扩展功能
文章信息
- 陈可可, 何漫漫, 王康, 严金平, 伊日布斯
- CHEN Ke-Ke, HE Man-Man, WANG Kang, YAN Jin-Ping, Irbis Chagan
- 厌氧菌Sunxiuqinia sp. SH-52外切型海藻酸裂解酶的异源表达及酶学特征
- Heterologous expression and characterization of exo-type alginate lyase from anaerobic bacterium Sunxiuqinia sp. SH-52
- 微生物学通报, 2019, 46(3): 531-540
- Microbiology China, 2019, 46(3): 531-540
- DOI: 10.13344/j.microbiol.china.180163
-
文章历史
- 收稿日期: 2018-03-09
- 接受日期: 2018-05-28
- 网络首发日期: 2018-06-12
海藻酸是由β-D-甘露糖醛酸(M)和它的C5差向异构体α-L-古罗糖醛酸(G)单体组成,通过1, 4-糖苷键连接成线性多糖,根据海藻酸分子的多聚体结构可以分为3种情况:单体M连续排列形成的聚甘露糖醛酸(PolyM);单体G连续排列形成的聚古罗糖醛酸(PolyG);M和G随机交替排列形成的杂聚古罗糖醛酸-甘露糖醛酸(PolyMG)。海藻酸因具有特殊的化学特性和生物活性在食品、医药和化工等领域具有广泛的应用价值[1],近年来其作为一种生产乙醇和化学试剂的可再生海洋生物质引起了关注[2]。
目前降解海藻酸的方法主要是在高温高压下使用强酸或强碱水解,但存在效率低、耗能高和污染环境等缺点[3],一定程度上限制了海藻酸产业的发展,而利用海藻酸裂解酶对海藻酸进行降解具有反应条件温和可控、偏好性高且降解效率高的优点[4]。海藻酸裂解酶可由海藻酸分解菌和一些海洋动植物产生[5-11],通过β-消除反应将海藻酸降解为一系列不饱和的海藻酸寡糖,按其降解方式分为内切型海藻酸裂解酶和外切型海藻酸裂解酶,分布于多糖裂解酶(PL) 5、6、7、14、15、17和18家族中[12]。
外切型海藻酸裂解酶在已报道的海藻酸裂解酶中相对较少[13],它可将海藻酸或海藻酸寡糖降解为单糖,在利用海藻酸为原料生产生物乙醇时,多糖降解是海洋生物质转化利用的前提条件[2]。在细菌细胞内,单糖首先通过非酶促作用进行转化,从而进入KDPG (2-酮-3-脱氧-6-磷酸葡萄糖酸)途径被细菌利用[14],单糖的产生是海藻酸完全降解的标志。通过搜索NCBI PubMed中的文献,自从在Sphingomonas sp. A1中首次发现外切型海藻酸裂解酶A1-IV后[15],陆续在15种细菌中发现了18种外切型海藻酸裂解酶,这些酶分别属于PL17[13, 15-20]、PL15[21-22]、PL6[23-24]和PL7家族[25-26]。
尽管已经表征了许多海藻酸裂解酶,但海藻酸分解菌往往拥有复杂多样的海藻酸裂解酶组分系统,其高效降解海藻酸的机制还不完全清楚,且目前商业化海藻酸裂解酶制剂的活性仍有较大的提升空间。因此,关于新菌株和酶的研究仍在持续进行[27-28]。此外,在线数据库,像碳水化合物活性酶数据库(CAZy)中有大量没有被完全表征或来源未知的海藻酸裂解酶序列,表明还有大量的海藻酸裂解酶需要表征并阐明其特性,因此通过基因工程手段实现海藻酸裂解酶基因在大肠杆菌、酵母等工程菌的异源表达,研究新型海藻酸裂解酶的酶学特性和降解特征等,为寻找具有应用价值的海藻酸裂解酶奠定基础。本文克隆和异源表达了来自厌氧海藻酸分解菌Sunxiuqinia sp. SH-52的海藻酸裂解酶基因SHA-4,对重组酶SHA-4的酶学性质及降解产物进行了研究,表明该酶将是扩展海藻酸裂解酶应用的潜在候选者。
1 材料与方法 1.1 材料 1.1.1 菌株与质粒厌氧海藻酸分解菌Sunxiuqinia sp. SH-52 (本实验室筛选保存);Escherichia coli JM109购自TaKaRa公司;Escherichia coli BL21购自北京转基因生物技术有限公司。载体pMD19-T和pGEX-4T-1购自TaKaRa公司。
1.1.2 主要试剂和仪器及培养基DNA、Protein marker购自TaKaRa公司;限制性内切酶、DNA聚合酶购自全式金及北京庄盟国际生物基因科技有限公司;所有试剂盒均购自北京索莱宝科技有限公司;IPTG (Isopropyl β-D-thiogalactoside)、GST琼脂糖凝胶亲和层析柱购自生工生物工程(上海)股份有限公司;海藻酸单糖购自青岛博智汇力生物科技有限公司;海藻酸钠购自西格玛奥德里奇(上海)贸易有限公司;PolyM和PolyG均为本实验室自制[29];其余试剂均为国产分析纯。
PCR仪,Applied Biosystem公司;凝胶成像系统,BioSense公司;控温摇床,上海申能博彩生物科技有限公司;超声波细胞粉碎机,宁波新芝生物科技股份有限公司。
LB培养基(g/L):蛋白胨10.0,酵母提取物5.0,NaCl 10.0。
1.2 方法 1.2.1 海藻酸裂解酶基因的克隆根据厌氧海藻酸分解菌Sunxiuqinia sp. SH-52海藻酸裂解酶SHA-4的基因序列和原核表达载体pGEX-4T-1上的多克隆位点引入酶切位点BamH I和Not I,设计SHA-4的PCR扩增引物。引物SHA-4-F:5′-GGATCCATGAAAAGCGCAATCAAC TATCTCA-3′;引物SHA-4-R:5′-GCGGCCGCTTATT GATTAACATGTGTCAAATTTTCG-3′。以传统法提取厌氧海藻酸分解菌Sunxiuqinia sp. SH-52的基因组DNA[30],作为模板PCR扩增海藻酸裂解酶SHA-4的基因片段。PCR反应体系(50 μL):10×EasyTaq buffer 5 μL,dNTPs (2.5 mmol/L) 4 μL,引物SHA-4-F和SHA-4-R (10 μmol/L)各1 μL,ddH2O 36 μL,基因组DNA 2 μL,EasyTaq DNA polymerase (5 U/μL) 1 μL。PCR反应条件:94 ℃ 5 min;94 ℃ 30 s,56 ℃ 30 s,72 ℃ 1.5 min,30个循环;72 ℃ 10 min;4 ℃保存。使用ClustalW软件在海藻酸裂解酶SHA-4和其它家族已表征的外切型海藻酸裂解酶之间进行多序列比对。系统进化树由MEGA 7.0通过邻接法构建,通过Bootstrap进行各分支的置信度检测,自展检测为1 000个重复。
