅, 等. 采用Illumina MiSeq测序技术分析断奶幼兔盲肠微生物群落的多样性[J]. 动物营养学报, 2015, 27(9): 2793-2802. DOI:10.3969/j.issn.1006-267x.2015.09.017 |
| [22] |
Guo W, Li Y, Wang LZ, et al. Evaluation of composition and individual variability of rumen microbiota in yaks by 16S rRNA high-throughput sequencing technology[J]. Anaerobe, 2015, 34: 74-79. DOI:10.1016/j.anaerobe.2015.04.010 |
| [23] |
Caporaso JG, Kuczynski J, Stombaugh J, et al. Qiime allows analysis of high-throughput community sequencing data[J]. Nature Methods, 2010, 7(5): 335-336. DOI:10.1038/nmeth.f.303 |
| [24] |
Schloss PD, Westcott SL, Ryabin T, et al. Introducing mothur: open-source, platform-independent, community- supported software for describing and comparing microbial communities[J]. Applied and Environmental Microbiology, 2009, 75(23): 7537-7541. DOI:10.1128/AEM.01541-09 |
| [25] |
Edgar RC. Search and clustering orders of magnitude faster than blast[J]. Bioinformatics, 2010, 26(19): 2460-2461. DOI:10.1093/bioinformatics/btq461 |
| [26] |
Caporaso JG, Bittinger K, Bushman FD, et al. Pynast: a flexible tool for aligning sequences to a template alignment[J]. Bioinformatics, 2010, 26(2): 266-267. DOI:10.1093/bioinformatics/btp636 |
| [27] |
Cole JR, Wang Q, Cardenas E, et al. The ribosomal database project: improved alignments and new tools for rRNA analysis[J]. Nucleic Acids Research, 2009, 37(Database issue): D141-D145. |
| [28] |
Quast C, Pruesse E, Yilmaz P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools[J]. Nucleic Acids Research, 2013, 41(D1): D590-D596. |
| [29] |
Lozupone C, Lladser ME, Knights D, et al. Unifrac: an effective distance metric for microbial community comparison[J]. The ISME Journal, 2011, 5(2): 169-172. DOI:10.1038/ismej.2010.133 |
| [30] |
Langille MG, Zaneveld J, Caporaso JG, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences[J]. Nature Biotechnology, 2013, 31(9): 814-821. DOI:10.1038/nbt.2676 |
| [31] |
Fu LL, Jiang BH, Liu JL, et al. Genome sequence analysis of a flocculant-producing bacterium, Paenibacillus shenyangensis[J]. Biotechnology Letters, 2016, 38(3): 447-453. DOI:10.1007/s10529-015-1990-2 |
| [32] |
Mao SY, Zhang ML, Liu JH, et al. Characterising the bacterial microbiota across the gastrointestinal tracts of dairy cattle: membership and potential function[J]. Scientific Reports, 2015, 5: 16116. DOI:10.1038/srep16116 |
| [33] |
Meale SJ, Li SC, Azevedo P, et al. Weaning age influences the severity of gastrointestinal microbiome shifts in dairy calves[J]. Scientific Reports, 2017, 7: 198. DOI:10.1038/s41598-017-00223-7 |
| [34] |
Sergeant MJ, Constantinidou C, Cogan TA, et al. Extensive microbial and functional diversity within the chicken cecal microbiome[J]. PLoS One, 2014, 9(3): e91941. DOI:10.1371/journal.pone.0091941 |
| [35] |
Durso LM, Harhay GP, Smith TP, et al. Animal-to-animal variation in fecal microbial diversity among beef cattle[J]. Applied and Environmental Microbiology, 2010, 76(14): 4858-4862. DOI:10.1128/AEM.00207-10 |
| [36] |
Chen YH, Penner GB, Li MJ, et al. Changes in bacterial diversity associated with epithelial tissue in the beef cow rumen during the transition to a high-grain diet[J]. Applied and Environmental Microbiology, 2011, 77(16): 5770-5781. DOI:10.1128/AEM.00375-11 |
| [37] |
