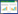扩展功能
文章信息
- 李阳, 赵丽婷, 顾正华, 李由然, 石贵阳, 丁重阳
- LI Yang, ZHAO Li-Ting, GU Zheng-Hua, LI You-Ran, SHI Gui-Yang, DING Zhong-Yang
- 灵芝多糖糖供体合成途径中关键酶的异源表达及其酶学性质
- Heterologous expression and characterization of the key enzymes involved in sugar donor synthesis of polysaccharide in Ganoderma lucidum
- 微生物学通报, 2019, 46(12): 3233-3247
- Microbiology China, 2019, 46(12): 3233-3247
- DOI: 10.13344/j.microbiol.china.190087
-
文章历史
- 收稿日期: 2019-01-28
- 接受日期: 2019-05-14
- 网络首发日期: 2019-05-31
2. 江南大学粮食发酵工艺与技术国家工程实验室 江苏 无锡 214122
2. National Engineering Laboratory for Cereal Fermentation Technology, Jiangnan University, Wuxi, Jiangsu 214122, China
灵芝多糖是一类结构复杂的天然大分子物质,也是灵芝发酵过程中的主要活性产物,具有抗肿瘤、降血糖血脂等功效。近年来,随着G. lucidum CGMCC 5.26菌株全基因组序列公布[1],通过发酵调控优化或菌种改良以促进多糖产量提高取得了一定的效果[2-3]。但由于灵芝多糖合成的代谢途径复杂且不清晰,其中涉及到的关键酶数量众多且特性不明,使得灵芝多糖产量在发酵水平的大幅提高仍存在障碍。因此需要全面清晰地了解灵芝多糖的生物合成过程,解析其中关键酶的性质与催化特点。
灵芝多糖的合成是一个极为复杂的生物过程,主要分为3个阶段,即糖供体(NDP-单糖)的合成、糖链的连接和多糖的胞外输出。目前灵芝中糖供体合成途径已基本清晰[4-6],且研究表明PGM、UGPG及PMI是其中的关键酶[7-9]。如图 1所示,PGM催化葡萄糖-6-磷酸生成葡萄糖-1-磷酸,进而在UGPG的作用下生成UDP-葡萄糖参与糖链的连接;PMI催化果糖-6-磷酸生成甘露糖-6-磷酸,再经过一系列转化最终也生成GDP-甘露糖参与糖链的连接。由于灵芝本体中提取这3种酶的工艺较为繁琐,而且提取率不高。因此,本研究基于灵芝全基因组序列,通过KEGG注释和BLAST注释分析获取目的基因片段,利用基因工程手段对PGM、UGPG和PMI进行异源表达并大量制备。首次对灵芝来源的三类酶的酶学性质展开探究,完善灵芝多糖糖供体合成途径关键酶酶学特性信息,不仅填补了灵芝中相关研究的空白,也为大幅促进灵芝多糖合成发酵策略的制定提供有效的依据。
|
| 图 1 灵芝多糖的糖供体合成途径 Figure 1 The biosynthetic pathway of sugar donors about polysaccharides in Ganoderma lucidum 注:PGM:磷酸葡萄糖变位酶;UGPG:UDP-葡萄糖焦磷酸化酶;PMI:磷酸甘露糖异构酶. Note: PGM: Phosphoglucomutase; UGPG: UDP-glucose pyrophosphorylase; PMI: Phosphomannose isomerase. |
|
|
大肠杆菌BL21(DE3)、大肠杆菌JM109和大肠杆菌质粒pET-28a(+)由中国高校工业微生物资源和信息中心(CICIM)保藏。实验涉及的菌株和质粒见表 1。
| 菌株和质粒 Strains and plasmids |
特性 Characteristics |
来源 Sources |
| Strains | ||
| DCQC-pgm | E. coli BL21(DE3), ∆pgm | This study |
| DCQC-ugpg | E. coli BL21(DE3), ∆ugpg | This study |
| DCQC-pmi | E. coli BL21(DE3), ∆pmi | This study |
| GLpgm-DCQC | DCQC-pgm harboring GLpgm-pET-28a(+) | This study |
| GLugpg-DCQC | DCQC-ugpg harboring GLugpg-pET-28a(+) | This study |
| GLpmi-DCQC | DCQC-pmi harboring GLpmi-pET-28a(+) | This study |
| Plasmids | ||
| pKD46 | AmpR, λ Red recombinase expression plasmid, ara-inducible expression, temperature sensitive replication | Laboratory stock |
| pKD13 | KanR, AmpR, oriR plasmid containing an Frt-Kan-Frt cassette | Laboratory stock |
| pCP20 | AmpR, CamR, repA (Ts), pSC101 based vector expressing the yeast Flp recombinase, temperature sensitive replication | Laboratory stock |
| GLpgm-pET-28a(+) | pET-28a(+) carrying pgm of GL | This study |
