扩展功能
文章信息
- 焦健, 田长富
- JIAO Jian, TIAN Chang-Fu
- 根瘤菌共生固氮能力的进化模式
- Evolution of rhizobial nodulation and nitrogen fixation
- 微生物学通报, 2019, 46(2): 388-397
- Microbiology China, 2019, 46(2): 388-397
- DOI: 10.13344/j.microbiol.china.180844
-
文章历史
- 收稿日期: 2018-10-29
- 接受日期: 2019-01-04
- 网络首发日期: 2019-01-09
生物固氮是原核微生物在固氮酶催化作用下将氮气还原为氨的过程。与工业固氮相比,生物固氮在常温常压下进行,消耗生物能ATP,被认为是绿色的固氮方式。如何利用好生物固氮,减少农业生产中化学氮肥的使用是世界性的研究热点和难点。农业上根瘤菌-豆科植物共生固氮体系每年固定的纯氮约为4 000万t,约占农业体系生物固氮总量的65%[1],是自然界效率最高的固氮体系,最具开发利用价值。美国、巴西、阿根廷、澳大利亚等国家种植豆科作物或牧草,通过接种根瘤菌剂已经实现了化学氮肥用量的大幅降低,甚至基本不使用化学氮肥[2]。但在我国,目前根瘤菌剂的应用仍面临诸多困难:首先,长期过量使用化学氮肥导致了抑制根瘤菌结瘤固氮的“氮阻遏”现象;另外,我国幅员辽阔且生态环境多样,在不同地区的豆科作物生产中农民会使用不同的商业栽培品种或农家品种;中国农业大学根瘤菌研究中心40年来的系统研究表明,与同一豆科植物共生的根瘤菌在不同生态环境下存在不同的优势菌种,而且大豆、花生、苜蓿、蚕豆等豆科植物的不同品种与根瘤菌菌株间存在显著的共生匹配差异[2-5]。这种共生特异性现象以及根瘤菌在不同土壤条件下的竞争存活能力差异是限制特定根瘤菌剂推广应用的重要瓶颈问题。要解决这些问题,就需要对根瘤菌-豆科植物共生固氮体系有更加全面和深入的认识。
根瘤菌是能够与豆科植物结瘤固氮的一类细菌的总称。在大多数时期,根瘤菌在土壤中营腐生生活。在条件适宜的情况下,根瘤菌与豆科植物展开分子“对话”,诱导植物形成特异化器官“根瘤”(在某些豆科植物上形成“茎瘤”),并在被侵染的植物细胞中大量增殖分化形成具有固氮能力的类菌体。类菌体被来自植物的所谓“共生体膜”所包被,共同组成共生体。固氮过程中,植物根瘤细胞通过豆血红蛋白对氧气浓度进行了精密调控,既保证了类菌体细胞的基本生命活动,又避免了氧气对固氮酶功能的破坏[6]。类菌体利用植物供给的碳源所产生的能量,将N2还原为NH3,后者在跨越共生体膜时变为植物可以直接利用的NH4+。当根瘤衰老时,其中定殖的大量根瘤菌细胞被释放到土壤中,在苜蓿、蚕豆、豌豆、花生等豆科植物根瘤中,成熟的类菌体基因组发生多倍化现象且不具备再繁殖的能力;而大豆、豇豆、扁豆、百脉根等豆科植物根瘤内部的类菌体没有发生上述终端分化,可以在根瘤破碎后生长繁殖;但所有豆科植物根瘤中都存在没有发生分化的内生根瘤菌细胞,它们可以随着根瘤的破碎而被释放出来并在土壤中生活[7-8]。
20世纪末的20年间,欧洲和美国的多个实验室用大量实验证明:绝大部分根瘤菌依赖关键的结瘤(nod)和固氮(nif)基因来完成与豆科植物的共生固氮过程,实现互利共生。但随着21世纪根瘤菌-豆科植物共生体系多样性和互作机制研究的进一步展开,人们不仅发现了一些不依赖nod基因的根瘤菌-豆科植物共生体系[9-11],还初步证实根瘤菌结瘤固氮效率的优化机制具有明显的菌种依赖性[12-15]。但是复杂多样的机制背后存在怎样的进化规律呢?本文将通过总结近年来国际上的代表性研究成果,对这一问题进行探讨。
1 关键共生基因的水平转移与根瘤菌的多点起源目前已经命名的根瘤菌归属于α-和β-变形杆菌纲18个属的200多个种,且新种属的数目还在随着多样性研究的深入持续增加[16]。与许多在特定生态位定殖的细菌不同,根瘤菌在系统发育树中的分布是分散的,并非严格集中在一个专门的系统分类单元。这种分散性不仅体现在纲、目等较高的分类水平上,甚至同一个属或种也可以同时包含根瘤菌和非根瘤菌。比如,甲基杆菌属(Methylobacterium)中除M. nodulans外其他几个已知种不是根瘤菌[17],贪铜菌属(Cupriavidus)同时包含根瘤菌C. taiwanensis和不具有结瘤固氮能力的其他物种[18]。由于目前大部分根瘤菌都分离自根瘤,一个根瘤往往只被一个根瘤菌克隆所侵染[19-21],因此自然界土壤中肯定还存在很多尚未被发现的根瘤菌物种。在基于种水平的rpoB高通量测序研究中,我们最近发现田间大豆、蚕豆和苜蓿根际土壤中存在很多已知根瘤菌属(Bradyrhizobium、Mesorhizobium、Rhizobium、Sinorhizobium等)的未知物种[3-4, 22],而关键结瘤固氮基因nod/nif是能够在同属甚至不同属的不同菌种间水平转移的[23-25]。所以,共生基因的水平转移是根瘤菌物种多样性形成的最重要原因。
在根瘤菌中,负责固氮酶复合体合成、加工与装配的nif基因以及负责结瘤因子合成与分泌的nod基因一般聚成几簇,集中分布在共生岛或共生质粒上[26-29]。与整体基因组相比,共生质粒和共生岛一般(G+C)mol%含量相对较低,并且含有大量插入序列元件,共生岛附近常常存在tRNA基因,这些特征表明共生基因易于发生水平基因转移;另外也有充分的实验证明这种转移是可以在自然界发生的[23, 25, 28]。nod和nif基因在基因组上的靠近对于它们的功能并不是必需的,这种“巧合”说明它们可能经历了近期的共转移[13]。
固氮的起源远早于结瘤,nif基因的分布也更加广泛,已经在细菌和古菌的14个门中发现了保守的nifHDK和nifENB[30]。系统发育分析表明,nifDK与16S rRNA基因的进化历史主要在较高分类水平(门)较为一致,在较低分类水平上具有不同进化特征的nif基因则反映了这些基因独立的水平转移或重组历史[31]。根瘤菌中通过水平基因转移而来的外源nif基因有时可能被基因组原有的nif基因所替换,因为有研究表明根瘤菌的nif基因往往与同属非根瘤菌的nif基因具有较近的亲缘关系[32-34]。
nod基因最早在何种根瘤菌中起源已经很难追溯。基于序列系统发育分析,推测参与结瘤因子骨架合成的nodABC以及负责结瘤因子硫化修饰的nodH起源于放线菌[35-37],参与结瘤因子分泌的nodIJ起源于β-变形菌纲[38],参与结瘤因子岩藻糖化和乙酰化修饰的nodZ和nolL由Bradyrhizobium扩散到α-变形菌纲的其他属细菌中[39]。系统发育分析表明,nod基因在根瘤菌之间发生水平转移的频次总体上与菌株的亲缘距离成反比:α-和β-变形杆菌纲间极罕见,属间较常见于与同一宿主结瘤的根瘤菌(例如与鹰嘴豆共生的不同属根瘤菌),属内菌株间的水平转移则十分频繁(例如与大豆共生的不同Sinorhizobium菌种)[13, 40-41]。这种频繁的nod基因水平转移现象可能是因为宿主植物的选择压力较强,在根瘤菌与其宿主长期共生互作的过程中,逐渐形成了一些典型的与宿主相关的生物型(Biovar)或共生型(Symbiovar)[40]。需要注意的是,并不是所有与同一宿主共生的根瘤菌都具有相同的nod基因型,例如与大豆共生的Bradyrhizobium和Sinorhizobium具有明显不同的nod基因进化历史,只有极少的属间出现nod基因转移现象[24, 41-42];苦参则是目前发现的共生混杂性最高的宿主之一,可以和具有不同结瘤因子结构的多个属种的根瘤菌共生[43-44]。
如前所述,根瘤菌在分类水平上具有巨大的多样性,而近年来快速发展的基因组学为研究这种多样性的形成原因提供了新的契机。目前已有大量根瘤菌完成基因组测序,其中仅在NCBI中提交基因组全图的根瘤菌就已达120株以上。通过比较分析α-和β-变形杆菌纲根瘤菌的基因组,发现没有根瘤菌共有且特有的基因;除了部分nod/nif基因相对保守外,其他共享的基因数目不足根瘤菌平均编码基因的10%,而且与物种树进化历史一致的核心基因数目不到核心基因组的15%[18, 45]。另一方面,对根瘤菌和豆科植物谷氨酰胺合成酶(GS)编码基因的序列分析表明,已知的各根瘤菌属在豆科植物出现之前已经分化[46]。所以,目前普遍接受根瘤菌的多点起源假说:不同分类地位的非根瘤菌曾多次、独立地以获得共生基因作为新起点进化为根瘤菌[12]。
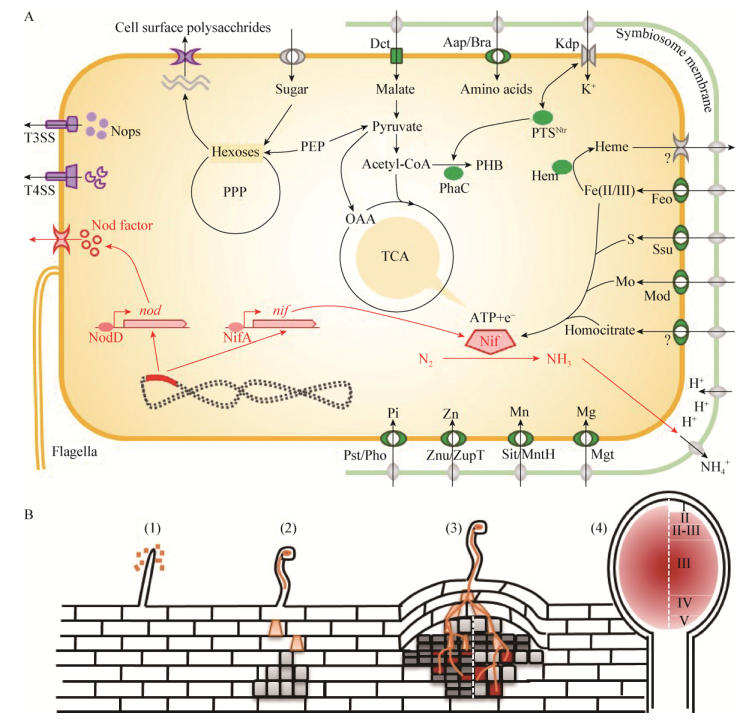