1.2.2 克隆和表达载体的构建使用TA克隆方法,SHA-4 PCR扩增产物经琼脂糖凝胶电泳鉴定、回收纯化后与pMD19-T连接,并热击转化至大肠杆菌JM109中。克隆得到的序列经测序验证后,对重组质粒pMD19-T-SHA-4进行BamH I和Not I双酶切并与相同酶切处理的pGEX-4T-1载体连接,转化至大肠杆菌BL21后筛选阳性转化子测序,同时进行双酶切鉴定,将鉴定正确的转化子命名为pGEX-4T-1-SHA-4。
1.2.3 重组蛋白的诱导表达及条件优化转化成功的阳性克隆子于LB液体培养基(含Amp终浓度为100 mg/L)中37 ℃、150 r/min培养过夜,按2%接种至50 mL LB液体培养基(含Amp终浓度为100 mg/L)中37 ℃、150 r/min培养至OD600达到0.4-0.5,对诱导条件进行优化,考察不同的诱导时间(0-24 h)、IPTG浓度(0.1-1.0 mmol/L)和诱导温度(16-37 ℃)对蛋白表达量及可溶性的影响,并通过12% SDS-PAGE检测确定最佳的诱导条件。
1.2.4 重组蛋白的纯化及活性测定用直径为0.45 μm的微孔滤膜过滤超声波破碎细胞的上清液,将此样品加入到预先平衡好的层析柱中,采用GST琼脂糖凝胶亲和层析柱纯化目的蛋白[31],纯化后进行12% SDS-PAGE分析并测定纯化效率。
酶活性的测定方法参照Yoon等的方法并在此基础上对缓冲液和底物稍作修改[32],改为20 mmol/L磷酸盐缓冲液(pH 7.0)和0.3%海藻酸钠。考马斯亮蓝法测定重组酶的蛋白质含量。
1.2.5 重组蛋白的酶学性质研究(1) 反应最适pH和pH稳定性:在不同pH的缓冲体系(pH 5.0-8.0磷酸盐缓冲液,pH 8.0-9.0 Tris-HCl)中以0.3%海藻酸钠为底物测定酶的最适反应pH,反应时间为10 min,将处于最适pH下的酶活力定义为100%。将酶液分别在不同pH缓冲液中保温30 min后在最适温度下反应10 min,测定酶的剩余活力,将剩余活力最高的pH下的酶活力设为100%。
(2) 反应最适温度和热稳定性:以0.3%海藻酸钠为底物,在最适pH条件下测定酶的最适反应温度,反应时间为10 min,将处于最适温度下的酶活力定义为100%。酶液分别在不同温度下保温30 min后在最适温度下反应10 min,测定酶的剩余活力,将剩余活力最高的温度下的酶活力设为100%。
(3) 金属离子对酶活性的影响:在酶反应体系中分别加入0.5 mmol/L不同金属离子化合物,以加入相同体积的水作为对照组,测定其酶活力,将对照组的酶活力定义为100%。
(4) 底物偏好性检测:在最适pH缓冲体系中分别以海藻酸钠、PolyG和PolyM为底物测定酶活力,反应时间为10 min,反应温度为最适温度,将此条件下以海藻酸钠为底物的酶活力定义为100%。
1.2.6 动力学分析为了测量20 mmol/L磷酸盐缓冲液(pH 7.0)中重组海藻酸裂解酶SHA-4的动力学参数,在试管中加入0.1 mL酶溶液和0.9 mL不同浓度的海藻酸钠,总体积为1.0 mL,混合物在最佳温度下反应30 min,测定反应前后的A235值。根据双倒数作图法(Lineweaver- Burk),以1/V为纵坐标,1/[S]为横坐标作图,通过横截距和纵截距求出米氏常数Km和Vmax[16]。
1.2.7 薄层层析(TLC)分析采用TLC法分析重组酶降解海藻酸的产物。将100 U的纯酶SHA-4加入500 μL含有0.3%的海藻酸钠缓冲液中,混匀后于37 ℃、pH 7.0的条件下反应,对不同时间的降解产物进行分析;采用Silica Gel 60 F254硅胶板,上样量为2 μL;点样完成后将硅胶板放入层析缸中,展开溶剂为正丁醇:醋酸:水(3:2:2,体积比);待展开完成后,取出硅胶板吹干,在硅胶板表面喷洒显色剂(1 mL 37.5% HCl,2 mL苯胺,10 mL 85% H3PO4,100 mL乙酸乙酯和2 g二苯胺)后放入120 ℃烘箱加热显色,拍照保存[33]。
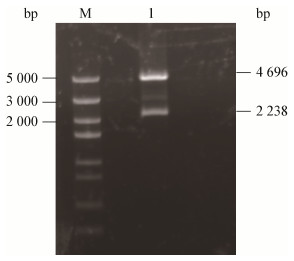
2 结果与分析 2.1 海藻酸裂解酶基因SHA-4的扩增与序列分析以厌氧海藻酸分解菌Sunxiuqinia sp. SH-52的基因组DNA为模板进行PCR扩增获得海藻酸裂解酶基因SHA-4 (图 1),片段大小为2.2 kb左右,与预期的2 238 bp相符。海藻酸裂解酶SHA-4的核苷酸序列在GenBank上的登录号是MH013392。使用MEGA 7.0对SHA-4与其它家族已表征的外切型海藻酸裂解酶基于氨基酸序列同源性构建系统进化树(图 2)。明显看到SHA-4与PL6家族的其它3种外切型海藻酸裂解酶形成一个深度支化的簇,因此该酶属于PL6家族。进行氨基酸序列比对发现在已经表征的海藻酸裂解酶中,SHA-4与PL6家族的海藻酸裂解酶OalS6有着最高的相似性,为45.34%。

|
| 图 1 SHA-4基因的PCR扩增产物电泳图 Figure 1 PCR amplification products of gene SHA-4 注:M:DNA marker;2:扩增产物. Note: M:DNA marker; 2: Amplification products. |
|
|

|
