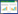扩展功能
文章信息
- 刘亚华, 李伟程, 余中节, 赵飞燕, 赵洁, 孙志宏, 张和平
- LIU Ya-Hua, LI Wei-Cheng, YU Zhong-Jie, ZHAO Fei-Yan, ZHAO Jie, SUN Zhi-Hong, ZHANG He-Ping
- 不同分离源植物乳杆菌的群体基因组分析
- Population genetics of Lactobacillus plantarum isolated from different environments
- 微生物学通报, 2019, 46(12): 3388-3401
- Microbiology China, 2019, 46(12): 3388-3401
- DOI: 10.13344/j.microbiol.china.190132
-
文章历史
- 收稿日期: 2019-02-24
- 接受日期: 2019-04-11
- 网络首发日期: 2019-05-15
植物乳杆菌是一种兼性异型发酵的乳酸菌,广泛存在于植物、乳制品、肉制品和胃肠道等多种生态环境中[1],具有维持肠道内菌群平衡、降胆固醇、降血脂和促进机体生长等多种益生功效,应用于食品工业及医疗保健等多个领域[2-5]。植物乳杆菌具有较高的经济价值,解析其基因组特征有助于在生产工业中的应用。
随着高通量测序的发展,越来越多的细菌基因组被测定,通过增加种群中研究个体数量能降低研究中的统计偏差,有助于提高研究的准确性。利用比较基因组学方法构建物种或种群的泛基因集(Pan-genome)和核心基因集(Core-genome),并以此为基础讨论物种或种群的直系同源序列,可以对物种或种群的群体遗传进行分析[6]。许多研究发现,乳酸菌基因组的多样性大多与其所在的生境有关。Broadbent等[7]通过比较基因组学对不同分离源的干酪乳杆菌(Lactobacillus casei)进行了分析,发现不同分离源的干酪乳杆菌基因组中存在环境特异性基因。OʼSullivan等[8]研究分离自不同生态环境的乳酸菌,发现与糖代谢、蛋白质水解和限制性修饰等功能相关的基因中存在环境特异性现象。然而,Martino等[9]通过对来自不同分离源的54株植物乳杆菌的比较基因组学研究发现,其与所在生态环境无关。该研究仅使用了54个菌株,而基因组数量的增加可能会对细菌群体遗传的认识有新的见解[10-12]。
为了进一步探究不同分离源的植物乳杆菌是否与其环境存在潜在的联系,本研究测定了2株分离自乳制品的植物乳杆菌全基因组序列,并从现有数据库中下载了124株菌的基因组序列,以126株菌的全基因组序列为研究对象,利用比较基因组学对植物乳杆菌的遗传进化关系及其功能基因进行分析,为全面解析植物乳杆菌功能特征及进化历程提供数据支持。
1 材料与方法 1.1 实验菌株测定的2株菌来自内蒙古农业大学乳酸菌菌种资源库(Lactic acid bacteria collection center,LABCC),分别为分离自蒙古前杭盖海尔汗苏木酸驼奶中的IMAU20970和分离自中国甘肃甘南州玛曲县阿尼玛乡酸牦牛奶中的IMAU80873。124株菌株基因组均下载自NCBI的GenBank数据库(http://www.ncbi.nlm.nih.gov/genbank/),它们具有明确的分离源,测序质量较好,菌株具体菌株信息如表 1所示。
| 菌株 Strain |
分离源 Habitat |
编码基因 CDSs |
基因组大小 Size (kb) |
(G+C)mol% (%) |
基因组 Genome |
拼接情况 Scaffolds |
GenBank登录号 GenBank accession No. |
| JDM1 | Plant | 2 904 | 3 197.8 | 44.7 | Complete genome | 1 | CP001617.1 |
| ST-Ⅲ | Plant | 3 020 | 3 307.9 | 44.5 | Complete genome | 1 | CP002222.1 |
| 16 | Plant | 3 076 | 3 361.0 | 44.3 | Complete genome | 1 | CP006033.1 |
| 5_2 | Plant | 2 991 | 3 237.7 | 44.7 | Complete genome | 1 | CP009236.1 |
| ZS2058 | Plant | 2 916 | 3 198.3 | 44.7 | Complete genome | 1 | CP012343.1 |
| CAUH2 | Plant | 3 010 | 3 274.6 | 44.5 | Complete genome | 1 | CP015126.1 |
| DSM.16365 | Plant | 3 042 | 3 350.3 | 44.9 | Complete genome | 1 | CP032751.1 |
| NC8 | Plant | 2 942 | 3 207.2 | 44.6 | Draft | 10 | AGRI00000000.1 |
| NCU116 | Plant | 3 066 | 3 354.7 | 44.4 | Complete genome | 1 | CP016071.1 |
| Nizo1839 | Plant | 2 902 | 3 197.0 | 45.1 | Draft | 72 | LUWB00000000.1 |
| Nizo2262 | Plant | 2 964 | 3 220.0 | 44.5 | Draft | 33 | LUWJ00000000.1 |
| Nizo2263 | Plant | 3 042 | 3 259.3 | 44.5 | Draft | 40 | LUWK00000000.1 |
| Nizo2264 | Plant | 2 819 | 3 087.3 | 45.3 | Draft | 32 | LUWL00000000.1 |
| Nizo2726 | Plant | 2 913 | 3 179.4 | 44.6 | Draft | 30 | LUWS00000000.1 |
| Nizo2741 | Plant | 3 090 | 3 346.5 | 44.3 | Draft | 49 | LUWT00000000.1 |
| Nizo2753 | Plant | 2 847 | 3 128.5 | 44.6 | Draft | 56 | LUWU00000000.1 |
| Nizo2766 | Plant | 2 905 | 3 169.1 | 44.4 | Draft | 90 | LUWW00000000.1 |
| Nizo2801 | Plant | 2 974 | 3 248.0 | 44.4 | Draft | 78 | LUWY00000000.1 |
| Nizo2806 | Plant | 2 932 | 3 188.8 | 44.6 | Draft | 39 | LUXA00000000.1 |
| Nizo2831 | Plant | 2 962 | 3 232.1 | 44.5 | Draft | 31 | LUXD00000000.1 |
| Nizo2891 | Plant | 3 205 | 3 469.2 | 44.1 | Draft | 78 | LUXG00000000.1 |
| Nizo3894 | Plant | 3 032 | 3 296.1 | 44.5 | Draft | 52 | LUXK00000000.1 |
| LY-78 | Plant | 2 835 | 3 128.8 | 44.8 | Complete genome | 1 | CP015308.1 |
| LP2 | Plant | 3 011 | 3 284.6 | 44.5 | Complete genome | 1 | CP020816.1 |
| KC3 | Plant | 3 026 | 3 330.0 | 44.6 | Complete genome | 1 | CP025586.1 |
| DSR_M2 | Plant | 2 991 | 3 250.8 | 44.6 | Complete genome | 1 | CP022294.1 |
| IDCC3501 | Plant | 2 976 | 3 242.7 | 44.5 | Complete genome | 1 | CP027349.1 |
| ATG-K6 | Plant | 2 999 | 3 262.5 | 44.5 | Complete genome | 1 | CP032464.1 |
| RI-139 | Plant | 3 104 | 3 328.1 | 44.3 | Draft | 78 | MKDS00000000.1 |
| RI-123 | Plant | 3 094 | 3 323.3 | 44.3 | Draft | 60 | MKDQ00000000.1 |