2 其他功能基因参与根表定殖、侵染以及胞内共生尽管根瘤菌的物种多样性极高,通过水平基因转移获得关键共生基因(nod/nif)并不能保证细菌具备有效的共生能力。例如,通过实验进化的方式获得共生基因后的非根瘤菌可以诱导相应豆科植物形成瘤状突起甚至正常形态且被侵染的根瘤,但是这些进化菌株仍不具备共生固氮能力[47-48],这与受体菌为根瘤菌的研究结果形成了鲜明对比[23, 25]。有关其他功能基因参与共生固氮的遗传学证据已有大量的积累,如图 1所示:在早期侵染结瘤阶段,除结瘤基因外负责细胞表面多糖(脂多糖/胞外多糖/荚膜多糖/环β-葡聚糖等)和效应蛋白合成与分泌的基因(三型分泌系统T3SS或四型分泌系统T4SS及其效应蛋白)参与根表定殖或侵染[14-15, 49-51];在共生固氮阶段,根瘤菌除了合成大量固氮酶还必须和宿主协调好碳(四碳二羧酸转运系统Dct)、氮(氨基酸吸收系统Aap/Bra)营养交换以及矿质元素(钼、铁、硫、磷、锌、镁、锰等离子的转运系统)的吸收,并克服高渗、低pH、免疫反应等不利条件[15, 52-57]。
|
| 图 1 与豆科植物共生过程中根瘤菌的相关代谢及调控途径 Figure 1 Rhizobial metabolism and regulation pathways involved in the establishment of symbiosis with legumes 注:A:共生基因nod/nif负责的结瘤因子和固氮酶合成途径用红色标记,其他功能基因负责的共生早期识别侵染(B1-B3)相关途径和共生固氮(B4)相关途径分别用紫色和绿色标记.其中许多途径仅在某些特定根瘤菌-豆科植物共生体系中是必需的.根瘤菌如何分泌血红素以及这种分泌对共生固氮的必要性尚未明确[58].许多根瘤菌缺失nifV基因,必须依靠植物提供高柠檬酸,但相关转运体系还未被鉴定[59]. B:典型的依赖于根毛侵染的结瘤过程:(1)吸附识别;(2)根毛卷曲形成侵染线,皮层去分化形成根瘤原基;(3)侵染线延伸将根瘤菌释放到根瘤原基细胞;(4)根瘤发育成熟,根瘤菌分化为具有固氮能力的类菌体.与定型瘤(B4左)相比,不定型瘤(B4右)因在形成和发育过程中具有持续的分生组织导致根瘤内有明显分区(B3-B4). Note: A: Nod factor synthesis and nitrogenase complex assembly mediated by key symbiosis genes nod and nif are labeled in red, while other pathways during recognition-infection at early stages (B1-B3) and symbiotic nitrogen fixation stage (B4) are labeled in purple and green, respectively. Many of these pathways are found to be necessary only for symbiosis between certain rhizobium-legume partners. How rhizobia secrete heme to and whether this secretion is necessary for effective symbiotic nitrogen fixation remain unclear[58]. It is necessary for many rhizobia to obtain homocitrate from host plants due to the absence of nifV gene in their genomes, however the transporter responsible for the uptake of this compound has not been identified[59]. B: Typical nodulation processes initiated by root hair infection: (1) attachment and recognition; (2) root hair curling, infection thread formation and cortex cell dedifferentiation into nodule primordia; (3) infection thread elongation and release of rhizobia into cells of nodule primordia; (4) nodule maturation along with rhizobial differentiation into nitrogen-fixing bacteroids. In contrast to the determinate nodules (left in B4), indeterminate nodules (right in B4) have distinct zones due to the presence of a persistent meristem (B3–B4). |
|
|
与关键共生基因nod/nif相比,其他功能基因对共生固氮的参与具有明显的共生体系特异性。一方面,一些共有直系同源基因在不同根瘤菌-豆科植物共生体系中的作用大小不同。例如Rhizobium leguminosarum不同菌株与豌豆和菜豆共生固氮需要氨基酸吸收通道Aap/Bra从宿主获得支链氨基酸来驱动,而这一工作机制在S. meliloti-苜蓿共生体系并不适用[60-62];形成终端分化类菌体的根瘤菌在根瘤中需要高亲和无机磷酸盐转运系统Pst,而在非终端分化的类菌体中低亲和转运子Pit就足以行使转运功能,维持正常的固氮过程[55, 63]。另一方面,根瘤菌可以利用系统发育分支特异的遗传机制共生固氮。例如在S. meliloti的根毛侵染过程中负责胞外多糖合成的基因(exo)发挥了至关重要的作用[64-66],这些基因在Sinorhizobium属中是保守的,而其他属根瘤菌中大都没有其直系同源基因[18, 45];Sinorhizobium和Bradyrhizobium分别调用了各自特有的调控系统(如FixT、RegSR)来参与控制nif基因和适应根瘤微氧环境相关基因的表达[6, 67-68];在一项实验进化学研究中,获得共生基因的病原菌Ralstoniaso lanacearum可以招募该种所特有的毒力调节途径实现结瘤以及胞内侵染效率的提高[69]。
利用几百个已知的与结瘤固氮相关的基因在不同属种根瘤菌基因组中的分布做聚类分析,发现聚群结果与根瘤菌属种的系统发育关系较为一致[45],说明根瘤菌祖先在获得共生基因之前可能已经具备了进化出共生能力的潜力,或者在长期的进化过程中结瘤固氮功能与受体基因组完成了有效整合。因此,根瘤菌基因组背景也是被选择的或经过筛选的。近年的基因组学分析发现,来自不同种属的根瘤菌都拥有较大的基因组且编码大量的转运子和调节基因,即使是与同一豆科植物共生的同种或同属根瘤菌也具有开放状态的泛基因组[28, 45, 70-71]。这些基因组特征有利于根瘤菌适应复杂多变的土壤环境和宿主品种以及周期性切换的自生/共生生活方式[45, 71-72]。
需要注意的是,过去几十年关于根瘤菌共生固氮机制的绝大多数研究都是针对与不同宿主共生的个别模式菌株展开的[14-15]。但农业生产上根瘤菌剂使用中的主要问题是与同一宿主共生的近缘根瘤菌菌种的竞争结瘤和固氮效率差异[73]。Sinorhizobium属快生型大豆根瘤菌是黄淮海和新疆等区域碱性土壤中与大豆共生的优势根瘤菌[45]。近年来,中国农业大学根瘤菌研究中心针对“大豆-Sinorhizobium”共生体系中的共生匹配差异机制展开了研究。通过对Tn5突变体库筛选和实验进化克隆重测序,发现三型分泌系统(T3SS)是限制S. fredii、S. sojae和Sinorhizobium sp. Ⅲ部分菌株与rj2(Rfg1)基因型栽培大豆结瘤的主要因素,通过破坏不匹配菌株T3SS编码基因即可使它们获得结瘤能力[5],这与之前报道的rj2(Rfg1)基因型大豆品种限制S. fredii USDA257结瘤的现象是一致的[74]。但是与rj2(Rfg1)大豆匹配的其他S. fredii菌株却可以编码完整的T3SS而不受限制[5]。说明不同菌株T3SS分泌的效应因子可能有差异,从而引起了宿主截然不同的响应。的确,不匹配菌株的效应蛋白编码基因nopP突变后,也可以获得与rj2(Rfg1)大豆结有效瘤的能力[5],而S. fredii匹配菌株具有不同的NopP蛋白序列。与此一致,在Bradyrhizobium中也发现了NopP序列的变异,而且决定了相应菌株能否与rj2(Rfg1)大豆结瘤[75]。但不是所有根瘤菌中都具有T3SS,例如与苜蓿共生的很多S. medicae和S. meliloti菌株是没有T3SS的[71]。