| 图 2 海藻酸裂解酶SHA-4的系统进化树 Figure 2 The phylogenetic tree of alginate lyase SHA-4 注:Bootstrap是从1 000个重复中获得的置信度值(以百分比表示);每个分支的右侧是海藻酸分解菌的种类和在GenBank上的登录号;标尺表示20%的序列差异. Note: Bootstraps are the confidence values (expressed as percentages) obtained from 1 000 replications; The right of each branch is the species of alginolytic bacteria and the accession number on GenBank; The scale indicates approximately 20% sequence difference. |
|
|
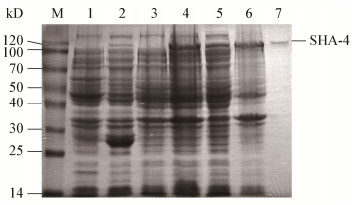
对测序完全正确的表达转化子进行双酶切验证,分别获得4.9 kb和2.3 kb的两条条带(图 3),此两条条带的大小和质粒及目的片段大小一致,结果表明重组表达质粒pGEX-4T-1-SHA-4构建成功。SDS-PAGE检测结果显示,与经IPTG诱导和未诱导的含空载体pGEX-4T-1的大肠杆菌细菌总蛋白以及未经诱导的含重组载体pGEX-4T-1-SHA-4的大肠杆菌总蛋白相比,经IPTG诱导后在102 kD (GST融合标签)左右出现新生条带,这与预期的海藻酸裂解酶SHA-4的理论值相符。此外,检测到破碎后的上清液及纯化的融合蛋白均具有海藻酸裂解酶活性,因此确定海藻酸裂解酶SHA-4成功诱导表达(图 4)。对该酶进行诱导条件优化后表明该重组酶的最佳IPTG浓度、最佳温度和最佳时间分别为0.1 mmol/L、28 ℃和6 h,纯化后SHA-4的比活力达到21 U/mg,纯化倍数为3.6 (表 1)。
|
| 图 3 重组海藻酸裂解酶表达载体的限制性内切酶分析 Figure 3 The restriction endonuclease analysis of the recombinant alginate lyase expression vector |
|
|
|
| 图 4 SDS-PAGE检测重组酶SHA-4的表达和纯化 Figure 4 Detection of recombinase SHA-4 expression and purification by SDS-PAGE 注:M:蛋白Marker;1:含pGEX-4T-1质粒的BL21未经IPTG诱导10 h的细菌总蛋白;2:含pGEX-4T-1质粒的BL21经IPTG诱导10 h的细菌总蛋白;3:含pGEX-4T-1-SHA-4质粒的BL21未经IPTG诱导10 h的细菌总蛋白;4:含pGEX-4T-1-SHA-4质粒的BL21经IPTG诱导10 h的细菌总蛋白;5:含pGEX-4T-1-SHA-4质粒的BL21经IPTG诱导的细菌总蛋白经超声破碎后的上清;6:含pGEX-4T-1-SHA-4质粒的BL21经IPTG诱导的细菌总蛋白经超声破碎后的沉淀;7:纯化的重组酶SHA-4. Note: M: Protein marker; 1: Total bacterial protein of BL21 containing pGEX-4T-1 plasmid without induction of IPTG for 10 h; 2: Total bacterial protein of BL21 containing pGEX-4T-1 plasmid induced by IPTG for 10 h; 3: Total bacterial protein of BL21 containing pGEX-4T-SHA-4 plasmid without induction by IPTG for 10 h; 4: Total bacterial protein of BL21 containing pGEX-4T-SHA-4 plasmid induced by IPTG for 10 h; 5: The supernatant after sonication with IPTG induced total protein of BL21 containing pGEX-4T-1-SHA-4 plasmid; 6: The precipitation after sonication with IPTG induced total protein of BL21 containing pGEX-4T-1-SHA-4 plasmid; 7: The purified recombinant enzyme SHA-4. |
|
|
| Step (SHA-4) | Cell extract | GST Purification |
| Total protein (mg) | 351.7 | 63.2 |
| Total activity (U) | 2 061.0 | 1 327.2 |
| Specific activity(U/mg) | 5.9 | 21.0 |
| Yield (%) | 100.0 | 64.3 |
| Purification (fold) | 1.0 | 3.6 |
重组酶的活性和稳定性与pH值的关系曲线如图 5所示。重组酶SHA-4的最适反应pH为7.5,中性条件下具有较高活性;该酶在pH 7.0时稳定性最高,中性及弱碱性pH 7.0-8.0条件下酶的稳定性较高,能保持60%以上的酶活力,表明酶SHA-4是中性偏弱碱性酶。

|
| 图 5 重组酶的最适pH和pH稳定性分析 Figure 5 Optimal pH and pH stability of the recombinant alginate lyase |
|
|