Evans NJ, Brown JM, Murray RD, et al. Characterization of novel bovine gastrointestinal tract treponema isolates and comparison with bovine digital dermatitis treponemes[J]. Applied and Environmental Microbiology, 2011, 77(1): 138-147. DOI:10.1128/AEM.00993-10 |
| [38] |
Turnbaugh PJ, Ley RE, Mahowald MA, et al. An obesity-associated gut microbiome with increased capacity for energy harvest[J]. Nature, 2006, 444(7122): 1027-1031. DOI:10.1038/nature05414 |
| [39] |
Guo XL, Xia XJ, Tang RY, et al. Real-time PCR quantification of the predominant bacterial divisions in the distal gut of Meishan and Landrace pigs[J]. Anaerobe, 2008, 14(4): 224-228. DOI:10.1016/j.anaerobe.2008.04.001 |
| [40] |
Gangarapu V, Yıldız K, Ince AT, et al. Role of gut microbiota: obesity and NAFLD[J]. Turkish Journal of Gastroenterology: the Official Journal of Turkish Society of Gastroenterology, 2014, 25(2): 133-140. |
| [41] |
Mouzaki M, Comelli EM, Arendt BM, et al. Intestinal microbiota in patients with nonalcoholic fatty liver disease[J]. Hepatology, 2013, 58(1): 120-127. DOI:10.1002/hep.26319 |
| [42] |
Liu RX, Hong J, Xu XQ, et al. Gut microbiome and serum metabolome alterations in obesity and after weight-loss intervention[J]. Nature Medicine, 2017, 23(7): 859. DOI:10.1038/nm.4358 |
| [43] |
de Jesús-Laboy KM, Godoy-Vitorino F, Piceno YM, et al. Comparison of the fecal microbiota in feral and domestic goats[J]. Genes, 2011, 3(1): 1-18. |
| [44] |
Wise MG, Siragusa GR. Quantitative analysis of the intestinal bacterial community in one-to three-week-old commercially reared broiler chickens fed conventional or antibiotic-free vegetable-based diets[J]. Journal of Applied Microbiology, 2010, 102(4): 1138-1149. |
| [45] |
Torok VA, Allison GE, Percy NJ, et al. Influence of antimicrobial feed additives on broiler commensal posthatch gut microbiota development and performance[J]. Applied and Environmental Microbiology, 2011, 77(10): 3380-3390. DOI:10.1128/AEM.02300-10 |
| [46] |
Suen G, Stevenson DM, Bruce DC, et al. Complete genome of the cellulolytic ruminal bacterium Ruminococcus albus 7[J]. Journal of Bacteriology, 2011, 193(19): 5574-5575. DOI:10.1128/JB.05621-11 |
| [47] |
Krause DO, Dalrymple BP, Smith WJ, et al. 16S rDNA sequencing of Ruminococcus albus and Ruminococcus flavefaciens: design of a signature probe and its application in adult sheep[J]. Microbiology, 1999, 145(7): 1797-1807. DOI:10.1099/13500872-145-7-1797 |
| [48] |
Gu SH, Chen DD, Zhang JN, et al. Bacterial community mapping of the mouse gastrointestinal tract[J]. PLoS One, 2013, 8(10): e74957. DOI:10.1371/journal.pone.0074957 |
| [49] |
Stevenson DM, Weimer PJ. Dominance of Prevotella and low abundance of classical ruminal bacterial species in the bovine rumen revealed by relative quantification real-time PCR[J]. Applied Microbiology and Biotechnology, 2009, 83(5): 987-988. DOI:10.1007/s00253-009-2033-5 |
| [50] |
de Oliveira MN, Jewell KA, Freitas FS, et al. Characterizing the microbiota across the gastrointestinal tract of a Brazilian Nelore steer[J]. Veterinary Microbiology, 2013, 164(3/4): 307-314. |
| [51] |
Bekele AZ, Koike S, Kobayashi Y. Genetic diversity and diet specificity of ruminal Prevotella revealed by 16S rRNA gene-based analysis[J]. FEMS Microbiology Letters, 2010, 305(1): 49-57. DOI:10.1111/fml.2010.305.issue-1 |
| [52] |