| GLugpg-pET-28a(+) | pET-28a(+) carrying ugpg of GL | This study |
| GLpmi-pET-28a(+) | pET-28a(+) carrying pmi of GL | This study |
| 注:GL:灵芝CGMCC 5.26菌株;Kan:卡那霉素;Amp:氨苄青霉素;Cam:氯霉素;R:抗性. Note: GL: G. lucidum strain CGMCC 5.26; Kan: Kanamycin; Amp: Ampicillin; Cam: Chloramphenicol; R: Resistance. |
||
研究中所用的引物及其序列见表 2。引物设计使用Clone manager 8.0软件。引物由金唯智生物科技有限公司(苏州)合成。
| 引物名称 Primers |
引物序列 Primers sequence (5′→3′) |
长度 Size (bp) |
| QCpgm-F | ATGGCAATCCACAATCGTGCAGGCCAACCTGCACAACAGAGTGATTTGATTAACGTCGCCATCCGTCGACCTGCAGTTC | 79 |
| QCpgm-R | TCAGAACTTCGCTAACAATCTCAACCGCTTCTTTCTCAATCTGCTTGCGATGTTCTTCAGTAGGCTGGAGCTGCTTCG | 78 |
| QCpmi-F | AAGCAGTTCACGAGTGCAGAATGCCGCCGGAGACATCGTTTCACTGCGTGATGTGATTGGATCCGTCGACCTGCAGTTC | 79 |
| QCpmi-R | TTACAGCTTGTTGTAAACACGCGCTAAACGGCCGTGGCCTTTGACAGTCACCGGTGATTCGTAGGCTGGAGCTGCTTCG | 79 |
| QCugpg-F | GCCGTTATCCCCGTTGCGGGATTAGGAACCAGGATGTTGCCGGCGACGAAAGCCATATCCGTCGACCTGCAGTTC | 75 |
| QCugpg-R | TTACTTCTTAATGCCCATCTCTTCTTCAAGCCAGGCTTTAAATTCCGTGCCAAGGTAGGCTGGAGCTGCTTCG | 73 |
| Vpgm-up | CTATCGTCGAAGCGAAACC | 19 |
| Vpgm-down | GTAGCGCATCAGGCAATTC | 19 |
| Vpmi-up | GCGCCAATAGAGTTAACGC | 19 |
| Vpmi-down | TACGCCTAGCGCAACAATG | 19 |
| Vugpg-up | CTGCTAATGTCGGCTGGTG | 19 |
| Vugpg-down | GATCGCTGACGGTATTGAC | 19 |
| K1 | AGGCTATTCGGCTATGACTG[10] | 20 |
| K2 | GGACAGGTCGGTCTTGACAA[10] | 20 |
| Kt | CGGCCACAGTCGATGAATCC[11] | 20 |
| pKD46-F | TTTGAGTTGTGGGTATCTGT[11] | 20 |
| pKD46-R | CTTGTATTATGGGTAGTTTCC[11] | 21 |
| GLpgm-F | GGAATTCCATATGTCGTACCAGGTCAAGGAG (NdeⅠ) | 31 |
| GLpgm-R | CGGAATTCCTACGTGATGACGGTCGGC (EcoRⅠ) | 27 |
| GLpmi-F | GGAATTCCATATGACGGCTGTCTTCAAGATC (NdeⅠ) | 31 |
| GLpmi-R | CGGAATTCTCATTTCACCTCGACAAATGC (EcoRⅠ) | 29 |
| GLugpg-F | GGAATTCCATATGCCCTCCGACTCCCTCATG (NdeⅠ) | 31 |
| GLugpg-R | CGGAATTCTTACAACTCAATGAGGTTC (EcoRⅠ) | 27 |
| 注:QCgene用于敲除基因的引物,下划线表示同源臂序列;Vgene用于验证的引物;GLgene用于扩增目的片段,下划线表示酶切位点序列. Note: Primers of “QCgene” are used for gene knockout, the underlined represents homologous arm sequences; Primers of “Vgene” are used for verifying inactivation of target genes; Primers of “GLgene” are used for amplification of the gene of interest, the underlined indicates restriction sites. |
||
LB培养基(g/L):酵母提取物5.0,胰蛋白胨10.0,氯化钠10.0,固体培养基另加琼脂粉20.0 g/L。抗性筛选时添加终浓度为100 mg/L的氨苄青霉素或30 mg/L的卡那霉素。
2×Taq PCR Master Mix,杭州宝赛生物科技有限公司;限制性内切酶EcoRⅠ、NdeⅠ及T4 DNA连接酶,Thermo Fisher Scientific公司;卡那霉素、氨苄青霉素与L-阿拉伯糖,以及酶活测定相关试剂,Sigma公司;PCR产物纯化试剂盒、质粒小量提取试剂盒和胶回收试剂盒,康宁生命科学(吴江)有限公司;2×Phanta Max Master Mix,南京诺唯赞生物科技有限公司;IPTG,生工生物工程(上海)股份有限公司。
核酸定量仪、电转仪,Eppendorf公司;酶标仪,TECAN公司。
1.2 方法 1.2.1 Red两步同源重组法敲除基因Datsenko等[12]提供的Red两步同源重组系统,借助质粒pKD46、pKD13和pCP20进行大肠杆菌相关基因的敲除,最终在敲除的目的基因位置留下100 bp左右的片段[13]。