此外,在Sinorhizobium属近缘大豆快生根瘤菌中,一个反硝化基因簇是含有该簇的菌株与大豆共生固氮所必需的[76],其中nap基因突变后,广谱结瘤菌株S. fredii CCBAU45436固氮能力显著降低(Nod+Fix-),而Sinorhizobium sp. Ⅲ CCBU05631不受影响;S. fredii与Sinorhizobium sp. Ⅲ的hemN1缺失突变株仍然能够固氮,但是都导致它们的共同宿主野大豆无法有效吸收根瘤菌固定的氮(Fix+/Eff-)。S. sojae CCBAU05684不含该基因簇,但由该菌编码的一个hemN1同源基因缺失后结瘤和固氮都受到影响(Nod+/-Fix-)[76]。对于广宿主菌株S. fredii CCBAU45436而言,其特有基因mdtA突变后与野大豆形成的根瘤不能有效固氮,但不具有该编码基因的菌株却可以与野大豆正常结瘤固氮;锌离子高亲和转运子的组分编码基因znuA缺失后与栽培大豆所结根瘤的固氮能力明显降低,但与野大豆所结根瘤的固氮能力仍维持在野生型水平[56, 77]。上述研究表明:同一株根瘤菌与不同宿主共生时会调用保守或特有的功能基因来参与结瘤固氮过程,不同根瘤菌与同一宿主共生时也有类似的现象。
3 结瘤固氮依赖于核心与附属基因的整合根瘤菌通过与豆科植物结瘤固氮实现互利共生,共生基因帮助根瘤菌获得共生能力的同时也使其自身获得了保留,因而也可以被视为一种共生[13]。从比较基因组学角度看,细菌基因组由核心基因和附属基因两部分组成:核心基因为一群近缘的菌株所共有,主要通过垂直遗传保持,负责必需的持家功能;而附属基因只存在于部分菌株,主要通过水平基因转移从环境中获取,一般与特定环境的适应相关[78]。所以,不同基因组背景的细菌适应相同或类似环境时采用的遗传机制就会有所差异。从这个角度讲,如果把根瘤菌与豆科植物建立共生的过程理解为对一系列特定环境的适应,结瘤固氮基因nod/nif仅是根瘤菌解决其中两个关键环节的“通用型”工具,根瘤菌还需要从基因组中调取许多“非通用型”工具来适应不同生态位,基因的保守程度体现这些工具的“通用程度”。有了相应的“工具”,还需要适时地对这些“工具”进行精密调控,才能最终实现根瘤菌对共生建立过程的不同阶段或对不同宿主的适应。
为了探究结瘤固氮功能与基因组背景的整合机制,我们结合比较基因组学与转录组学分析了2株S. fredii菌株的核心与附属基因在不同条件下的表达特征,发现基因的表达水平和在各条件下的表达变异程度以及在共表达网络中的连接度都与基因的保守程度成正比[77]。同时我们还发现,在共生条件下特异上调的基因中保守性较差的附属基因有显著的富集,而与nif基因共表达的除了来自于共生质粒的基因外,还有许多非共生质粒编码的基因。这一结果说明许多参与共生固氮的其他功能基因很可能与关键共生基因在转录调控水平上进行了整合,从而实现协调表达以适应共生固氮的需求。
从根本上讲,基因的转录调控多是由反式作用因子与顺式作用元件的相互作用介导的。不同细菌中核心与附属基因在调控网络中的整合依赖于许多特异调控途径的建立。比如,虽然根瘤菌中固氮基因的表达都受到NifA调控,但NifA自身编码基因的调控方式因菌而异[6]。在S. meliloti中nifA基因受FixL-FixJ双组分系统调节[79],而在B. japonicum中nifA受到氧化还原调控系统RegS-RegR调节[80]。T3SS参与多种病原菌或共生菌与宿主的互作[81],根瘤菌中与T3SS相关的结构基因和效应蛋白编码基因的表达依赖其正调控因子TtsI,而ttsI的表达可以被依赖植物类黄酮信号分子的NodD所激活[82]。这些例子说明已知的参与共生固氮过程的核心模块之间,以及它们与不同根瘤菌基因组背景中的基因调控网络之间都需要进行适宜的整合。
目前发现的参与结瘤固氮过程的转录因子多为局部调控因子,而最近有关多效转录调控因子MucR的研究表明,这一α-变形菌纲根瘤菌中十分保守的蛋白在包括共生在内的多种环境适应中发挥重要的调控功能[56]。进一步的ChIP-seq研究发现,该蛋白在基因组中主要通过优先结合AT-rich序列抑制非保守附属基因的表达,扮演了外来基因沉默子的角色。序列比对分析表明,MucR与其他已知外源基因沉默子如HNS、Lsr2、Rok等有着截然不同的物种分布和彼此独立的进化起源。外源基因被沉默可以避免它们的无序表达对宿主细胞造成潜在的毒性,这样外源基因就有机会保留下来——这个机制可能在原核生物的核心与附属基因整合过程中发挥了重要作用[83]。
值得注意的是,根瘤菌是一类兼性共生细菌,可以在土壤中营腐生生活,在侵染豆科植物根部之前有一个庞大的群体[15]。已有实验进化学研究发现:有些根瘤菌在根际可能处于一个容易发生突变的超突变状态[84],这有利于形成潜在的与特定宿主匹配的核心或附属基因突变体[69]。此外,我们最近的研究发现Sinorhizobium属大豆根瘤菌共生质粒上富集的可转座的插入序列(Insertion sequences)能够插入到共生质粒上的T3SS基因簇(包括分泌系统结构基因和效应因子编码基因nopP),从而介导了不匹配菌株向匹配菌株的进化[5]。所以,已有核心/附属基因调控网络的突变也是结瘤固氮功能可否正常发挥的重要进化途径,这对于根瘤菌适应不断变化的宿主植物品种至关重要。
4 总结水平基因转移在根瘤菌的进化中扮演着重要角色。某些具有特定基因组背景的非根瘤菌细菌可以通过这一方式获得关键共生基因,同时可能伴随着部分原有基因的突变或丢失,并进化为具有初步结瘤甚至固氮能力的根瘤菌。在此之后的长期进化过程中,根瘤菌继续通过水平基因转移的方式从环境中不断获得附属基因,并将其整合到核心调控网络中实现协调表达,从而适应特定的宿主和土壤环境。本质上,核心与附属基因的整合依赖许多顺式作用元件和反式作用因子介导的调控途径的重塑,其中外源基因沉默子可在全局水平上抑制附属基因的无序表达,因而在该过程中发挥着重要作用。伴随根瘤菌的适应性分化,必然导致不同菌株在生物地理分布、共生匹配性和共生固氮效率方面产生差异。基于这一认识,当前的根瘤菌剂菌种筛选工作并没有捷径,需要根据土壤条件和豆科植物栽培品种通过科学筛选获得匹配的高效菌株。
| [1] |
Herridge DF, Peoples MB, Boddey RM. Global inputs of biological nitrogen fixation in agricultural systems[J]. Plant and Soil, 2008, 311(1/2): 1-18. |
| [2] |
Chen WX, Wang ET. China Rhizobia[M]. Beijing: Science Press, 2011. (in Chinese) 陈文新, 汪恩涛. 中国根瘤菌[M]. 北京: 科学出版社, 2011. |
| [3] |
Xiong HY, Zhang XX, Guo HJ, et al. The epidemicity of facultative microsymbionts in faba bean rhizosphere soils[J]. Soil Biology and Biochemistry, 2017, 115: 243-252. DOI:10.1016/j.soilbio.2017.08.032 |
| [4] |
Wang XL, Cui WJ, Feng XY, et al. Rhizobia inhabiting nodules and rhizosphere soils of alfalfa: a strong selection of facultative microsymbionts[J]. Soil Biology and Biochemistry, 2018, 116: 340-350. DOI:10.1016/j.soilbio.2017.10.033 |