| RI-140 | Plant | 3 089 | 3 312.6 | 44.3 | Draft | 85 | MKDT00000000.1 |
| NF92 | Plant | 3 066 | 3 372.8 | 44.4 | Draft | 77 | RDQI00000000.1 |
| 4_3 | Plant | 3 043 | 3 320.4 | 44.3 | Draft | 105 | AYTU00000000.1 |
| CGMCC1.2437 | Plant | 2 962 | 3 212.3 | 44.5 | Draft | 48 | AZEJ00000000.1 |
| FMNP01 | Plant | 3 017 | 3 313.6 | 44.5 | Draft | 4 | JPSU00000000.1 |
| IPLA88 | Plant | 3 001 | 3 254.1 | 44.4 | Draft | 208 | ASJE00000000.1 |
| JBE245 | Plant | 2 993 | 3 262.6 | 44.5 | Complete genome | 1 | CP014780.1 |
| Lp90 | Plant | 3 065 | 3 324.1 | 44.3 | Draft | 33 | JIBX00000000.1 |
| Nizo1840 | Plant | 2 846 | 3 204.4 | 44.9 | Draft | 189 | LUWC00000000.1 |
| 19.1 | Drosophila | 3 594 | 3 651.1 | 43.9 | Draft | 42 | LUXM00000000.1 |
| NAB2 | Drosophila | 3 595 | 3 550.8 | 44.1 | Draft | 44 | LUXO00000000.1 |
| NAB1 | Drosophila | 3 494 | 3 454.7 | 44.2 | Draft | 44 | LUXN00000000.1 |
| CNW10 | Drosophila | 2 945 | 3 194.7 | 44.5 | Draft | 83 | LUWN00000000.1 |
| ER | Drosophila | 3 074 | 3 452.8 | 44.3 | Draft | 37 | LUXL00000000.1 |
| DmCS_001 | Drosophila | 2 945 | 3 194.7 | 44.5 | Draft | 83 | JOJT00000000.1 |
| BDGP2 | Drosophila | 3 297 | 3 581.6 | 44.2 | Complete genome | 1 | CP023174.1 |
| WJL1 | Drosophila | 3 246 | 3 503.1 | 44.2 | Draft | 102 | AUTE00000000.1 |
| WJL2 | Drosophila | 3 229 | 3 477.5 | 44.2 | Draft | 13 | LKLZ00000000.1 |
| WCFS1 | Mammals | 3 063 | 3 348.6 | 44.5 | Complete genome | 1 | AL935263.2 |
| CMPG5300 | Mammals | 3 233 | 3 507.8 | 44.2 | Draft | 7 | CM002918.1 |
| Nizo2258 | Mammals | 2 874 | 3 131.3 | 44.4 | Draft | 142 | LUWG00000000.1 |
| Nizo2259 | Mammals | 3 177 | 3 430.6 | 44.3 | Draft | 109 | LUWH00000000.1 |
| Nizo3892 | Mammals | 3 055 | 3 324.9 | 44.4 | Draft | 45 | LUXI00000000.1 |
| NRCC1 | Mammals | 2 694 | 2 995.1 | 44.9 | Draft | 134 | LUSM00000000.1 |
| AG30 | Mammals | 3 068 | 3 442.9 | 44.3 | Draft | 48 | JHWA00000000.1 |
| Lp1610 | Mammals | 3 070 | 3 298.7 | 44.4 | Draft | 27 | LDEL00000000.1 |
| Lp1612 | Mammals | 2 979 | 3 240.5 | 44.4 | Draft | 25 | LDEM00000000.1 |
| ZJ316 | Gut | 2 972 | 3 299.8 | 44.4 | Complete genome | 1 | CP004082.1 |
| HFC8 | Gut | 3 065 | 3 405.7 | 44.3 | Complete genome | 1 | CP012650.1 |
| LZ95 | Gut | 3 026 | 3 322.5 | 44.5 | Complete genome | 1 | CP012122.1 |
| 43_3 | Gut | 3 039 | 3 294.5 | 44.4 | Draft | 171 | LOMH00000000.1 |
| LP91 | Gut | 2 671 | 2 925.6 | 45.1 | Draft | 145 | AXDQ00000000.1 |
| 90sk | Gut | 3 058 | 3 371.5 | 44.3 | Draft | 47 | JXAX00000000.1 |
| CIP104448 | Gut | 2 938 | 3 237.3 | 44.4 | Draft | 178 | JSUW00000000.1 |
| TIFN101 | Gut | 2 801 | 3 079.4 | 44.5 | Draft | 174 | JSUX00000000.1 |
| E2C2 | Gut | 3 361 | 3 603.6 | 44.0 | Draft | 121 | LSST00000000.1 |
| E2C5 | Gut | 3 370 | 3 615.2 | 44.0 | Draft | 121 | LTCD00000000.1 |
| Nizo2256 | Gut | 2 753 | 2 997.7 | 44.6 | Draft | 87 | LUWE00000000.1 |
| Nizo1838 | Gut | 2 787 | 3 034.1 | 44.5 | Draft | 100 | LUWA00000000.1 |
| Nizo3893 | Gut | 3 020 | 3 342.8 | 44.3 | Draft | 239 | LUXJ00000000.1 |
| Nizo2257 | Gut | 2 878 | 3 156.7 | 44.4 | Draft | 145 | LUWF00000000.1 |
| Nizo1837 | Gut | 3 058 | 3 309.7 | 44.4 | Draft | 48 | LTAU00000000.1 |
| Nizo2260 | Gut | 3 058 | 3 310.2 | 44.4 | Draft | 42 | LUWI00000000.1 |
| TL2766 | Gut | 3 037 | 3 303.7 | 44.4 | Draft | 33 | LZXZ00000000.1 |
| SNU.Lp177 | Gut | 2 931 | 3 204.8 | 45.0 | Draft | 101 | LGIM00000000.1 |
| 8RA_3 | Gut | 3 040 | 3 301.9 | 44.4 | Draft | 18 | LBDF00000000.1 |
| EBKLp545 | Gut | 3 012 | 3 306.5 | 44.3 | Draft | 138 | PZPN00000000.1 |
| MPL16 | Gut | 2 937 | 3 278.5 | 43.6 | Draft | 166 | LUHN00000000.1 |
| plantarum | Gut | 2 939 | 3 246.6 | 44.6 | Complete genome | 1 | CP021086.1 |
| B21 | Meat products | 3 039 | 3 310.7 | 44.4 | Complete genome | 1 | CP010528.1 |
| MF1298 | Meat products | 3 272 | 3 576.4 | 44.2 | Complete genome | 1 | CP013149.1 |
| RI-113 | Meat products | 3 183 | 3 463.0 | 44.3 | Complete genome | 1 | CP017406.1 |