根据NCBI提供的E. coli BL21(DE3)基因组信息,设计敲除盒片段引物QCpgm-F/QCpgm-R、QCugpg-F/QCugpg-R、QCpmi-F/QCpmi-R,以pKD13质粒为模板,分别用2×Taq酶扩增得到大肠杆菌中pgm、ugpg、pmi三个基因的敲除盒片段。扩增产物经琼脂糖凝胶电泳回收纯化,电转化入含有pKD46质粒的E. coli BL21(DE3)中,挑取转化子进行PCR验证。验证引物Vpgm-up/Vpgm-down、Vugpg-up/Vugpg-down、Vpmi-up/Vpmi-down的设计分别取自基因pgm、ugpg、pmi的上下游区域,同时利用K1/down、K2/up进行交叉验证[10],验证成功后,用引物V-up/V-down扩增目的条带,送金唯智生物科技有限公司(苏州)进行测序验证。pKD46-F/ pKD46-R用于验证质粒pKD46的导入和丢失[11]。
1.2.2 基因克隆与序列分析以G. lucidum CGMCC 5.26菌株的cDNA为模板,用引物GLpgm-F/GLpgm-R、GLugpg-F/GLugpg-R、GLpmi-F/GLpmi-R分别扩增目的基因gl-pgm (GenBank登录号为GL24280)、gl-ugpg (GenBank登录号为GL25739)、gl-pmi (GenBank登录号为GL22193)的序列。PCR反应条件:95 ℃ 3 min;95 ℃ 15 s,55 ℃ 15 s,72 ℃ 1 min,30个循环;72 ℃ 10 min,16 ℃保温。PCR反应体系(100 μL):模板(100 ng/μL) 1 μL,上、下游引物(10 μmol/L)各2 μL,2×Phanta Max Master Mix 50 μL,ddH2O 45 μL。将扩增片段两端加dATP后胶回收获得纯化产物。纯化产物与pMD18-T载体连接并转化至E. coli JM109。挑取的阳性转化子,经EcoRⅠ、NdeⅠ双酶切验证正确后,将相应的重组质粒分别命名为pMD18-GLpgm、pMD18-GLpmi和pMD18-GLugpg,并送往金唯智生物科技有限公司(苏州)测序。将3个酶基因的测序结果运用Expasy中的ProtParam预测酶的蛋白质分子量及等电点等。
1.2.3 重组载体构建将测序无误的3个目的基因片段分别与表达载体pET-28a(+)构建重组载体:GLpgm-pET-28a(+)、GLugpg-pET-28a(+)和GLpmi-pET-28a(+),并分别转化至相应的E. coli BL21(DE3)缺失菌株感受态细胞中,将阳性菌株分别命名为GLpgm-QCDC、GLugpg-QCDC、GLpmi-QCDC。同时将pET-28a(+)空载质粒导入3个缺失菌株感受态细胞中,重组菌株分别命名为28a-DCQCpgm、28a-DCQCugpg和28a-DCQCpmi。
1.2.4 酶的诱导表达与分离纯化接种重组菌株GLpgm-QCDC、GLugpg-QCDC和GLpmi-QCDC至含有30 mg/L卡那霉素的液体LB培养基中,以菌株28a-DCQCpgm、28a-DCQCugpg、28a-DCQCpmi为对照。37 ℃、220 r/min培养过夜后以1%的接种量接种至新鲜无菌的LB培养基中,在OD600达到0.6−0.8时,加入诱导剂IPTG使其终浓度为0.5 mmol/L,25 ℃、150 r/min培养10 h。12 000 r/min离心5 min收集菌体,用1×PBS溶液洗涤3次并重悬菌体,进行超声波破碎,破碎条件:300 W,超声1 s,间隔2 s,循环300次。破碎液在4 ℃、12 000 r/min离心20 min,收集的上清液即为粗酶液。
将粗酶液利用Co-NTA重力亲和层析柱进行纯化。结合缓冲液:1×PBS,0.5 mol/L NaCl,20 mmol/L咪唑,pH 7.4。洗脱缓冲液:1×PBS,0.5 mol/L NaCl,300 mmol/L咪唑,pH 7.4。收集酶液,利用SDS-PAGE检测蛋白纯度及分子量大小。用2 L的1×PBS缓冲液低温透析3次后获得纯化后酶液。
1.2.5 酶活力检测、蛋白测定和电泳分析酶活测定:在30 ℃下,将酶活测定体系(220 μL)预热10 min后,加入30 μL的酶液开始反应,340 nm下测定反应体系中每分钟增加或减少1 μmol的NAD(P)H所需要的酶量定义为1个酶活单位U。
PGM酶活测定体系[14]:50 mmol/L三乙醇胺缓冲液(pH 7.2),5 mmol/L MgCl2,50 μmol/L葡萄糖-1, 6-二磷酸,0.4 mmol/L NADP+,4 U葡萄糖-6-磷酸脱氢酶,1.4 mmol/L α-葡萄糖-1-磷酸。UGPG酶活测定体系[15]:50 mmol/L Tris-HCl缓冲液(pH 7.8),16 mmol/L MgCl2,0.4 mmol/L UDP-葡萄糖,0.4 mmol/L NADP+,4 U葡萄糖-6-磷酸脱氢酶,2.1 U α-磷酸葡萄糖变位酶,4 mmol/L焦磷酸。PMI酶活测定体系[16]:50 mmol/L MOPS缓冲液(pH 7.0),1 mmol/L CoCl2,10 mmol/L甘露糖-6-磷酸,1 mmol/L NADP+,4 U葡萄糖-6-磷酸脱氢酶,4 U磷酸葡萄糖异构酶。
蛋白质浓度测定采用Bradford法[17]。SDS-PAGE电泳参照Sch gger等的方法[18]。