| [5] |
Zhao R, Liu LX, Zhang YZ, et al. Adaptive evolution of rhizobial symbiotic compatibility mediated by co-evolved insertion sequences[J]. The ISME Journal, 2017, 12(1): 101-111. |
| [6] |
Dixon R, Kahn D. Genetic regulation of biological nitrogen fixation[J]. Nature Reviews Microbiology, 2004, 2(8): 621-631. DOI:10.1038/nrmicro954 |
| [7] |
Oono R, Schmitt I, Sprent JI, et al. Multiple evolutionary origins of legume traits leading to extreme rhizobial differentiation[J]. New Phytologist, 2010, 187(2): 508-520. DOI:10.1111/j.1469-8137.2010.03261.x |
| [8] |
Montiel J, Downie JA, Farkas A, et al. Morphotype of bacteroids in different legumes correlates with the number and type of symbiotic NCR peptides[J]. Proceedings of the National Academy of Sciences of the United States of America, 2017, 114(19): 5041-5046. |
| [9] |
Giraud E, Moulin L, Vallenet D, et al. Legumes symbioses: absence of nod genes in photosynthetic bradyrhizobia[J]. Science, 2007, 316(5829): 1307-1312. DOI:10.1126/science.1139548 |
| [10] |
Miché L, Moulin L, Chaintreuil C, et al. Diversity analyses of Aeschynomene symbionts in Tropical Africa and Central America reveal that nod-independent stem nodulation is not restricted to photosynthetic bradyrhizobia[J]. Environmental Microbiology, 2010, 12(8): 2152-2164. |
| [11] |
Okazaki S, Kaneko T, Sato S, et al. Hijacking of leguminous nodulation signaling by the rhizobial type Ⅲ secretion system[J]. Proceedings of the National Academy of Sciences of the United States of America, 2013, 110(42): 17131-17136. DOI:10.1073/pnas.1302360110 |
| [12] |
Masson-Boivin C, Giraud E, Perret X, et al. Establishing nitrogen-fixing symbiosis with legumes: how many rhizobium recipes?[J]. Trends in Microbiology, 2009, 17(10): 458-466. DOI:10.1016/j.tim.2009.07.004 |
| [13] |
Remigi P, Zhu J, Young JPW, et al. Symbiosis within symbiosis: evolving nitrogen-fixing legume symbionts[J]. Trends in Microbiology, 2016, 24(1): 63-75. |
| [14] |
Masson-Boivin C, Sachs JL. Symbiotic nitrogen fixation by rhizobia—the roots of a success story[J]. Current Opinion in Plant Biology, 2018, 44: 7-15. DOI:10.1016/j.pbi.2017.12.001 |
| [15] |
Poole P, Ramachandran V, Terpolilli J. Rhizobia: from saprophytes to endosymbionts[J]. Nature Reviews Microbiology, 2018, 16(5): 291-303. DOI:10.1038/nrmicro.2017.171 |
| [16] |
Shamseldin A, Abdelkhalek A, Sadowsky MJ. Recent changes to the classification of symbiotic, nitrogen-fixing, legume-associating bacteria: a review[J]. Symbiosis, 2017, 71(2): 91-109. DOI:10.1007/s13199-016-0462-3 |
| [17] |
Jourand P, Giraud E, Béna G, et al. Methylobacterium nodulans sp. nov., for a group of aerobic, facultatively methylotrophic, legume root-nodule-forming and nitrogen-fixing bacteria[J]. International Journal of Systematic and Evolutionary Microbiology, 2004, 54(6): 2269-2273. DOI:10.1099/ijs.0.02902-0 |
| [18] |
Amadou C, Pascal G, Mangenot S, et al. Genome sequence of the β-rhizobium Cupriavidustaiwanensis and comparative genomics of rhizobia[J]. Genome Research, 2008, 18(9): 1472-1483. DOI:10.1101/gr.076448.108 |
| [19] |
Pobigaylo N, Szymczak S, Nattkemper TW, et al. Identification of genes relevant to symbiosis and competitiveness in Sinorhizobium meliloti using signature-tagged mutants[J]. Molecular Plant-Microbe Interactions, 2008, 21(2): 219-231. DOI:10.1094/MPMI-21-2-0219 |
| [20] |
Shimoda Y, Shinpo S, Kohara M, et al. A large scale analysis of protein-protein interactions in the nitrogen-fixing bacterium Mesorhizobiumloti[J]. DNA Research, 2008, 15(1): 13-23. DOI:10.1093/dnares/dsm028 |
| [21] |