| TMW1.25 | Meat products | 3 054 | 3 351.9 | 44.3 | Complete genome | 1 | CP017354.1 |
| TMW1.708 | Meat products | 2 928 | 3 246.5 | 44.5 | Complete genome | 1 | CP017374.1 |
| LPL-1 | Meat products | 2 917 | 3 200.6 | 44.6 | Complete genome | 1 | CP021997.1 |
| Nizo2877 | Meat products | 2 979 | 3 231.8 | 44.5 | Draft | 2 | LKHZ00000000.1 |
| Nizo2484 | Meat products | 3 141 | 3 386.1 | 44.2 | Draft | 70 | LUWO00000000.1 |
| Nizo2485 | Meat products | 3 107 | 3 348.6 | 44.3 | Draft | 62 | LUWP00000000.1 |
| Nizo2494 | Meat products | 3 032 | 3 303.2 | 44.5 | Draft | 57 | LUWQ00000000.1 |
| Nizo2855 | Meat products | 2 979 | 3 223.1 | 44.5 | Draft | 37 | LUXE00000000.1 |
| Nizo2457 | Meat products | 3 064 | 3 339.1 | 44.4 | Draft | 73 | LUWM00000000.1 |
| RI-422 | Meat products | 3 095 | 3 327.0 | 44.3 | Draft | 55 | MKDK00000000.1 |
| A6 | Meat products | 3 084 | 3 368.6 | 44.3 | Draft | 66 | LRUO00000000.1 |
| Lp820 | Meat products | 2 963 | 3 195.5 | 44.5 | Draft | 44 | NQNI00000000.1 |
| CRL681 | Meat products | 3 126 | 3 370.2 | 44.3 | Draft | 28 | QOSF00000000.1 |
| RI-408 | Meat products | 2 810 | 3 073.5 | 44.6 | Draft | 123 | MKDH00000000.1 |
| RI-393 | Meat products | 3 087 | 3 325.3 | 44.3 | Draft | 74 | MKDB00000000.1 |
| 19L3 | Dairy products | 3 115 | 3 289.3 | 44.4 | Draft | 241 | AWTS00000000.1 |
| 2165 | Dairy products | 3 108 | 3 180.0 | 44.5 | Draft | 192 | AVFI00000000.1 |
| 2025 | Dairy products | 2 820 | 3 334.3 | 44.4 | Draft | 164 | AVFJ00000000.2 |
| CRL1506 | Dairy products | 2 966 | 3 228.1 | 44.6 | Draft | 14 | LNCP00000000.1 |
| LZ206 | Dairy products | 2 891 | 3 263.7 | 44.5 | Complete genome | 1 | CP015966.1 |
| LZ227 | Dairy products | 3 082 | 3 425.3 | 44.3 | Complete genome | 1 | CP015857.1 |
| Nizo2029 | Dairy products | 2 996 | 3 243.0 | 44.5 | Draft | 96 | LUWD00000000.1 |
| Nizo2776 | Dairy products | 2 767 | 3 124.6 | 44.5 | Draft | 190 | LUWX00000000.1 |
| Nizo2802 | Dairy products | 3 094 | 3 372.5 | 44.4 | Draft | 89 | LUWZ00000000.1 |
| UCMA3037 | Dairy products | 2 836 | 3 108.3 | 44.5 | Draft | 68 | APHP00000000.1 |
| Nizo3400 | Dairy products | 2 914 | 3 267.4 | 44.4 | Draft | 154 | LUXH00000000.1 |
| NL42 | Dairy products | 3 051 | 3 353.1 | 44.4 | Draft | 52 | JZSB00000000.1 |
| IMAU20970 | Dairy products | 3 137 | 3 341.7 | 44.7 | Draft | 251 | RCDK00000000 |
| IMAU80873 | Dairy products | 3 036 | 3 193.8 | 44.5 | Draft | 157 | RCDJ000000000 |
| P-8 | Dairy products | 2 939 | 3 246.6 | 44.6 | Complete genome | 1 | CP005942.2 |
| CCFM605 | Dairy products | 3 102 | 3 314.6 | 44.4 | Draft | 55 | QPQR00000000.1 |
| CGMCC12436 | Dairy products | 3 010 | 3 259.9 | 44.5 | Draft | 53 | QPQS00000000.1 |
| 10CH | Dairy products | 3 013 | 3 311.1 | 44.5 | Complete genome | 1 | CP023728.1 |
| RI-191 | Dairy products | 2 978 | 3 256.7 | 44.4 | Draft | 72 | MKDA00000000.1 |
| RI-086 | Dairy products | 2 826 | 3 084.7 | 44.6 | Draft | 90 | MKDP00000000.1 |
| Lp998 | Dairy products | 3 014 | 3 274.1 | 44.5 | Draft | 112 | FMBQ00000000.1 |
| 1 | Dairy products | 3 023 | 3 287.1 | 44.4 | Draft | 41 | FKLQ00000000.1 |
| TJA26B | Dairy products | 3 019 | 3 243.5 | 44.5 | Draft | 122 | QXEU00000000.1 |
| D13 | Dairy products | 3 080 | 3 343.3 | 44.4 | Draft | 58 | NIGG00000000.1 |
| RI-048 | Dairy products | 2 940 | 3 185.1 | 44.5 | Draft | 94 | MJHG00000000.1 |
| RI-029 | Dairy products | 2 884 | 3 123.8 | 44.6 | Draft | 113 | MJHE00000000.1 |
| JMCC0013 | Dairy products | 3 028 | 3 292.4 | 44.3 | Draft | 83 | PEKI00000000.1 |
| WLPL04 | Dairy products | 2 881 | 3 141.6 | 44.7 | Complete genome | 1 | CP023771.1 |
| LL441 | Dairy products | 2 773 | 3 124.6 | 44.5 | Draft | 170 | LWKN00000000.1 |
126株菌按分离源划分为6组,其中39株为植物分离株,9株为果蝇分离株,22株为哺乳动物肠道分离株,9株为哺乳动物口腔等部位分离株,18株为肉制品分离株,29株为乳制品分离株。
1.2 主要试剂和仪器基因组DNA提取试剂盒,北京天根生化科技有限公司;MRS培养基,Oxoid公司。电热恒温培养箱,北京一恒科技有限公司;超微量紫外分光光度计,NanoDrop公司。