1.2.6 纯化酶的酶性质研究(1) 最适反应pH及其稳定性的研究:在30 ℃下,分别测定不同pH (5.0−10.5)条件下的纯化酶活力,以最高酶活力为100%计算相对酶活力,确定最适反应pH。在30 ℃下,将酶液置于不同pH缓冲液中保温2 h,在最适pH下测定纯化酶活力,以最初酶活力为100%计算相对酶活力,考察纯化酶的pH稳定性。(2)最适反应温度及其稳定性的研究:在最适pH下,分别测定不同温度(20−70 ℃)下纯化酶活力,以最高酶活力为100%计算相对酶活力,确定最适反应温度。在最适pH下,将酶液置于不同温度中保温2 h,并在最适反应条件下测定纯化酶活力,以最初酶活力为100%计算相对酶活力,考察纯化酶的温度稳定性。(3)金属离子对纯化酶的影响:酶活反应体系中分别加入终浓度1 mmol/L金属离子(Na+、K+、Ca2+、Co2+、Fe2+、Zn2+、Al3+、Ni2+、Fe3+、Cu2+、Mn2+、Mg2+、Cd2+、Ag+),最适反应条件下测定纯化酶活力,以未处理情况下的酶活力为100%计算相对酶活力,考察金属离子对纯化酶的激活或抑制作用。(4)酶动力学常数的测定:配制不同浓度的反应底物,在最适反应条件下测定纯化酶活力,重复3次,利用双倒数法求得Vmax和Km值[19]。
2 结果与分析 2.1 靶标基因缺失菌株的构建为了避免原始大肠杆菌宿主菌株内相关酶的酶活对纯化酶活力的检测存在影响,构建了3株相关基因缺失的菌株DCQC-pgm、DCQC-ugpg和DCQC-pmi,并且通过摇瓶试验证实了靶标基因缺失菌株与原始菌株生长特性上没有明显差异。
以pKD13质粒为模板,扩增并胶回收得到敲除盒片段pgm、ugpg和pmi的纯化产物,大小均约为1 400 bp (图 2)。将敲除盒片段分别电转E. coli BL21(DE3)(pKD46)的感受态细胞中,转化子分别经Vpgm-up/Vpgm-down、Vugpg-up/Vugpg-down和Vpmi-up/Vpmi-down菌落PCR验证,分别得到1 900、1 952和1 912 bp的条带(图 2)。而以E. coli BL21(DE3)(pKD46)为对照,分别得到2 128、1 441和1 675 bp的片段(图 2);同时用K1/down、K2/up、K1/Kt三组引物进行交叉验证,其中K1/down分别得到长度为957、1 243、1 028 bp,K2/up分别得到1 050、811、994 bp的核酸条带,K1/Kt均得到581 bp的条带(图 2),该结果表明Frt-Kan-Frt敲除盒片段正确整合到靶基因位置。pKD46质粒丢失后用pKD46-F/pKD46-R引物进行菌落PCR,无产物表明质粒pKD46丢失成功(图 2)。再将质粒pCP20导入含Frt-Kan-Frt敲除盒片段的菌株中,转化子经Vpgm-up/Vpgm-down、Vugpg-up/Vugpg-down、Vpmi-up/Vpmi-down分别菌落PCR后,得到长度为490、640、600 bp的片段(图 2),结果表明相应菌株中的抗性基因被敲除,测序结果也证实了相应基因的缺失,即成功构建了基因缺失菌株DCQC-pgm、DCQC-ugpg和DCQC-pmi。

|
| 图 2 E. coli BL21(DE3)中pgm (A)、ugpg (B)、pmi (C)的基因敲除PCR鉴定电泳图谱 Figure 2 PCR identification of pgm (A)、ugpg (B)、pmi (C) gene knockout in E. coli BL21(DE3) 注:M:核酸标准分子质量;1:QCgene-F/QCgene-R扩增产物;2:Frt-Kan-Frt片段整合到靶基因位点,Vgene-up/Vgene-down验证;3:E. coli BL21(DE3)为对照;4,5,6:分别由引物K1/down、K2/up、K1/Kt验证;7:pKD46-F/pKD46-R验证;8:导入pCP20后抗性片段丢失,Vgene-up/Vgene-down验证. Note: M: DNA Marker; 1: PCR product of primer "QCgene-F/QCgene-R"; 2: The fragment Frt-Kan-Frt is integrated into the target gene locus, verification by primer "Vgene-up/Vgene-down"; 3: E. coli BL21(DE3) is used for control; 4, 5, 6: Verification by primers "K1/down", "K2/up", "K1/Kt", respectively; 7: Verification by primer "pKD46-F/pKD46-R"; 8: Resistance fragment is lost after introduction of pCP20, verification by primer "Vgene-up/Vgene-down". |
|
|
以灵芝的cDNA为模板进行PCR扩增,获得gl-pgm、gl-ugpg和gl-pmi片段大小分别约为1 701、1 515和1 284 bp (图 3)。经过ProtParam预测,gl-pgm编码566个氨基酸,分子量为61.4 kD,理论pI为6.21;gl-ugpg编码504个氨基酸,分子量为56.8 kD,理论pI为6.98;gl-pmi编码427个氨基酸,分子量为46.1 kD,理论pI为5.29。

|
| 图 3 片段gl-pgm、gl-ugpg、gl-pmi的琼脂糖凝胶电泳分析 Figure 3 Agarose gel electrophoresis of the cloned gl-pgm, gl-ugpg and gl-pmi gene fragments 注:M:核酸标准分子质量;1:gl-pgm扩增产物;2:gl-ugpg扩增产物;3:gl-pmi扩增产物. Note: M: DNA Marker; 1: PCR product of gl-pgm; 2: PCR product of gl-ugpg; 3: PCR product of gl-pmi. |
|
|
将测序无误的片段分别从pMD18-GLpgm、pMD18-GLugpg、pMD18-GLpmi上酶切回收得到纯化片段,分别与线性化大肠杆菌的线性化载体pET-28a(+)连接,获得的重组质粒用EcoRⅠ、NdeⅠ双酶切得到5 369 bp的pET-28a(+)载体片段和相应大小的目的片段(图 4)。