Wang D, Wang YC, Wu LJ, et al. Construction and pilot screening of a signature-tagged mutant library of Sinorhizobium fredii[J]. Archives of Microbiology, 2016, 198(2): 91-99. |
| [22] |
Zhang XX, Guo HJ, Jiao J, et al. Pyrosequencing of rpoB uncovers a significant biogeographical pattern of rhizobial species in soybean rhizosphere[J]. Journal of Biogeography, 2017, 44(7): 1491-1499. |
| [23] |
Sullivan JT, Ronson CW. Evolution of rhizobia by acquisition of a 500-kb symbiosis island that integrates into a phe-tRNA gene[J]. Proceedings of the National Academy of Sciences of the United States of America, 1998, 95(9): 5145-5149. DOI:10.1073/pnas.95.9.5145 |
| [24] |
Zhang XX, Guo HJ, Wang R, et al. Genetic divergence of Bradyrhizobium strains nodulating soybeans as revealed by multilocus sequence analysis of genes inside and outside the symbiosis island[J]. Applied and Environmental Microbiology, 2014, 80(10): 3181-3190. DOI:10.1128/AEM.00044-14 |
| [25] |
Ling J, Wang H, Wu P, et al. Plant nodulation inducers enhance horizontal gene transfer of Azorhizobiumcaulinodans symbiosis island[J]. Proceedings of the National Academy of Sciences of the United States of America, 2016, 113(48): 13875-13880. DOI:10.1073/pnas.1615121113 |
| [26] |
Galibert F, Finan TM, Long SR, et al. The composite genome of the legume symbiont Sinorhizobiummeliloti[J]. Science, 2001, 293(5530): 668-672. DOI:10.1126/science.1060966 |
| [27] |
Kaneko T, Nakamura Y, Sato S, et al. Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110[J]. DNA Research, 2002, 9(6): 189-197. DOI:10.1093/dnares/9.6.189 |
| [28] |
Young JPW, Crossman LC, Johnston AWB, et al. The genome of Rhizobiumleguminosarum has recognizable core and accessory components[J]. Genome Biology, 2006, 7(4): R34. |
| [29] |
Wang SS, Hao BH, Li JR, et al. Whole-genome sequencing of Mesorhizobiumhuakuii 7653R provides molecular insights into host specificity and symbiosis island dynamics[J]. BMC Genomics, 2014, 15: 440. DOI:10.1186/1471-2164-15-440 |
| [30] |
Dos Santos PC, Fang Z, Mason SW, et al. Distribution of nitrogen fixation and nitrogenase-like sequences amongst microbial genomes[J]. BMC Genomics, 2012, 13: 162. DOI:10.1186/1471-2164-13-162 |
| [31] |
Henson BJ, Hartman L, Watson LE, et al. Evolution and variation of the nifD and hupL elements in the heterocystous cyanobacteria[J]. International Journal of Systematic and Evolutionary Microbiology, 2011, 61(12): 2938-2949. DOI:10.1099/ijs.0.028340-0 |
| [32] |
Bontemps C, Elliott GN, Simon MF, et al. Burkholderia species are ancient symbionts of legumes[J]. Molecular Ecology, 2010, 19(1): 44-52. |
| [33] |
Menna P, Hungria M. Phylogeny of nodulation and nitrogen-fixation genes in Bradyrhizobium: supporting evidence for the theory of monophyletic origin, and spread and maintenance by both horizontal and vertical transfer[J]. International Journal of Systematic and Evolutionary Microbiology, 2011, 61(12): 3052-3067. DOI:10.1099/ijs.0.028803-0 |
| [34] |
Okubo T, Piromyou P, Tittabutr P, et al. Origin and evolution of nitrogen fixation genes on symbiosis islands and plasmid in Bradyrhizobium[J]. Microbes and Environments, 2016, 31(3): 260-267. DOI:10.1264/jsme2.ME15159 |
| [35] |
Persson T, Battenberg K, Demina IV, et al. Candidatus Frankia datiscae Dg1, the actinobacterial microsymbiont of Datiscaglomerata, expresses the canonical nod genes nodABC in symbiosis with its host plant[J]. PLoS One, 2015, 10(5): e0127630. DOI:10.1371/journal.pone.0127630 |
| [36] |
van Nguyen T, Wibberg D, Battenberg K, et al. An assemblage of Frankia Cluster Ⅱ strains from California contains the canonical nod genes and also the sulfotransferase gene nodH[J]. BMC Genomics, 2016, 17: 796. DOI:10.1186/s12864-016-3140-1 |