重组酶的活性和稳定性与温度的关系曲线如图 6所示。重组酶SHA-4的最适温度为37 ℃,在36-40 ℃范围内酶活性较高,当温度升高到45 ℃时,酶活力迅速下降,仅为峰值的67%;探究热稳定性可知,该酶在35 ℃保温30 min后剩余酶活力为89%,在37 ℃保温30 min剩余酶活力为80%,在40 ℃保温30 min后酶活力丧失了54%,在55 ℃保温30 min后剩余的酶活力为19%,由此可知该酶在高于40 ℃时热稳定性较差。

|
| 图 6 重组酶的最适温度和热稳定性分析 Figure 6 Optimal temperature and temperature stability of the recombinant alginate lyase |
|
|
金属离子对重组酶活性的影响结果如图 7所示,对于重组酶SHA-4而言,Hg2+对酶活具有完全抑制作用,Na+对酶活具有明显抑制作用,Mg2+对酶活基本无影响,而Cu2+对酶活具有明显的促进作用,使其活性提高了168%。

|
| 图 7 金属离子对重组酶活性的影响 Figure 7 Effects of metal ions on the activity of the recombinant alginate lyase 注:*:与对照组相比具有显著差异性的实验组,P < 0.05.数据处理使用SPSS 22.0软件的方差分析(One-way ANOVA). Note: *: There is a significant difference in the experimental group compared with the control group, P < 0.05. Data processing (One-way ANOVA) using SPSS 22.0 software. |
|
|
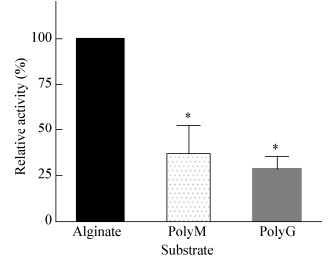
重组酶SHA-4的底物偏好性结果如图 8所示,SHA-4对3种底物均可降解,但对海藻酸钠的酶活性明显高于PolyG和PolyM,因此重组酶SHA-4对海藻酸钠具有底物偏好性。
|
| 图 8 重组酶SHA-4的底物偏好性 Figure 8 Substrate preference of recombinase SHA-4 注:*:具有显著差异性,P < 0.05.数据处理使用SPSS 22.0软件的方差分析(One-way ANOVA). Note: *: There is a significant difference, P < 0.05. Data processing One-way ANOVA using SPSS 22.0 software. |
|
|
使用Lineweaver-Burk的双倒数图计算Km和Vmax值,重组酶SHA-4的最大反应速度Vmax为8.7 mg/(mL·min),米氏常数Km为2.5 mg/mL。
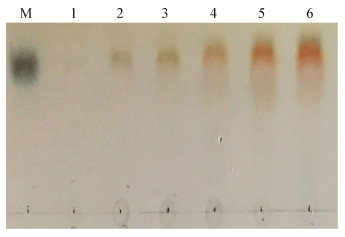
2.5 重组酶的降解产物分析如图 9所示,不同时间的降解产物均为单个斑点,而且和海藻酸单糖标品所处位置相同,表明该酶是一种外切型海藻酸裂解酶,在降解海藻酸的过程中仅产生海藻酸单糖,而不产生其它低聚糖产物。
|
| 图 9 TLC分析SHA-4降解海藻酸的产物 Figure 9 TLC analysis the alginate degraded products by SHA-4 注:M:海藻酸单糖;1:0 min;2:10 min;3:30 min;4:1 h;5:5 h;6:12 h. Note: M: Alginate-monosaccharide; 1: 0 min; 2: 10 min; 3: 30 min; 4: 1 h; 5: 5 h; 6: 12 h. |
|
|
本实验室前期筛选分离到一株厌氧海藻酸分解菌,并将其命名为Sunxiuqinia sp. SH-52。系统进化分析发现与该菌株亲缘关系最近的是Sunxiuqinia elliptica DQHS-4T,但两菌株16S rRNA基因序列的相似度只有93%,并且在细胞形态、需氧性和底物利用等方面具有显著的差异,因此该菌株可能是一株新型海藻酸分解菌。与好氧菌相比,厌氧微生物可以直接发酵底物生成乙醇,并且具有利用单位底物生成的ATP较好氧菌少、底物的转化效率高等优点,因此厌氧海藻酸分解菌更有利于海藻生物质生产生物能源[34]。
SHA-4海藻酸裂解酶属于PL6家族,进行氨基酸相似性分析表明SHA-4与其它已知的海藻酸裂解酶序列相似性较低,NCBI在线分析蛋白质保守结构域发现,在N端包含一个Chondroitinase B结构域,因此在结构上SHA-4不同于其它已知的海藻酸裂解酶。值得注意的是,尽管PL6家族的其它海藻酸裂解酶也含有Chondroitinase B结构域,它们却是内切型海藻酸裂解酶[35-36]。
利用紫外法测得重组酶SHA-4的比活力达到21.0 U/mg。对该酶降解海藻酸的产物进行分析表明SHA-4是外切型海藻酸裂解酶。目前异源表达的外切型海藻酸裂解酶中,酶的比活力最大的是来自Halomonas sp.的OalY2,为36.2 U/mg[13],Hirayama等对来自Agrobacterium tumefaciens的Atu3025、Sphingomonas sp. A1的A1-IV和Saccharophagus degradans 2-40的Alg17C分别进行异源表达,纯化这3种海藻酸裂解酶后在相同条件下进行酶活测定,结果表明A1-IV的比活力最高,为13.5±1.4 U/mg,而Alg17C最低,为2.3±0.5 U/mg[37],相较而言本研究得到了一种酶活性较高的外切型海藻酸裂解酶SHA-4。