1.3 菌株培养与基因组DNA的提取将冻干菌粉取适量接种于5 mL MRS液体培养基中,在37 ℃下静置培养24 h,按2%接种量接种于5 mL液体MRS培养基中,继代培养3代,3 000 r/min离心10 min收集菌体。采用基因组DNA提取试剂盒提取基因组DNA,操作步骤按照试剂盒说明书进行,使用超微量紫外分光光度计检验DNA样品的纯度和质量。
1.4 基因组测序和组装基因组DNA提取后进行质量鉴定,在浓度和纯度达到测序要求后由上海美吉生物医药科技有限公司进行全基因组测序。采用Illumina HiSeq 4000测序平台完成全基因组测序,参照文献[13]构建文库,每株菌分别构建PE150的文库。将获取的原始数据进行质量评估,经过低质量Reads的过滤、去除接头和引物后,获得高质量的Reads序列,然后运用软件SOAPdenovo V1.06[14]对高质量的Reads进行拼接,采用GapCloser[15]软件对内部的Gap进行填充和单个碱基的矫正,最终完成基因组的组装。
1.5 比较基因组学分析 1.5.1 基因组单拷贝基因串联分析及基因差异可视化利用Anvi’o 5.2.0软件[16]中anvi-get-sequences- for-hmm-hits模块提取基因组单拷贝基因,利用anvi-gen-phylogenomic-tree模块基于Maximum Likelihood模型构建单拷贝基因串联系统发育树。利用Anvi’o软件中anvi-pan-genome模块构建菌株泛基因集存在与缺失,并绘制可视化图。
1.5.2 构建核心基因集和附属基因集分析基于Prokka软件[17]对菌株基因组进行基因预测后,采用Roary软件[18]识别核心基因集和附属基因集(Accessory gene),其中以编码蛋白氨基酸相似性大于80%的原则识别核心基因。使用MEGA 7.0软件[19]利用邻接(Neighbor-Joining)法构建系统进化树。使用R语言软件(V3.3.2)中的Prcomp函数进行主成分分析(Principal component analysis,PCA),比较不同环境菌株附属基因差异。
1.5.3 功能基因注释将126株菌株蛋白序列与COG (Clusters of orthologous groups,直系同源基因簇数据库)进行BLASTp (E-value < 1e–10,Identity值≥40%)比对,完成蛋白序列功能注释。同时应用Server-RAST Annotation Server (http://rast.nmpdr.org/)工具对126株菌的基因组进行功能注释。
1.5.4 环境特异性基因分析基于RAST注释结果计算环境特异性基因拷贝数。如果一个基因在某个环境出现频次显著高于其在所有环境分离株出现频次,那么该基因具有环境特异性,称为环境特异性基因。为了确定结果是否具有统计学意义,采用Pearson’s chi-squared检验(SPSS 17.0软件),将检验结果P < 0.05的基因认定为环境特异性基因[20]。
1.5.5 数据统计分析用t检验比较两两菌株之间的基因组大小和编码基因数量差异。采用Mann-Whitney检验不同分组间功能基因差异。
1.6 菌株核酸登录号植物乳杆菌IMAU20970和IMAU80873基因组序列已提交至GenBank数据库,登录号分别为RCDK00000000和RCDJ000000000。
2 结果与讨论 2.1 基因组基本特征分析测定的菌株IMAU20970组装后得到251个Scaffolds,基因组长度为3 341.7 kb,编码3 184个基因,(G+C)mol%含量为44.7%;IMAU80873组装后得到157个Scaffolds,基因组长度为3 193.8 kb,编码3 072个基因,(G+C)mol%含量为44.5%。两株菌的基因组大小和编码基因数量与其他乳制品分离株基本一致。
126株植物乳杆菌的平均基因组大小为3 284.3±125.6 kb,平均编码基因数量为3 017.8±148.3个,平均(G+C)mol%为44.5%±0.2%。通过比较不同分离源基因组大小和编码基因数量发现(图 1),果蝇分离株的基因组大小显著高于植物、哺乳动物肠道、肉制品和乳制品分离株(P < 0.05),其编码基因数量显著高于其他环境分离株(P < 0.05)。乳制品分离株的基因组大小和编码基因数量显著低于肉制品和果蝇分离株(P < 0.05)。植物、哺乳动物口腔等部位、哺乳动物肠道和肉制品分离株间基因组大小和编码基因数量均无显著差异(P > 0.05)。

|
| 图 1 植物乳杆菌基因组大小(A)和编码基因数量(B) Figure 1 Genomic size (A) and number of predicted genes (B) of L. plantarum stains isolated from different environments 注:小写字母完全不同的,表示分组间有差异显著(P < 0.05),有相同小写字母的,表示分组间无差异显著(P > 0.05). Note: The lowercase letters were completely different indicates significant differences (P < 0.05) among groups, the same lowercase letters indicated that there was no significant difference (P > 0.05) among groups. |
|
|
植物乳杆菌生长环境的高度多样性被认为与其拥有较大基因组有关[21]。较大的基因组可能编码更多参与代谢和胁迫耐受能力的基因,使其具有较强的环境适应能力[22-23]。果蝇作为昆虫类动物,其饮食结构和所在的生存环境可能与其他分离源不同,果蝇分离株具有较大的基因组和较多的编码基因可能是为了适应果蝇这种独特的宿主环境而发生了适应性进化。
2.2 基因组单拷贝基因串联和核心基因系统发育比较分析基于138个单拷贝基因串联系统发育树结果如图 2所示,系统发育树共分为三大分支,分别为A、B和C分支。分支A中包括18株乳制品分离株(占乳制品分离株的62.1%),本实验测定的2株乳制品分离株均分布在该分支中,16株植物分离株(占植物分离株的41%),1株果蝇分离株,其余菌株分离自肉制品、哺乳动物肠道和哺乳动物口腔等部位;分支B包括6株果蝇分离株(占果蝇分离株的66.7%),17株植物分离株(占植物分离株的43.6%),11株肉制品分离株(占肉制品分离株的61.1%),4株乳制品分离株,其余菌株分离自哺乳动物肠道和哺乳动物口腔等部位;分支C包括7株乳制品分离株,2株果蝇分离株,其余不同分离源的菌株混合分布在该分支中。

|
| 图 2 单拷贝基因串联系统发育树 Figure 2 Phylogenetic tree based on common single copy genes tandem |
|
|
基于1 076个核心基因构建的系统发育树主要分为5个分支(图 3),分别为B1、B2、B3、B4和B5分支。B1主要包括13个乳源菌株(占乳制品分离株的44.8%),1株果蝇分离株,其余分离源均匀分布在该分支;B2分支包括6株植物源分离株,其余菌株分离自肉制品、哺乳肠道和哺乳动物口腔等部位;B3和B4分支中不同分离源的菌株呈均匀分布;B5分支包括5株果蝇分离株(占果蝇分离株的55.6%),其余分离源菌株均匀分布在该分支。核心基因系统发育树与基因组单拷贝基因串联系统发育树具有一定相似性,两个系统发育树中乳制品和果蝇分离株均呈现分离源聚集性,而植物、肉制品、哺乳肠道和哺乳动物口腔等部位分离株未呈现出明显分离源聚集性。核心基因几乎包括所有个体均有的功能特性,其数目较单拷贝基因数目多,这可能会导致个别菌株与单拷贝基因串联系统发育树存在一定差异[24-25]。物种在不同环境中的进化历程不同,细菌基因组对特定环境的适应和进化会积累与该环境特征相关的遗传变异信息[26-27]。果蝇和乳制品环境与其他分离源的环境相比可能较为独特,导致菌株间的遗传进化存在明显区别。
|
| 图 3 核心基因系统发育树 Figure 3 Phylogenetic tree constructed based on the core genes of L. plantarum strains |
|
|
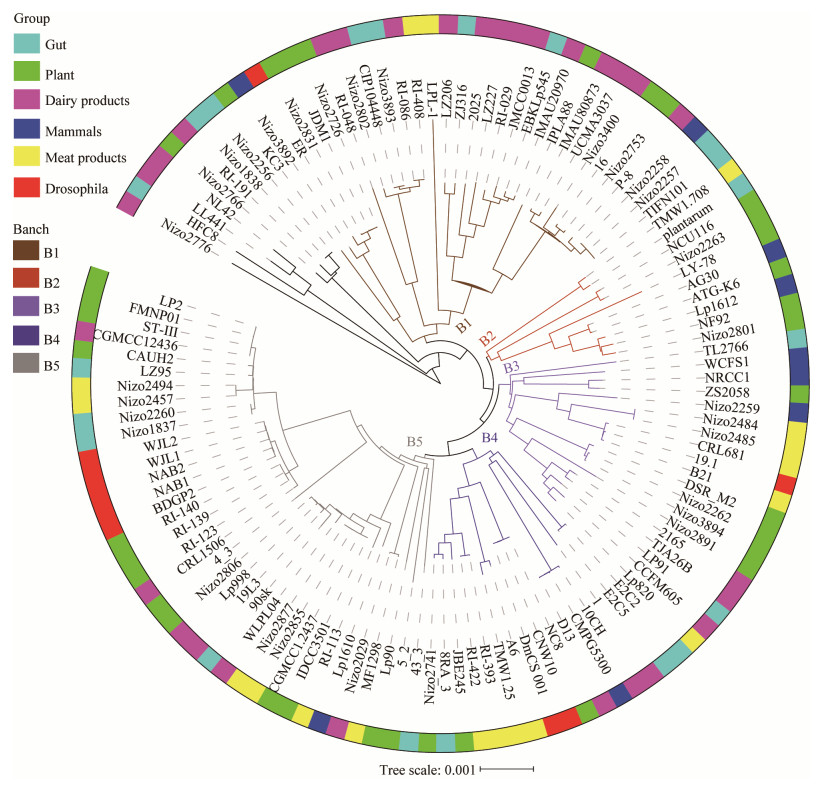
利用Anvi’o软件对126株菌泛基因的存在与缺失进行了差异可视化,结果如图 4所示,不同环境分离源菌株未呈现出明显规律,这与之前Martino等[9]研究结果一致。
|
| 图 4 植物乳杆菌泛基因集存在与缺失分析 Figure 4 Analysis of L. plantarum strains based on pan-genome presence-absence 注:每一圈代表一株菌株基因组的基因分布,黑色代表该基因存在,灰色代表该基因缺失. Note: Each ring represents a L. plantarum strain and each layer displays the gene distribution, black color stands for gene presence, whereas grey color represents gene absence. |