| [37] |
Ktari A, Nouioui I, Furnholm T, et al. Permanent draft genome sequence of Frankia sp. NRRL B-16219 reveals the presence of canonical nod genes, which are highly homologous to those detected in Candidatus Frankia Dg1 genome[J]. Standards in Genomic Sciences, 2017, 12: 51. DOI:10.1186/s40793-017-0261-3 |
| [38] |
Aoki S, Ito M, Iwasaki W. From β-to α-proteobacteria: the origin and evolution of rhizobial nodulation genes nodIJ[J]. Molecular Biology and Evolution, 2013, 30(11): 2494-2508. DOI:10.1093/molbev/mst153 |
| [39] |
Ormeño-Orrillo E, Servín-Garcidueñas LE, Imperial J, et al. Phylogenetic evidence of the transfer of nodZ and nolL genes from Bradyrhizobium to other rhizobia[J]. Molecular Phylogenetics and Evolution, 2013, 67(3): 626-630. |
| [40] |
Rogel MA, Ormeño-Orrillo E, Romero EM. Symbiovars in rhizobia reflect bacterial adaptation to legumes[J]. Systematic and Applied Microbiology, 2011, 34(2): 96-104. DOI:10.1016/j.syapm.2010.11.015 |
| [41] |
Guo HJ, Wang ET, Zhang XX, et al. Replicon-dependent differentiation of symbiosis-related genes in Sinorhizobium strains nodulating Glycinemax[J]. Applied and Environmental Microbiology, 2014, 80(4): 1245-1255. |
| [42] |
Barcellos FG, Menna P, Da Silva Batista JS, et al. Evidence of horizontal transfer of symbiotic genes from a Bradyrhizobium japonicum inoculant strain to indigenous diazotrophs Sinorhizobium (Ensifer) fredii and Bradyrhizobiumelkanii in a Brazilian Savannah soil[J]. Applied and Environmental Microbiology, 2007, 73(8): 2635-2643. |
| [43] |
Jiao YS, Liu YH, Yan H, et al. Rhizobial diversity and nodulation characteristics of the extremely promiscuous legume Sophora flavescens[J]. Molecular Plant-Microbe Interactions, 2015, 28(12): 1338-1352. DOI:10.1094/MPMI-06-15-0141-R |
| [44] |
Liu YH, Jiao YS, Liu LX, et al. Nonspecific symbiosis between Sophoraflavescens and different rhizobia[J]. Molecular Plant-Microbe Interactions, 2018, 31(2): 224-232. DOI:10.1094/MPMI-05-17-0117-R |
| [45] |
Tian CF, Zhou YJ, Zhang YM, et al. Comparative genomics of rhizobia nodulating soybean suggests extensive recruitment of lineage-specific genes in adaptations[J]. Proceedings of the National Academy of Sciences of the United States of America, 2012, 109(22): 8629-8634. DOI:10.1073/pnas.1120436109 |
| [46] |
Turner SL, Young JPW. The glutamine synthetases of rhizobia: phylogenetics and evolutionary implications[J]. Molecular Biology and Evolution, 2000, 17(2): 309-319. |
| [47] |
Hirsch AM, Wilson KJ, Jones JD, et al. Rhizobium meliloti nodulation genes allow Agrobacteriumtumefaciens and Escherichiacoli to form pseudonodules on alfalfa[J]. Journal of Bacteriology, 1984, 158(3): 1133-1143. |
| [48] |
Marchetti M, Capela D, Glew M, et al. Experimental evolution of a plant pathogen into a legume symbiont[J]. PLoS Biology, 2010, 8(1): e1000280. DOI:10.1371/journal.pbio.1000280 |
| [49] |
Kawaharada Y, Kelly S, Nielsen MW, et al. Receptor-mediated exopolysaccharide perception controls bacterial infection[J]. Nature, 2015, 523(7560): 308-312. DOI:10.1038/nature14611 |
| [50] |
Gibson KE, Kobayashi H, Walker GC. Molecular determinants of a symbiotic chronic infection[J]. Annual Review of Genetics, 2008, 42: 413-441. DOI:10.1146/annurev.genet.42.110807.091427 |
| [51] |
Hubber A, Vergunst AC, Sullivan JT, et al. Symbiotic phenotypes and translocated effector proteins of the Mesorhizobium loti strain R7A VirB/D4 type Ⅳ secretion system[J]. Molecular Microbiology, 2004, 54(2): 561-574. |
| [52] |