对海藻酸裂解酶SHA-4的酶学性质进行研究表明,该酶的最适温度和pH分别为37 ℃和7.5,与来自Vibrio splendidus的OalC类似,是一种反应条件较为温和的酶[18]。金属离子对SHA-4的酶活影响表明,Cu2+和Mn2+具有促进作用,而Na+等盐离子却具有抑制作用。这一特性与已报道的一些酶不同,大多海藻酸裂解酶在一定浓度盐离子存在时活性都会增加,例如同样来自PL6家族的OlaS6和来自Azotobacter chroococcum的海藻酸裂解酶在不同浓度Na+处理下活性有明显的增加[23, 38],而来自海洋细菌Halomonas sp.的海藻酸裂解酶OalY1和OalY2在缺少Na+时,酶活性很低,当浓度达到0.5 mol/L时酶活达到最高,其中OalY1的活性提高了约30倍[13]。研究SHA-4的底物偏好性发现该酶对海藻酸的底物偏好性大于PolyM和PolyG。海藻酸多糖是由4种二元组M-M、M-G、G-M和G-G组成,而不同海藻酸裂解酶对4种二元组之间糖苷键的剪切能力不同,因此导致对PolyG、PolyM和海藻酸的偏好性不同,例如来自Haliotis tuberculata的内切型海藻酸裂解酶偏好性剪切M-M和G-M[29],而来自Chlorostoma rustica的海藻酸裂解酶偏好性剪切G-G[39]。由于天然海藻酸中主要存在G-M或M-G二元组,因此SHA-4是一种偏好性剪切M-G或G-M的海藻酸裂解酶。有学者对来自PL6家族的海藻酸裂解酶的底物偏好性进行了研究,该家族的海藻酸裂解酶对PolyMG或和PolyG具有底物偏好性,其中已表征的外切型海藻酸裂解酶都仅偏好性降解PolyG[23-24],而本研究表征的外切型海藻酸裂解酶则对PolyMG具有偏好性。
本研究获得的重组酶SHA-4属于PL6家族,是该家族发现的第一个偏好性降解PolyMG的外切型海藻酸裂解酶,作为工具酶有很好的应用前景,可应用于制备海洋药物、保健食品、化工原料和食品添加剂。在海藻酸裂解酶的不断深入研究中,本研究有助于重组酶的规模化生产和海藻酸降解机理等相关功能的研究。
| [1] |
Pawar SN, Edgar KJ. Alginate derivatization: a review of chemistry, properties and applications[J]. Biomaterials, 2012, 33(11): 3279-3305. DOI:10.1016/j.biomaterials.2012.01.007 |
| [2] |
Enquist-Newman M, Faust AME, Bravo DD, et al. Efficient ethanol production from brown macroalgae sugars by a synthetic yeast platform[J]. Nature, 2014, 505(7482): 239-243. DOI:10.1038/nature12771 |
| [3] |
Qin GK, Zhang YZ, Chen XL, et al. Research advances on alginate lyases[J]. China Biotechnology, 2004, 42(2): 26-29,33. (in Chinese) 秦国奎, 张玉忠, 陈秀兰, 等. 海藻酸盐裂解酶研究进展[J]. 中国生物工程杂志, 2004, 42(2): 26-29,33. |
| [4] |
Zhu BW, Yin H. Alginate lyase: review of major sources and classification, properties, structure-function analysis and applications[J]. Bioengineered, 2015, 6(3): 125-131. DOI:10.1080/21655979.2015.1030543 |
| [5] |
Sawant SS, Salunke BK, Kim BS. A rapid, sensitive, simple plate assay for detection of microbial alginate lyase activity[J]. Enzyme and Microbial Technology, 2015, 77: 8-13. DOI:10.1016/j.enzmictec.2015.05.003 |
| [6] |
Ertesvåg H, Erlien F, Skjåk-Braek G, et a1. Biochemical properties and substrate specificities of a recombinantly produced Azotobacter vinelandii alginate lyase[J]. Journal of Bacteriology, 1998, 180(15): 3779-3784. |
| [7] |
Shimizu E, Ojima T, Nishita K. cDNA cloning of an alginate lyase from abalone, Haliotis discus hannai[J]. Carbohydrate Research, 2003, 338(24): 2841-2852. DOI:10.1016/j.carres.2003.08.009 |
| [8] |
Wakabayashi M, Sakatoku A, Noda F, et al. Isolation and characterization of Microbulbifer species 6532A degrading seaweed thalli to single cell detritus particles[J]. Biodegradation, 2012, 23(1): 93-105. DOI:10.1007/s10532-011-9489-6 |
| [9] |