|
|
细菌对环境的适应过程可能存在基因的获得或缺失,研究发现细菌的附属基因的分布模式在一定程度上反映了菌株基因组的特异性[28]。本研究将泛基因集中核心基因和稀有基因(存在频率≤5%)剔除,进行了基于4 058个附属基因存在与缺失分布结果的主成分分析,结果如图 5所示。果蝇分离株主要集中在图片右下角,乳制品分离株主要集中在图片右上角,同时两者各自呈现明显聚集趋势。而肉制品、植物、哺乳动物肠道和口腔等部位分离株分布较为离散且相互重叠,没有明显聚集趋势。这一结果和泛基因集存在与缺失结果存在差异,这可能是由于泛基因集数目较多(13 471个基因),导致规律性较弱。但该结果与系统发育分析结果一致,表明果蝇和乳制品分离株可能与其他分离源菌株存在差异。菌株对环境的适应可能会获得有利于其在该环境下生存的环境特异性基因。果蝇和乳制品分离株的附属基因可能为了适应这种独特环境不断进化,导致它们具有一定分离源特异性[29-30]。

|
| 图 5 菌株附属基因聚类分析 Figure 5 PCA and clustering of accessory genes 注:基于菌株附属基因存在与否的“1/0”矩阵做主成分分析聚类图. Note: PCA and clustering of L. plantarum by using accessory gene presence-absence matrix ("1/0" matrix). |
|
|
为了解不同分离源菌株在功能基因上的差异,基于COG数据库对126菌株基因组进行了注释。通过COG数据库注释结果发现(图 6),果蝇分离株参与碳水化合物(G)、信号转导机制(T)、转录(K)和氨基酸转运和代谢(E)的功能基因显著高于其他分离源(P < 0.05)。而乳制品分离株复制、重组和修复(L)及细胞周期调控、细胞分裂、染色体分配(D)的基因显著高于其他分离源(P < 0.05)。其他分离源之间功能基因数量无显著差异(P > 0.05)。

|
| 图 6 不同分离源植物乳杆菌COG数据库注释结果 Figure 6 Annotation of L. plantarum strains based on COG database 注:红色表示每一行的基因比平均值多,而蓝色代表比平均值少. *:P < 0.05,**:P < 0.01. COG功能类别:A:RNA加工和修饰RNA;B:染色质结构和动力学;C:能量生产和转换;D:细胞周期调控,细胞分裂,染色体分配;E:氨基酸转运和代谢;F:核苷酸转运和代谢;G:碳水化合物转运和代谢;H:辅酶转运和代谢;I:脂质转运和代谢;J:翻译,核糖体结构和生物合成;K:转录;L:复制,重组和修复;M:细胞壁/细胞膜/胞外被膜生物合成;N:细胞运动;O:翻译后修饰,蛋白质转换,伴侣;P:机离子转运和代谢;Q:次级代谢产物的生物合成,转运和代谢;R:一般功能预测;S:功能未知;T:信号转导机制;U:胞内运输,分泌和囊泡运输;V:防御机制;W:胞外结构;Y:细胞核结构;Z:细胞骨架. Note: Red indicates groups with more genes than average values in each row, whereas blue indicates those with less. Significance was denoted by * and ** for P < 0.05 and P < 0.01. COG function category: A: Processing and modification; B: Chromatin structure and dynamics; C: Energy production and conversion; D: Cell cycle control, cell division, chromosome partitioning; E: Amino acid transport and metabolism; F: Nucleotide transport and metabolism; G: Carbohydrate transport and metabolism; H: Coenzyme transport and metabolism; I: Lipid transport and metabolism; J: Translation, ribosomal structure and biogenesis; K: Transcription; L: Replication, recombination and repair; M: Cell wall/ membrane/envelope biogensi; N: Cell motility; O: Posttranslational modification, protein turnover, chaperones; P: Inorganic ion transport and metabolism; Q: Secondary netabolites biosynthesis, transport and catabolism; R: General function prediction only; S: Function unknown; T: Signal transduction mechanisms; U: Intracellular trafficking, secretion, and vesicular transport; V: Defense mechanisms; W: Extracellular structures; Y: Nuclear structure; Z: Cytoskeleton. |
|
|
126株植物乳杆菌中参与碳水化合物代谢的编码基因数量最多(15.85%)。碳水化合物是生物体生长发育的主要能量来源,研究表明,乳酸菌通过获得与环境相关的碳水化合物或丢失非必要碳水化合物基因特异性适应环境[30-31]。果蝇分离株具有较多的碳水化合物基因,这可能是由于其营养环境较为独特,导致菌株参与碳水化合物的基因发生了适应性进化[32]。乳制品分离株具有较多参与细胞周期调控的功能基因,有学者发现细菌参与细胞周期调控的相关基因与其适应环境变化密切相关[33],乳制品环境中可能由于某些特殊的环境压力导致菌株的功能基因较为活跃。
2.4.2 不同分离源菌株基于RAST数据库功能基因分析通过RAST数据库注释结果发现(图 7),果蝇分离株参与碳水化合物(Carbohydrates)和氨基酸及其衍生物(Amino acids and derivatives)代谢的基因显著高于其他分离源(P < 0.000 1和P < 0.05)。乳制品分离株膜转运(Membrane transport)和硫代谢(Sulfur metabolism)显著高于其他分离源(P < 0.05)。肉制品分离株参与脂肪酸、脂肪和类异戊二烯(Fatty acids,lipids and isoprenoids)显著高于其他分离源(P < 0.05)。其他分离源之间功能基因数量无显著差异(P > 0.05)。果蝇分离株参与碳水化合物代谢和氨基酸及其衍生物的基因最多,这一结果与COG注释结果一致。乳制品分离株拥有较多的硫代谢基因,可能与其丰富的营养环境有关[34]。

|
| 图 7 不同分离源植物乳杆菌RAST数据库注释结果 Figure 7 Annotation of L. plantarum strains based on RAST database 注:红色表示每一行的基因比平均值多,而蓝色代表比平均值少. *:P < 0.05,**:P < 0.01,***:P < 0.001. Note: Red indicates groups with more genes than average values in each row, whereas blue indicates those with less. Significance was denoted by *, ** and *** for P < 0.05, P < 0.01 and P < 0.001. |
|
|
为了探究不同分离源菌株是否存在环境特异性基因,基于RAST注释结果对环境特异性基因进行了计算。126株菌中共发现50个环境特异性基因,果蝇分离株的环境特异性基因最多,共34个;乳制品分离株的环境特异性基因有7个,其余分离源菌株环境特异性基因较少,具体信息见表 2。
| 分离源 Habitat |
环境特异性基因 Number of habitat-specific genes |
主要基因功能 Major gene function |