Oldroyd GE, Murray JD, Poole PS, et al. The rules of engagement in the legume-rhizobial symbiosis[J]. Annual Review of Genetics, 2011, 45: 119-144. DOI:10.1146/annurev-genet-110410-132549 |
| [53] |
Hood G, Ramachandran V, East AK, et al. Manganese transport is essential for N2-fixation by Rhizobium leguminosarum in bacteroids from galegoid but not phaseoloid nodules[J]. Environmental Microbiology, 2017, 19(7): 2715-2726. DOI:10.1111/emi.2017.19.issue-7 |
| [54] |
Davies BW, Walker GC. Disruption of sitA compromises Sinorhizobiummeliloti for manganese uptake required for protection against oxidative stress[J]. Journal of Bacteriology, 2007, 189(5): 2101-2109. DOI:10.1128/JB.01377-06 |
| [55] |
Hu Y, Jiao J, Liu LX, et al. Evidence for phosphate starvation of rhizobia without terminal differentiation in legume nodules[J]. Molecular Plant-Microbe Interactions, 2018, 31(10): 1060-1068. DOI:10.1094/MPMI-02-18-0031-R |
| [56] |
Jiao J, Wu LJ, Zhang BL, et al. MucR is required for transcriptional activation of conserved ion transporters to support nitrogen fixation of Sinorhizobiumfredii in soybean nodules[J]. Molecular Plant-Microbe Interactions, 2016, 29(5): 352-361. DOI:10.1094/MPMI-01-16-0019-R |
| [57] |
Zipfel C, Oldroyd GED. Plant signalling in symbiosis and immunity[J]. Nature, 2017, 543(7645): 328-336. DOI:10.1038/nature22009 |
| [58] |
O'Brian MR. Heme synthesis in the rhizobium-legume symbiosis: a palette for bacterial and eukaryotic pigments[J]. Journal of Bacteriology, 1996, 178(9): 2471-2478. DOI:10.1128/jb.178.9.2471-2478.1996 |
| [59] |
Hakoyama T, Niimi K, Watanabe H, et al. Host plant genome overcomes the lack of a bacterial gene for symbiotic nitrogen fixation[J]. Nature, 2009, 462(7272): 514-517. DOI:10.1038/nature08594 |
| [60] |
Lodwig EM, Hosie AHF, Bourdès A, et al. Amino-acid cycling drives nitrogen fixation in the legume-Rhizobium symbiosis[J]. Nature, 2003, 422(6933): 722-726. DOI:10.1038/nature01527 |
| [61] |
Prell J, White JP, Bourdes A, et al. Legumes regulate Rhizobium bacteroid development and persistence by the supply of branched-chain amino acids[J]. Proceedings of the National Academy of Sciences of the United States of America, 2009, 106(30): 12477-12482. DOI:10.1073/pnas.0903653106 |
| [62] |
Prell J, Bourdès A, Kumar S, et al. Role of symbiotic auxotrophy in the Rhizobium-legume symbioses[J]. PLoS One, 2010, 5(11): e13933. DOI:10.1371/journal.pone.0013933 |
| [63] |
Yuan ZC, Zaheer R, Finan TM. Regulation and properties of PstSCAB, a high-affinity, high-velocity phosphate transport system of Sinorhizobiummeliloti[J]. Journal of Bacteriology, 2006, 188(3): 1089-1102. DOI:10.1128/JB.188.3.1089-1102.2006 |
| [64] |
Leigh JA, Signer ER, Walker GC. Exopolysaccharide-deficient mutants of Rhizobiummeliloti that form ineffective nodules[J]. Proceedings of the National Academy of Sciences of the United States of America, 1985, 82(18): 6231-6235. DOI:10.1073/pnas.82.18.6231 |
| [65] |
Niehaus K, Kapp D, Pühler A. Plant defence and delayed infection of alfalfa pseudonodules induced by an exopolysaccharide (EPS Ⅰ)-deficient Rhizobiummeliloti mutant[J]. Planta, 1993, 190(3): 415-425. |
| [66] |
González JE, Reuhs BL, Walker GC. Low molecular weight EPS Ⅱ of Rhizobiummeliloti allows nodule invasion in Medicagosativa[J]. Proceedings of the National Academy of Sciences of the United States of America, 1996, 93(16): 8636-8641. DOI:10.1073/pnas.93.16.8636 |
| [67] |
Garnerone A, Cabanes D, Foussard M, et al. Inhibition of the FixL Sensor Kinase by the FixT Protein in Sinorhizobium meliloti[J]. The Journal of Biological Chemistry, 1999, 274(45): 32500-32506. DOI:10.1074/jbc.274.45.32500 |