|
| 图 4 pET-28a(+) (A)和重组载体(B)经EcoRⅠ、NdeⅠ双酶切后琼脂糖凝胶电泳分析 Figure 4 Agarose gel electrophoresis of pET-28a(+) and recombinant plasmids were digested by EcoRⅠ and NdeⅠ 注:M:核酸标准分子质量;1:pET-28a(+)质粒;2:pET-28a(+)-GLpgm重组质粒;3:pET-28a(+)-GLugpg重组质粒;4:pET-28a(+)-GLpmi重组质粒. Note: M: DNA Marker; 1: pET-28a(+); 2: Recombinant plasmids pET-28a(+)-GLpgm; 3: Recombinant plasmids pET-28a(+)-GLugpg; 4: Recombinant plasmids pET-28a(+)-GLpmi. |
|
|
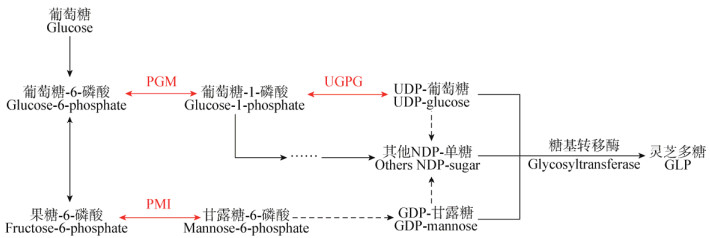
以基因缺失菌株DCQC-pgm、DCQC-ugpg和DCQC-pmi为对照,将重组菌株GLpgm-DCQC、GLpmi-DCQC和GLugpg-DCQC经IPTG诱导结束后,取破碎菌体上清测定目的酶活力并进行SDS-PAGE电泳(图 5)。发现含有目的基因重组载体的重组菌株出现了较空载体明显的蛋白条带,其分子量大小分别如图 5中的A2、B4、C6,与预测的理论分子量大小接近,说明目的酶均在相应靶标基因缺失的菌株中实现表达。
|
| 图 5 重组菌株表达产物SDS-PAGE电泳图 Figure 5 SDS-PAGE analysis of proteins expressed in recombinant strains 注:M:标准蛋白Marker;1、3、5:缺陷菌株DCQC-pgm、DCQC-ugpg、DCQC-pmi细胞破碎上清;2、4、6:重组菌株GLpgm-DCQC、GLugpg-DCQC、GLpmi-DCQC细胞破碎上清. Note: M: Standard protein marker; 1, 3, 5: The supernatant of the defecient strains DCQC-pgm, DCQC-ugpg and DCQC-pmi, respectively; 2, 4, 6: The supernatant of the recombinant strains GLpgm-DCQC, GLugpg-DCQC and GLpmi-DCQC, respectively. |
|
|
将3个目的酶的粗酶液经过Co-NTA纯化后均得到单一的条带(图 6),表明获得纯化的酶,可用于进一步的酶学性质研究。纯化酶在常规酶活测定条件下,GLpgm、GLugpg、GLpmi比酶活分别为4.75、6.26、13.68 U/mg。

|
| 图 6 纯化酶的SDS-PAGE电泳图 Figure 6 SDS-PAGE analysis of purified enzymes 注:M:标准蛋白Marker;1、3、5:重组菌株GLpgm-DCQC、GLugpg-DCQC、GLpmi-DCQC粗酶液;2、4、6:GLpgm、GLugpg、GLpmi纯化酶液. Note: M: Standard protein marker; 1, 3, 5: The crude mixtures containing the GLpgm, GLugpg and GLpmi, respectively; 2, 4, 6: The purified enzymes of GLpgm, GLugpg and GLpmi, respectively. |
|
|
将纯化后的GLpgm、GLugpg和GLpmi置于不同pH缓冲液中测定酶活力,得到pH-酶活力关系(图 7)。

|
| 图 7 pH对酶活力及其稳定性的影响 Figure 7 Effects of pH on activities and stabilities of purified enzymes 注:A、B:pH-GLpgm酶活力、稳定性;C、D:pH-GLugpg酶活力、稳定性;E、F:pH-GLpmi酶活力、稳定性. Note: A, B: pH-Enzyme of GLpgm activity and stability; C, D: pH-Enzyme of GLugpg activity and stability; E, F: pH-Enzyme of GLpmi activity and stability. |
|
|
GLpgm最适反应pH为8.5,当10.0 < pH < 6.5时,酶活力基本丧失。将GLpgm置于不同pH缓冲溶液中于30 ℃保温2 h,结果表明该酶在pH 6.5−8.0间稳定性较好,均能保持80%以上的酶活力;在pH高于9.0的条件下,GLpgm基本失活。
GLugpg的最适pH为7.5,当9.5 < pH < 6.0时,酶活力基本丧失。将GLugpg置于不同缓冲溶液中30 ℃保温2 h,结果表明该酶在pH 7.0−8.0间稳定性较好,能保持以上70%的酶活力。
GLpmi的最适pH为7.5,当10.0 < pH < 5.0时,酶活力基本丧失。将GLpmi置于不同缓冲溶液中30 ℃保温2 h,结果表明该酶在pH 6.0−9.0间稳定性较好,2 h以后仍能保持60%以上的酶活力。
2.6.2 最适反应温度和热稳定性将纯化后的GLpgm、GLugpg和GLpmi置于不同温度条件下测定酶活力,得到温度-酶活力关系(图 8)。

|