| Plant | 4 | N-acetylneuraminate lyase (EC 4.1.3.3) Predicted sialic acid transporter Alpha-L-Rha alpha-1, 3-L-rhamnosyltransferase Alpha-L-Rha alpha-1, 3-L-rhamnosyltransferase (EC 2.4.1.-) |
| Drosophila | 34 | Fructose-1, 6-bisphosphatase (EC 3.1.3.11) Chitinase (EC 3.2.1.14) Multiple sugar ABC transporter, ATP-binding protein Beta-glucosidase (EC 3.2.1.21) Multiple sugar ABC transporter, substrate-binding protein Glucan 1, 6-alpha-glucosidase (EC 3.2.1.70) Transcriptional regulator of rhamnose utilization, AraC family |
| Mammals | 2 | Enoyl-[acyl-carrier-protein] reductase [NADPH] (EC 1.3.1.10) Prephenate dehydratase (EC 4.2.1.51) |
| Gut | 3 | Quinolinate phosphoribosyltransferase (EC 2.4.2.19) D-Lactate dehydrogenase (EC 1.1.2.5) PTS system, mannitol-specific IIA component (EC 2.7.1.69) |
| Meat products | 1 | Glycolate dehydrogenase (EC 1.1.99.14) |
| Dairy products | 7 | Programmed cell death toxin YdcE, Programmed cell death toxin MazF, Programmed cell death antitoxin MazE Para-aminobenzoate synthase, amidotransferase component (EC 2.6.1.85) CRISPR-associated protein Cas2 |
果蝇分离株的22个环境特异性基因参与碳水化合物代谢,主要参与低聚果糖、果糖、L-鼠李糖和几丁质代谢。研究发现植物乳杆菌基因组中存在参与糖代谢的“生活适应岛”,该区域可能通过频繁获得或丢失与生境相关的功能基因,从而适应生存环境[35-36]。果蝇主要以腐烂的水果、植物体和酵母菌为食,而水果和植物中富含低聚果糖,几丁质是酵母菌细胞壁的主要成分[37-38],植物乳杆菌可能为了适应这种营养环境获得了相应的碳水化合物基因[39]。乳制品分离株2个环境特异性基因mazE和mazF与mazEF毒素-抗毒素系统有关,1个环境特异性基因与CRISPR系统有关。这些基因均负责防御外来DNA的入侵,如噬菌体[40-41]。乳制品中营养成分丰富,常常受到噬菌体感染,在这过程中,细菌逐渐进化了多种机制用以抵抗噬菌体入侵[42]。有研究报道乳酸菌可通过获得具有噬菌体抗性的外源DNA片段帮助其在牛奶中的生长和竞争,而植物乳杆菌该功能基因的存在可能更有利于其在乳制品环境中生存[43-44]。
植物源分离株的4个环境特异性基因参与唾液酸代谢和鼠李糖代谢。肉制品分离株1个环境特异性基因参与乙醇酸盐代谢。哺乳动物口腔等部位分离株2个环境特异性基因分别参与脂肪酸和氨基酸代谢。哺乳动物肠道分离株的3个环境特异性基因分别参与NAD和NADP辅助因子生物合成、乳糖和甘露醇代谢。然而,这些不同分离源菌株中的环境特异性基因与环境潜在的联系尚不清楚,还需进一步深入研究。
3 结论本研究对来自不同分离源的126株植物乳杆菌的全基因组序列进行了比较基因组学分析。结果发现,果蝇分离株基因组大小和基因数量显著高于植物、肉制品、乳制品、哺乳动物肠道和其他部位分离源。基于单拷贝基因串联和核心基因系统发育树分析均发现,果蝇和乳制品分离株分别集中分布在某一分支中,其余分离源均匀分布在系统发育树各个分支中。附属基因分析结果与系统发育树分析结果一致。功能基因注释结果发现,果蝇分离株的环境特异性基因参与低聚果糖和几丁质代谢,乳制品分离株的环境特异性基因参与mazEF毒素-抗毒素系统和CRISPR系统。本研究发现果蝇和乳制品分离株存在环境特异性,而其他分离源菌株没有呈现明显的环境特异性。果蝇和乳制品的环境与其他分离源相比可能较为独特,果蝇和乳制品分离株可能为了适应自身的生长环境发生了适应性进化。本研究结果为了解植物乳杆菌的进化历程提供了参考。
| [1] |
Siezen RJ, van Hylckama Vlieg JET. Genomic diversity and versatility of Lactobacillus plantarum, a natural metabolic engineer[J]. Microbial Cell Factories, 2011, 10(Suppl 1): S3. DOI:10.1186/1475-2859-10-S1-S3 |
| [2] |
Zeng D, Tang YR, Ni XQ, et al. Study on the effects of Lactobacillus plantarum F22 on liver and intestinal microflora[J]. Acta Nutrimenta Sinica, 2010, 32(4): 370-374. (in Chinese) 曾东, 唐雨蕊, 倪学勤, 等. 植物乳杆菌F22对肝脏和肠道微生物菌群的影响研究[J]. 营养学报, 2010, 32(4): 370-374. |
| [3] |
Connelly P. Lactobacillus plantarum-a literature review of therapeutic benefits[J]. Journal of the Australian Traditional-Medicine Society, 2008, 14(2): 79-82. |
| [4] |
Bao Y, Wang ZL, Zhang Y, et al. Effect of Lactobacillus plantarum P-8 on lipid metabolism in hyperlipidemic rat model[J]. European Journal of Lipid Science and Technology, 2012, 114(11): 1230-1236. DOI:10.1002/ejlt.201100393 |
| [5] |
Gao PF, Ma C, Sun Z, et al. Feed-additive probiotics accelerate yet antibiotics delay intestinal microbiota maturation in broiler chicken[J]. Microbiome, 2017, 5: 91. DOI:10.1186/s40168-017-0315-1 |
| [6] |
Sun ZH. Application of population genomics in the research of lactic acid bacteria[J]. Journal of Chinese Institute of Food Science and Technology, 2017, 17(8): 12-18. (in Chinese) 孙志宏. 群体基因组学在乳酸菌研究中的应用[J]. 中国食品学报, 2017, 17(8): 12-18. |
| [7] |
Broadbent JR, Neeno-Eckwall EC, Stahl B, et al. Analysis of the Lactobacillus casei supragenome and its influence in species evolution and lifestyle adaptation[J]. BMC Genomics, 2012, 13: 533. DOI:10.1186/1471-2164-13-533 |
| [8] |
O'Sullivan O, O'Callaghan J, Sangrador-Vegas A, et al. Comparative genomics of lactic acid bacteria reveals a niche-specific gene set[J]. BMC Microbiology, 2009, 9: 50. DOI:10.1186/1471-2180-9-50 |
| [9] |