| [68] |
Emmerich R, Strehler P, Hennecke H, et al. An imperfect inverted repeat is critical for DNA binding of the response regulator RegR of Bradyrhizobiumjaponicum[J]. Nucleic Acids Research, 2000, 28(21): 4166-4171. DOI:10.1093/nar/28.21.4166 |
| [69] |
Capela D, Marchetti M, Clérissi C, et al. Recruitment of a lineage-specific virulence regulatory pathway promotes intracellular infection by a plant pathogen experimentally evolved into a legume symbiont[J]. Molecular Biology and Evolution, 2017, 34(10): 2503-2521. DOI:10.1093/molbev/msx165 |
| [70] |
Galardini M, Pini F, Bazzicalupo M, et al. Replicon-dependent bacterial genome evolution: the case of Sinorhizobium meliloti[J]. Genome Biology and Evolution, 2013, 5(3): 542-558. |
| [71] |
Sugawara M, Epstein B, Badgley BD, et al. Comparative genomics of the core and accessory genomes of 48 Sinorhizobium strains comprising five genospecies[J]. Genome Biology, 2013, 14(2): R17. |
| [72] |
diCenzo GC, Checcucci A, Bazzicalupo M, et al. Metabolic modelling reveals the specialization of secondary replicons for niche adaptation in Sinorhizobiummeliloti[J]. Nature Communications, 2016, 7: 12219. |
| [73] |
Brockwell J, Bottomley PJ. Recent advances in inoculant technology and prospects for the future[J]. Soil Biology and Biochemistry, 1995, 27(4/5): 683-697. |
| [74] |
Yang SM, Tang F, Gao MQ, et al. R gene-controlled host specificity in the legume-rhizobia symbiosis[J]. Proceedings of the National Academy of Sciences of the United States of America, 2010, 107(43): 18735-18740. |
| [75] |
Sugawara M, Takahashi S, Umehara Y, et al. Variation in bradyrhizobial NopP effector determines symbiotic incompatibility with Rj2-soybeans via effector-triggered immunity[J]. Nature Communications, 2018, 9: 3139. DOI:10.1038/s41467-018-05663-x |
| [76] |
Liu LX, Li QQ, Zhang YZ, et al. The nitrate-reduction gene cluster components exert lineage-dependent contributions to optimization of Sinorhizobium symbiosis with soybeans[J]. Environmental Microbiology, 2017, 19(12): 4926-4938. DOI:10.1111/1462-2920.13948 |
| [77] |
Jiao J, Ni M, Zhang BL, et al. Coordinated regulation of core and accessory genes in the multipartite genome of Sinorhizobium fredii[J]. PLoS Genetics, 2018, 14(5): e1007428. |
| [78] |
Young JPW. Bacteria are smartphones and mobile genes are apps[J]. Trends in Microbiology, 2016, 24(12): 931-932. DOI:10.1016/j.tim.2016.09.002 |
| [79] |
Bobik C, Meilhoc E, Batut J. FixJ: a major regulator of the oxygen limitation response and late symbiotic functions of Sinorhizobium meliloti[J]. Journal of Bacteriology, 2006, 188(13): 4890-4902. DOI:10.1128/JB.00251-06 |
| [80] |
Bauer E, Kaspar T, Fischer HM, et al. Expression of the fixR-nifA operon in Bradyrhizobiumjaponicum depends on a new response regulator, RegR[J]. Journal of Bacteriology, 1998, 180(15): 3853-3863. |
| [81] |
Galán JE, Collmer A. Type Ⅲ secretion machines: bacterial devices for protein delivery into host cells[J]. Science, 1999, 284(5418): 1322-1328. DOI:10.1126/science.284.5418.1322 |
| [82] |
Deakin WJ, Broughton WJ. Symbiotic use of pathogenic strategies: rhizobial protein secretion systems[J]. Nature Reviews Microbiology, 2009, 7(4): 312-320. DOI:10.1038/nrmicro2091 |
| [83] |
Singh K, Milstein JN, Navarre WW. Xenogeneic silencing and its impact on bacterial genomes[J]. Annual Review of Microbiology, 2016, 70: 199-213. |
| [84] |
Remigi P, Capela D, Clerissi C, et al. Transient hypermutagenesis accelerates the evolution of legume endosymbionts following horizontal gene transfer[J]. PLoS Biology, 2014, 12(9): e1001942. |
 2019, Vol. 46
2019, Vol. 46