Sim SJ, Baik KS, Park SC, et al. Characterization of alginate lyase gene using a metagenomic library constructed from the gut microflora of abalone[J]. Journal of Industrial Microbiology & Biotechnology, 2012, 39(4): 585-593. |
| [10] |
An QD, Zhang GL, Wu HT, et al. Properties of an alginate-degrading Flavobacterium sp. strain LXA isolated from rotting algae from coastal China[J]. Canadian Journal of Microbiology, 2008, 54(4): 314-320. DOI:10.1139/W08-013 |
| [11] |
Kobayashi T, Uchimura K, Miyazaki M, et al. A new high-alkaline alginate lyase from a deep-sea bacterium Agarivorans sp[J]. Extremophiles, 2009, 13(1): 121-129. DOI:10.1007/s00792-008-0201-7 |
| [12] |
Xu QQ, Zhu YX, Wang KP, et al. Cloning and identification of alginate lyases genes from Vibrio alginolyticus ATCC 17749[J]. Journal of East China University of Science and Technology (Natural Science Edition), 2013, 39(1): 29-34,41. (in Chinese) 胥晴晴, 朱云霞, 王克平, 等. 解藻酸弧菌株ATCC 17749中海藻酸裂解酶基因的克隆与鉴定[J]. 华东理工大学学报:自然科学版, 2013, 39(1): 29-34,41. |
| [13] |
Yang XM, Li SY, Wu Y, et al. Cloning and characterization of two thermo- and salt-tolerant oligoalginate lyases from marine bacterium Halomonas sp[J]. FEMS Microbiology Letters, 2016, 363(9): fnw079. DOI:10.1093/femsle/fnw079 |
| [14] |
Gacesa P, Leaves GS, Weightman AJ. Genetic analysis of Klebsiella pneumoniae alginate lyase by transposon Tn10 mutagenesis[J]. Hydrobiologia, 1987, 151(1): 571-575. |
| [15] |
Hashimoto W, Miyake O, Ochiai A, et al. Molecular identification of Sphingomonas sp. A1 alginate lyase (A1-IV') as a member of novel polysaccharide lyase family 15 and implications in alginate lyase evolution[J]. Journal of Bioscience and Bioengineering, 2005, 99(1): 48-54. DOI:10.1263/jbb.99.48 |
| [16] |
Kim HT, Chung JH, Wang DM, et al. Depolymerization of alginate into a monomeric sugar acid using Alg17C, an exo-oligoalginate lyase cloned from Saccharophagus degradans 2-40[J]. Applied Microbiology and Biotechnology, 2012, 93(5): 2233-2239. DOI:10.1007/s00253-012-3882-x |
| [17] |
Ryu M, Lee EY. Saccharification of alginate by using exolytic oligoalginate lyase from marine bacterium Sphingomonas sp. MJ-3[J]. Journal of Industrial and Engineering Chemistry, 2011, 17(5/6): 853-858. |
| [18] |
Jagtap SS, Hehemann JH, Polz MF, et al. Comparative biochemical characterization of three exolytic oligoalginate lyases from Vibrio splendidus reveals complementary substrate scope, temperature, and pH adaptations[J]. Applied and Environmental Microbiology, 2014, 80(14): 4207-4214. DOI:10.1128/AEM.01285-14 |
| [19] |