Martino ME, Bayjanov JR, Caffrey BE, et al. Nomadic lifestyle of Lactobacillus plantarum revealed by comparative genomics of 54 strains isolated from different habitats[J]. Environmental Microbiology, 2016, 18(12): 4974-4989. DOI:10.1111/1462-2920.13455 |
| [10] |
Kim EB, Marco ML. Nonclinical and clinical Enterococcus faecium strains, but not Enterococcus faecalis strains, have distinct structural and functional genomic features[J]. Applied and Environmental Microbiology, 2014, 80(1): 154-165. DOI:10.1128/AEM.03108-13 |
| [11] |
Chavda KD, Chen L, Fouts DE, et al. Comprehensive genome analysis of carbapenemase-producing Enterobacter spp.: new insights into phylogeny, population structure, and resistance mechanisms[J]. mBio, 2016, 7(6): e02093-16. |
| [12] |
He QW, Hou QC, Wang YJ, et al. Comparative genomic analysis of Enterococcus faecalis: insights into their environmental adaptations[J]. BMC Genomics, 2018, 19(1): 527. DOI:10.1186/s12864-018-4887-3 |
| [13] |
Sinha R, Stanley G, Gulati GS, et al. Index switching causes "spreading-of-signal" among multiplexed samples in illumina HiSeq 4000 DNA sequencing[J]. BioRxiv, 2017. DOI:10.1101/125724 |
| [14] |
Luo RB, Liu BH, Xie YL, et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler[J]. GigaScience, 2012, 1(1): 18. DOI:10.1186/2047-217X-1-18 |
| [15] |
Patel S, Lu ZX, Jin XZ, et al. Comparison of three assembly strategies for a heterozygous seedless grapevine genome assembly[J]. BMC Genomics, 2018, 19: 57. DOI:10.1186/s12864-018-4434-2 |
| [16] |
Eren AM, Esen ÖC, Quince C, et al. Anvi'o: an advanced analysis and visualization platform for 'omics data[J]. PeerJ, 2015, 3: e1319. DOI:10.7717/peerj.1319 |
| [17] |
Seemann T. Prokka: rapid prokaryotic genome annotation[J]. Bioinformatics, 2014, 30(14): 2068-2069. DOI:10.1093/bioinformatics/btu153 |
| [18] |
Page AJ, Cummins CA, Hunt M, et al. Roary: rapid large-scale prokaryote pan genome analysis[J]. Bioinformatics, 2015, 31(22): 3691-3693. DOI:10.1093/bioinformatics/btv421 |
| [19] |
Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets[J]. Molecular Biology and Evolution, 2016, 33(7): 1870-1874. DOI:10.1093/molbev/msw054 |
| [20] |
Wang YJ, Hou QC, Li WC, et al. Comparison of the virulence genes of Enterococcus faecalis isolated from different environments[J]. Microbiology China, 2018, 45(12): 2695-2707. (in Chinese) 王彦杰, 侯强川, 李伟程, 等. 不同分离源粪肠球菌的毒力基因比较[J]. 微生物学通报, 2018, 45(12): 2695-2707. |
| [21] |
Liu CJ, Wang R, Gong FM, et al. Complete genome sequences and comparative genome analysis of Lactobacillus plantarum strain 5-2 isolated from fermented soybean[J]. Genomics, 2015, 106(6): 404-411. DOI:10.1016/j.ygeno.2015.07.007 |
| [22] |
Dini-Andreote F, Andreote FD, Araújo WL, et al. Bacterial genomes: habitat specificity and uncharted organisms[J]. Microbial Ecology, 2012, 64(1): 1-7. |
| [23] |
Darmon E, Leach DRF. Bacterial genome instability[J]. Microbiology and Molecular Biology Reviews, 2014, 78(1): 1-39. DOI:10.1128/MMBR.00035-13 |
| [24] |
Gabaldón T, Dessimoz C, Huxley-Jones J, et al. Joining forces in the quest for orthologs[J]. Genome Biology, 2009, 10(9): 403. DOI:10.1186/gb-2009-10-9-403 |
| [25] |
Zhou DS, Yang RF. Comparative and evolutionary genomics in bacterial systems[J]. Journal of Microbiology, 2003, 23(5): 31-34, 43. (in Chinese) 周冬生, 杨瑞馥. 细菌比较基因组学和进化基因组学[J]. 微生物学杂志, 2003, 23(5): 31-34, 43. DOI:10.3969/j.issn.1005-7021.2003.05.010 |
| [26] |
Papizadeh M, Rohani M, Nahrevanian H, et al. Probiotic characters of Bifidobacterium and Lactobacillus are a result of the ongoing gene acquisition and genome minimization evolutionary trends[J]. Microbial Pathogenesis, 2017, 111: 118-131. DOI:10.1016/j.micpath.2017.08.021 |
| [27] |
Cremonesi P, Chessa S, Castiglioni B. Genome sequence and analysis of Lactobacillus helveticus[J]. Frontiers in Microbiology, 2013, 3: 435. |
| [28] |