| 图 8 温度对酶活力及其稳定性的影响 Figure 8 Effects of temperature on activities and stabilities of purified enzymes 注:A、B:温度-GLpgm酶活力、稳定性;C、D:温度-GLugpg酶活力、稳定性;E、F:温度-GLpmi酶活力、稳定性. Note: A, B: Temperature-Enzyme of GLpgm activity and stability; C, D: Temperature-Enzyme of GLugpg activity and stability; E, F: Temperature-Enzyme of GLpmi activity and stability. |
|
|
GLpgm的最适反应温度为35 ℃,反应温度高于60 ℃时酶活力大大下降。将GLpgm置于不同温度下保温2 h后,在20 ℃时酶活力保持90%以上,稳定性良好,但高于45 ℃时,酶活力下降到20%以下。
GLugpg的最适反应温度为40 ℃,当温度高于60 ℃时,酶活力基本丧失。将GLugpg置于不同温度保温2 h,温度低于30 ℃时残余酶活保留80%−90%,即30 ℃以下热稳定性较好。
GLpmi的最适反应温度为30 ℃,当温度高于55 ℃时,酶活力基本丧失。将GLpmi置于不同温度保温2 h,温度在30 ℃以下时,残余酶活均保留90%以上,具有良好的稳定性。
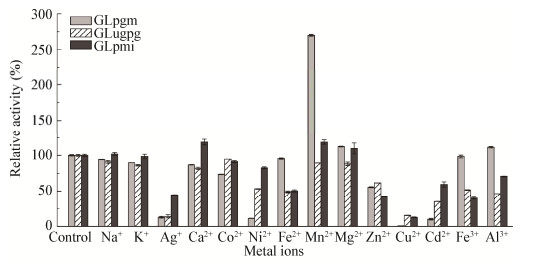
2.6.3 金属离子对酶活力的影响测定14种金属离子在浓度为1 mmol/L下对不同酶活力的影响(图 9)。对于3种纯化酶,Ag+和Cu2+均具有较强的抑制作用;Mn2+、Mg2+对GLpgm和GLpmi均有促进作用,尤其是Mn2+对GLpgm的促激活作用最强。另外,Ni2+和Cd2+对GLpgm的抑制作用也较强;而各种金属离子对GLugpg均没有明显的激活作用;金属离子Na+和Ca2+对GLpmi的酶活力均有激活作用。
|
| 图 9 金属离子对酶活力的影响 Figure 9 Effects of metal ions on the stability of purified enzymes |
|
|
GLpgm以底物α-葡萄糖-1-磷酸(0.1−5.0 mmol/L)、GLugpg以底物UDP-葡萄糖(0.1−5.0 mmol/L)和GLpmi以底物甘露糖-6-磷酸(1−10 mmol/L),在最适反应条件下测定酶活力,分别采用Linewaeaver-Burk双倒数进行作图(图 10),计算得到的酶促动力学参数见表 3。

|
| 图 10 纯化酶的Linewaeaver-Burk作图 Figure 10 Linewaeaver-Burk plot of purified enzymes 注:图A、B、C分别为GLpgm、GLugpg和GLpmi的Linewaeaver-Burk双倒数图. Note: Figure A, B and C are the Linewaeaver-Burk polt of GLpgm, GLugpg and GLpmi, respectively. |
|
|
| 酶 Enzymes |
Vmax (μmol/(L·min)) |
Km (mmol/L) |
kcat/
Km (mmol/(L·s)) |
| GLpgm | 322.58 | 0.68 | 196.08 |
| GLugpg | 1 250.00 | 0.25 | 818.60 |
| GLpmi | 1 250.00 | 0.50 | 1 105.22 |
目前关于PGM、UGPG和PMI酶学性质的研究多集中在细菌与植物来源,在食用真菌中的相关报道较少。从表 4中可知,不同来源的PGM、UGPG和PMI其最适反应pH基本上在7.0−8.0内,最适反应温度在30−40 ℃之间,物种间差异并不明显。本研究结果基本在此范围内,但GLpgm的最适反应pH为8.5,稍高于其他来源的PGM,在碱性环境下具备良好的催化特性,在一定程度上为拓宽该酶的应用提供指导意义。
| 酶 Enzymes |
来源 Sources |
生物体 Organisms |
最适pH Optimum pH |
最适温度 Optimum temperature |
促进剂 Activators |
抑制剂 Inhibitors |
Km
(mmol/L) |
kcat/Km
(mmol/(L·s)) |
| PGM | Bacteria | E. coli[20] | 9.0 | 0.05 | 54 | |||
| P. mucilaginosus[21] | 7.5 | 40 | Mg2+/Zn2+ | 0.24 | 47.5 | |||
| S. sanxanigenens[22] | 7.6/8.0 | 35 | 0.2−1.0 | 10−52 | ||||
| F. oxysporum[23] | 7.0 | 45 | Ca2+/Co2+/Ni2+/Mn2+/Mg2+ | 0.1 | 7.38 | |||
| L. lactis[24] | 7.2 | 30 | Cd2+/Zn2+ | 0.016 8 | ||||
| Fungi | S. cerevisiae[25] | 7.5 | 30 | Mg2+ | Cu2+/Zn2+ | 0.060/0.026/0.120 | ||
| Plants | S. oleracea[26] | 7.8−7.9 | ||||||
| S. tuberosum[27] | 7.5/7.9 | |||||||
| Zea mays L.[28] | 8.0 | 35 | Mg2+ | Mn2+ | 0.02 | |||
| Pisum sativum L.[29] | 7.9 | 35 | Mg2+/Mn2+ | Mn2+ | 0.018 | |||
| Animals | O. cuniculus[30] | 7.5 | Mg2+ | Ag+/Cu2+ | 0.063 | |||
| Homo sapiens[31] | 7.4−7.6 | 0.016−0.020 | ||||||
| UGPG | Bacteria | E. coli[32] | 7.6−7.8 | 37 | Co2+/Mg2+/Mn2+ | |||