Shin JW, Lee OK, Park HH, et al. Molecular characterization of a novel oligoalginate lyase consisting of AlgL- and heparinase II/III-like domains from Stenotrophomonas maltophilia KJ-2 and its application to alginate saccharification[J]. Korean Journal of Chemical Engineering, 2015, 32(5): 917-924. DOI:10.1007/s11814-014-0282-1 |
| [20] |
Wang LN, Li SY, Yu WG, et al. Cloning, overexpression and characterization of a new oligoalginate lyase from a marine bacterium, Shewanella sp[J]. Biotechnology Letters, 2015, 37(3): 665-671. DOI:10.1007/s10529-014-1706-z |
| [21] |
Ochiai A, Yamasaki M, Mikami B, et al. Crystal structure of exotype alginate lyase Atu3025 from Agrobacterium tumefaciens[J]. Journal of Biological Chemistry, 2010, 285(32): 24519-24528. DOI:10.1074/jbc.M110.125450 |
| [22] |
Mori T, Takahashi M, Tanaka R, et al. Falsirhodobacter sp. alg1 harbors single homologs of endo and exo-type alginate lyases efficient for alginate depolymerization[J]. PLoS On, 2016, 11(5): e0155537. DOI:10.1371/journal.pone.0155537 |
| [23] |
Li SY, Wang LN, Han F, et al. Cloning and characterization of the first polysaccharide lyase family 6 oligoalginate lyase from marine Shewanella sp. Kz7[J]. Journal of Biochemistry, 2016, 159(1): 77-86. DOI:10.1093/jb/mvv076 |
| [24] |
Mathieu S, Henrissat B, Labre F, et al. Functional exploration of the polysaccharide lyase family PL6[J]. PLoS One, 2016, 11(7): e0159415. DOI:10.1371/journal.pone.0159415 |
| [25] |
Yagi H, Fujise A, Itabashi N, et al. Purification and characterization of a novel alginate lyase from the marine bacterium Cobetia sp. NAP1 isolated from brown algae[J]. Bioscience, Biotechnology, and Biochemistry, 2016, 80(12): 2338-2346. DOI:10.1080/09168451.2016.1232154 |
| [26] |
Lundqvist L. Structural and interaction studies of polysaccharides by NMR spectroscopy[D]. Uppsala: Doctoral Dissertation of Swedish University of Agricultural Sciences, 2015
|
| [27] |
Xu F, Dong F, Wang P, et al. Novel molecular insights into the catalytic mechanism of marine bacterial alginate lyase AlyGC from polysaccharide lyase family 6[J]. Journal of Biological Chemistry, 2017, 292(11): 4457-4468. DOI:10.1074/jbc.M116.766030 |
| [28] |
Zhu BW, Sun YD, Ni FQ, et al. Characterization of a new endo-type alginate lyase from Vibrio sp. W13[J]. International Journal of Biological Macromolecules, 2015, 75: 330-337. DOI:10.1016/j.ijbiomac.2015.01.053 |
| [29] |
Heyraud A, Colin-Morel P, Girond S, et al. HPLC analysis of saturated or unsaturated oligoguluronates and oligomannuronates. application to the determination of the action pattern of Haliotis tuberculata alginate lyase[J]. Carbohydrate Research, 1996, 291: 115-126. |