Bakshi U, Sarkar M, Paul S, et al. Assessment of virulence potential of uncharacterized Enterococcus faecalis strains using pan genomic approach-identification of pathogen-specific and habitat-specific genes[J]. Scientific Reports, 2016, 6: 38648. DOI:10.1038/srep38648 |
| [29] |
Song YQ, He QW, Zhang JC, et al. Genomic variations in probiotic Lactobacillus plantarum P-8 in the human and rat gut[J]. Frontiers in Microbiology, 2018, 9: 893. DOI:10.3389/fmicb.2018.00893 |
| [30] |
El Kafsi H, Binesse J, Loux V, et al. Lactobacillus delbrueckii ssp. lactis and ssp. bulgaricus: a chronicle of evolution in action[J]. BMC Genomics, 2014, 15(1): 407. DOI:10.1186/1471-2164-15-407 |
| [31] |
Smokvina T, Wels M, Polka J, et al. Lactobacillus paracasei comparative genomics: towards species pan-genome definition and exploitation of diversity[J]. PLoS One, 2013, 8(7): e68731. DOI:10.1371/journal.pone.0068731 |
| [32] |
Goh YJ, Goin C, O'Flaherty S, et al. Specialized adaptation of a lactic acid bacterium to the milk environment: the comparative genomics of Streptococcus thermophilus LMD-9[J]. Microbial Cell Factories, 2011, 10(Suppl 1): S22. DOI:10.1186/1475-2859-10-S1-S22 |
| [33] |
Chen T. Research on construction algorithm of gene regulation network[D]. Shanghai: Master's Thesis of Fudan University, 2009 (in Chinese) 陈婷.基因调控网络的构建算法研究[D].上海: 复旦大学硕士学位论文, 2009 http://cdmd.cnki.com.cn/Article/CDMD-10246-2009183874.htm |
| [34] |
Weimer B, Seefeldt K, Dias B. Sulfur metabolism in bacteria associated with cheese[J]. Antonie Van Leeuwenhoek, 1999, 76(1-4): 247-261. |
| [35] |
Cai H, Thompson R, Budinich MF, et al. Genome sequence and comparative genome analysis of Lactobacillus casei: insights into their niche-associated evolution[J]. Genome Biology and Evolution, 2009, 1: 239-257. DOI:10.1093/gbe/evp019 |
| [36] |
Kleerebezem M, Boekhorst J, van Kranenburg R, et al. Complete genome sequence of Lactobacillus plantarum WCFS1[J]. Proceedings of the National Academy of Sciences of the United States of America, 2003, 100(4): 1990-1995. DOI:10.1073/pnas.0337704100 |
| [37] |
Jovanovic-Malinovska R, Kuzmanova S, Winkelhausen E. Oligosaccharide profile in fruits and vegetables as sources of prebiotics and functional foods[J]. International Journal of Food Properties, 2014, 17(5): 949-965. DOI:10.1080/10942912.2012.680221 |
| [38] |
Leisner JJ, Vogensen FK, Kollmann J, et al. α-Chitinase activity among lactic acid bacteria[J]. Systematic and Applied Microbiology, 2008, 31(2): 151-156. DOI:10.1016/j.syapm.2008.03.003 |
| [39] |
Elena Martino M, Joncour P, Leenay R, et al. Bacterial adaptation to the host's diet is a key evolutionary force shaping Drosophila-Lactobacillus symbiosis[J]. Cell Host & Microbe, 2018, 24(1): 109-119. |
| [40] |
Hazan R, Engelberg-Kulka H. Escherichia coli mazEF-mediated cell death as a defense mechanism that inhibits the spread of phage P1[J]. Molecular Genetics and Genomics, 2004, 272(2): 227-234. |
| [41] |
Barrangou R, Fremaux C, Deveau H, et al. CRISPR provides acquired resistance against viruses in prokaryotes[J]. Science, 2007, 315(5819): 1709-1712. DOI:10.1126/science.1138140 |
| [42] |
Zhao J. Population genetics and functional genomics of Streptococcus thermophilus isolated from natural fermented dairy products[D]. Hohhot: Doctoral Dissertation of Inner Mongolia Agricultural University, 2018 (in Chinese) 赵洁.自然发酵乳中嗜热链球菌群体遗传学和功能基因组学研究[D].呼和浩特: 内蒙古农业大学博士学位论文, 2018 http://cdmd.cnki.com.cn/Article/CDMD-10129-1018881781.htm |
| [43] |
Xi Y, Wang ZY, Zhang HP, et al. A review on the anti-phage mechanism of lactic acid bacteria[J]. Journal of Dairy Science and Technology, 2015, 38(6): 22-26. (in Chinese) 习羽, 汪政煜, 张和平, 等. 乳酸菌抗噬菌体相关机理的研究进展[J]. 乳业科学与技术, 2015, 38(6): 22-26. |
| [44] |
Zhang WY, Meng H, Zhang HP. Progress on the genomics of lactic acid bacteria — a review[J]. Acta Microbiologica Sinica, 2008, 48(9): 1270-1275. (in Chinese) 张文羿, 孟和, 张和平. 乳酸菌基因组学研究进展[J]. 微生物学报, 2008, 48(9): 1270-1275. DOI:10.3321/j.issn:0001-6209.2008.09.023 |
 2019, Vol. 46
2019, Vol. 46