Stevenson DM, Weimer PJ. Dominance of Prevotella and low abundance of classical ruminal bacterial species in the bovine rumen revealed by relative quantification real-time PCR[J]. Applied Microbiology and Biotechnology, 2007, 75(1): 165-174. DOI:10.1007/s00253-006-0802-y |
| [53] |
Mullins CR, Mamedova LK, Carpenter AJ, et al. Analysis of rumen microbial populations in lactating dairy cattle fed diets varying in carbohydrate profiles and Saccharomyces cerevisiae fermentation product[J]. Journal of Dairy Science, 2013, 96(9): 5872-5881. DOI:10.3168/jds.2013-6775 |
| [54] |
Frey JC, Pell AN, Berthiaume R, et al. Comparative studies of microbial populations in the rumen, duodenum, ileum and faeces of lactating dairy cows[J]. Journal of Applied Microbiology, 2010, 108(6): 1982-1993. |
| [55] |
Rhee KJ, Sethupathi P, Driks A, et al. Role of commensal bacteria in development of gut-associated lymphoid tissues and preimmune antibody repertoire[J]. Journal of Immunology, 2004, 172(2): 1118-1124. DOI:10.4049/jimmunol.172.2.1118 |
| [56] |
Surana NK, Kasper DL. Moving beyond microbiome-wide associations to causal microbe identification[J]. Nature, 2017, 552(7684): 244-247. DOI:10.1038/nature25019 |
| [57] |
Pryde SE, Duncan SH, Hold GL, et al. The microbiology of butyrate formation in the human colon[J]. FEMS Microbiology Letters, 2002, 217(2): 133-139. DOI:10.1111/fml.2002.217.issue-2 |
| [58] |
Sunkara LT, Achanta M, Schreiber NB, et al. Butyrate enhances disease resistance of chickens by inducing antimicrobial host defense peptide gene expression[J]. PLoS One, 2011, 6(11): e27225. DOI:10.1371/journal.pone.0027225 |
| [59] |
Mo SJ, Kim BS, Yun SJ, et al. Genome sequencing of Clostridium butyricum, DKU-01, isolated from infant feces[J]. Gut Pathogens, 2015, 7(1): 1-7. DOI:10.1186/s13099-015-0049-1 |
| [60] |
Bian GR, Ma L, Su Y, et al. The microbial community in the feces of the white rhinoceros (Ceratotherium simum) as determined by barcoded pyrosequencing analysis[J]. PLoS One, 2013, 8(7): e70103. DOI:10.1371/journal.pone.0070103 |
| [61] |
Zeng Y, Dong Z, Zhang Y, et al. Characterization of the cellulolytic bacteria communities along the gastrointestinal tract of Chinese Mongolian sheep by using PCR-DGGE and real-time PCR analysis[J]. World Journal of Microbiology and Biotechnology, 2015, 31(7): 1103-1113. DOI:10.1007/s11274-015-1860-z |
| [62] |
Daly K, Proudman CJ, Duncan SH, et al. Alterations in microbiota and fermentation products in equine large intestine in response to dietary variation and intestinal disease[J]. British Journal of Nutrition, 2012, 107(7): 989-995. DOI:10.1017/S0007114511003825 |
| [63] |
Zhu LF, Wu Q, Dai JY, et al. Evidence of cellulose metabolism by the giant panda gut microbiome[J]. Proceedings of the National Academy of Science of the United States of America, 2011, 108(43): 17714-17719. DOI:10.1073/pnas.1017956108 |
| [64] |
Wang Y, Liu JH, Yin YY, et al. Rumen microbial and fermentation characteristics are affected differently by acarbose addition during two nutritional types of simulated severe subacute ruminal acidosis in vitro[J]. Anaerobe, 2017, 47: 39-46. DOI:10.1016/j.anaerobe.2017.04.003 |
| [65] |
Jiao JZ, Wang PP, He ZX, et al. In vitro evaluation on neutral detergent fiber and cellulose digestion by post-ruminal microorganisms in goats[J]. Journal of the Science of Food & Agriculture, 2013, 94(9): 1745-1752. |
 2019, Vol. 46
2019, Vol. 46