| [30] |
Shen DX, Feng ZC, Du J. The comparison of bacterial DNA extraction methods[J]. Central Plains Medical Journal, 2004, 31(10): 20-22. (in Chinese) 沈德新, 封志纯, 杜江. 细菌DNA提取方法比较[J]. 中原医刊, 2004, 31(10): 20-22. |
| [31] |
Ameirika, Sha HX, Hwang JS. Identification of a target protein of Hydra actinoporin-like toxin-1 (HALT-1) using GST affinity purification and SILAC-based quantitative proteomics[J]. Toxicon, 2017, 133: 153-161. DOI:10.1016/j.toxicon.2017.05.007 |
| [32] |
Yoon HJ, Hashimoto W, Miyake O, et al. Overexpression in Escherichia coli, purification, and characterization of Sphingomonas sp. A1 alginate lyases[J]. Protein Expression and Purification, 2000, 19(1): 84-90. |
| [33] |
Hu T, Li CX, Zhao X, et al. Preparation and characterization of guluronic acid oligosaccharides degraded by a rapid microwave irradiation method[J]. Carbohydrate Research, 2013, 373: 53-58. DOI:10.1016/j.carres.2013.03.014 |
| [34] |
Qian L, Tang LW, Huang SS, et al. Research progress of bioethanol from alginate fermentation[J]. China Biotechnology, 2013, 33(1): 122-127. (in Chinese) 钱龙, 唐丽薇, 黄庶识, 等. 海藻酸转化生物乙醇研究进展[J]. 中国生物工程杂志, 2013, 33(1): 122-127. |
| [35] |
Lee SI, Choi SH, Lee EY, et al. Molecular cloning, purification, and characterization of a novel polyMG-specific alginate lyase responsible for alginate MG block degradation in Stenotrophomas maltophilia KJ-2[J]. Applied Microbiology and Biotechnology, 2012, 95(6): 1643-1653. DOI:10.1007/s00253-012-4266-y |
| [36] |
Maki H, Mori A, Fujiyama K, et al. sequence analysis and expression in Escherichia coli of a gene encoding an alginate lyase from Pseudomonas sp. OS-ALG-9[J]. Journal of General Microbiology, 1993, 139(5): 987-993. DOI:10.1099/00221287-139-5-987 |
| [37] |
Hirayama M, Hashimoto W, Murata K, et al. Comparative characterization of three bacterial exo-type alginate lyases[J]. International Journal of Biological Macromolecules, 2016, 86: 519-524. DOI:10.1016/j.ijbiomac.2016.01.095 |
| [38] |
Peciña A, Pascual A, Paneque A. Cloning and Expression of the algL Gene, Encoding the Azotobacter chroococcum alginate lyase: purification and characterization of the enzyme[J]. Journal of Bacteriology, 1999, 181(5): 1409-1414. |
| [39] |
Sun LP, Xue CH, Xu JC, et al. Purification and charaterization of alginate lyase from Chlorostoma rustica[J]. Journal of Fishery Sciences of China, 2004, 11(3): 266-271. (in Chinese) 孙丽萍, 薛长湖, 许家超, 等. 锈凹螺褐藻胶裂解酶的分离纯化及性质研究[J]. 中国水产科学, 2004, 11(3): 266-271. DOI:10.3321/j.issn:1005-8737.2004.03.015 |
 2019, Vol. 46
2019, Vol. 46