| B. bifidum[33] | 6.5 | 37 | Mg2+ | |||||
| S. paucimobilis[34] | 0.007 5 | |||||||
| A. xylinum[35] | 3.22 | |||||||
| Fungi | S. cerevisiae[36] | 8.0 | 30 | Co2+/Mg2+/Mn2+ | 0.035 | |||
| G. lucidum[3] | 7.5 | 0.774 | 273 | |||||
| Plants | G. chouae[37] | 7.5 | 40 | Cu2+/Zn2+ | 0.001 67 | 11 980 | ||
| S. japonica[37] | 8.0 | 37 | Ca2+/Cu2+ | 0.004 33 | 4 610 | |||
| S. tuberosum[38-39] | 8.0/8.5 | 30/37 | 0.18/0.12 | |||||
| S. vulgare[40] | 9.0 | 37 | Co2+/Mg2+/Mn2+ | Co2+/Mn2+ | 0.05 | |||
| Nostoc sp.[41] | 7.5 | 37 | 3.04 | 2.09 | ||||
| Apocynaceae[42] | 7.2 | 37 | Mg2+ | 773.8 | 0.211 | |||
| Animals | Homo sapiens[43] | 7.8 | 37 | Co2+/Mg2+/Mn2+/Ca2+ | 0.14/0.18 | |||
| G. gallus[44] | 7.6 | 25 | Co2+/Mg2+/Mn2+ | 0.050−0.066 | ||||
| PMI | Bacteria | E. coli[45-46] | 7.4/7.1 | 22/25 | 1.210/0.354 | |||
| B. cepacia[47] | 7.6 | 32 | Mg2+/Ca2+/Mn2+/Co2+ | Zn2+ | 12.4 | |||
| P. aeruginosa[48] | 7.0 | 25 | Ca2+/Co2+ | 1.18 | ||||
| B. cereus[49] | 7.0 | 35 | Ni2+/Ca2+/Zn2+/Cu2+/Mg2+ | |||||
| S. chungbukense[50] | 8 | 37 | Ca2+/Co2+ | |||||
| Fungi | C. albicans[51] | Ag+/Cd2+/Zn2+ | 0.2/0.8/1.0/4.0 | |||||
| S. cerevisiae[52] | 7.1 | 25 | Ca2+/Zn2+ | Co2+ | 0.121 | |||
| A. fumigatus[53] | 7.6 | 37 | ||||||
| Plants | A. thaliana[54] | 7.5 | 25 | Cd2+/Zn2+ | 0.041 3/0.372 0 | |||
| A. konjac[55] | 6.5−7.0 | Co2+ | 0.73 | |||||
| S. japonica[56] | 8.5 | 15 | ||||||
| Animals | P. magellanicus[57] | 7.6 | ||||||
| Homo sapiens[58] | 0.23/0.25 |
Mn2+和Mg2+对多种来源的PGM和PMI酶活均有一定的促进作用,本研究中GLpgm和GLpmi的酶活也受到这两种金属离子的激活;Turnquist等[59]研究了多种来源的UGPG,发现其活性也会受到Mn2+和Mg2+的促进作用,而本研究中金属离子均对GLugpg表现出抑制作用,推测GLugpg对金属离子的依赖性并不高,可能是其特殊的酶活性结合位点造成的。从表 4中可看出,细菌来源的PMI酶活会受到Ni2+和Co2+的促进,而真菌和植物来源的PMI酶活却受到Ni2+和Co2+抑制,同样的,本实验中灵芝来源的GLpmi酶活也受到这两种金属离子的抑制。
不同来源的PGM、UGPG和PMI的Km值存在较大差异,本研究中GLpgm的Km值为0.68 mmol/L,高于细菌、真菌、植物和动物来源的PGM;有研究[3]显示灵芝来源的UGPG的Km值为0.774 mmol/L,而本研究中的GLugpg的Km值仅为0.25 mmol/L,且催化效率也优于同属其他菌株,但其Km值普遍比植物来源的酶高;从表 4可看出,细菌中PMI的Km值普遍高于真菌、植物和动物来源的PMI,本研究中GLpmi的Km值为0.50 mmol/L,与真菌C. albicans[51]和植物A. konjac[54]中的PMI相近。另外,本研究中GLpgm、GLugpg和GLpmi均具有较高的催化效率,其中GLpgm的催化效率优于大多数细菌来源的PGM。
本研究对灵芝多糖前体合成途径中关键酶PGM、UGPG和PMI的酶学性质研究结果显示,在反应pH、温度、金属离子作用方面,其与植物和真菌来源的这3种酶较为相似,且催化效率上表现更为优异。本研究对灵芝多糖糖供体合成途径关键酶酶学特性进行了完善,将有利于全面清晰地了解灵芝多糖的合成过程,并进一步为高效发酵调控策略的制定提供理论依据。
| [1] |
Chen S, Xu J, Liu C, et al. Genome sequence of the model medicinal mushroom Ganoderma lucidum[J]. Nature Communications, 2012, 3: 9913. |
| [2] |
Xu JW, Ji SL, Li HJ, et al. Increased polysaccharide production and biosynthetic gene expressions in a submerged culture of Ganoderma lucidum by the overexpression of the homologous α-phosphoglucomutase gene[J]. Bioprocess and Biosystems Engineering, 2015, 38(2): 399-405. DOI:10.1007/s00449-014-1279-1 |
| [3] |
Li MJ, Chen TX, Gao T, et al. UDP-glucose pyrophosphorylase influences polysaccharide synthesis, cell wall components, and hyphal branching in Ganoderma lucidum via regulation of the balance between glucose-1-phosphate and UDP-glucose[J]. Fungal Genetics and Biology, 2015, 82: 251-263. DOI:10.1016/j.fgb.2015.07.012 |
| [4] |
Liu GQ, Zhao Y, Wang XL, et al. Biosynthesis and fermentation control of polysaccharides from Ganoderma lucidum[J]. Mycosystema, 2011, 30(2): 198-205. (in Chinese) 刘高强, 赵艳, 王晓玲, 等. 灵芝多糖的生物合成和发酵调控[J]. 菌物学报, 2011, 30(2): 198-205. |
| [5] |
Wang Q. Effect of polysacchairdes composition and the related enzymatic analysis in Ganoderma lucidum submerged culture[D]. Wuxi: Master's Thesis of Jiangnan University, 2013 (in Chinese) 王琼.灵芝菌丝体培养中多糖组分的变化与相关酶活性分析[D].无锡: 江南大学硕士学位论文, 2013 |
| [6] |
Ma ZB, Ye C, Deng WW, et al. Reconstruction and analysis of a genome-scale metabolic model of Ganoderma lucidum for improved extracellular polysaccharide production[J]. Frontiers in Microbiology, 2018, 9: 3076-3089. DOI:10.3389/fmicb.2018.03076 |
| [7] |
Qiao SK. The effect of fermentation conditions on mycelium morphology and polysaccharides biosynthesis of Ganoderma lucidum in submerged culture[D]. Wuxi: Master's Thesis of Jiangnan University, 2014 (in Chinese) 乔双逵.液态发酵过程发酵条件对灵芝形态及灵芝多糖合成影响的研究[D].无锡: 江南大学硕士学位论文, 2014 http://cdmd.cnki.com.cn/Article/CDMD-10295-1014380450.htm |
| [8] |
Li J. The effect of three monosaccharide carbon sources on polysaccharides synthesis of Ganoderma lucidum[D]. Wuxi: Master's Thesis of Jiangnan University, 2015 (in Chinese) 李洁.三种单糖碳源对灵芝多糖合成影响的研究[D].无锡: 江南大学硕士学位论文, 2015 http://cdmd.cnki.com.cn/Article/CDMD-10295-1015427301.htm |
| [9] |
Tang YJ, Zhong JJ. Exopolysaccharide biosynthesis and related enzyme activities of the medicinal fungus, Ganoderma lucidum, grown on lactose in a bioreactor[J]. Biotechnology Letters, 2002, 24(12): 1023-1026. DOI:10.1023/A:1015677313598 |
| [10] |
Serra-Moreno R, Acosta S, Hernalsteens JP, et al. Use of the lambda red recombinase system to produce recombinant prophages carrying antibiotic resistance genes[J]. BMC Molecular Biology, 2006, 7: 31. DOI:10.1186/1471-2199-7-31 |
| [11] |
Li X, Li YX, Dai JJ. Usage of two-step red homologous recombination method to knockout the gene of Escherichia coli[J]. China Animal Husbandry & Veterinary Medicine, 2017, 44(7): 1934-1940. (in Chinese) 李鑫, 李亚芯, 戴建君. Red两步同源重组法在大肠杆菌基因敲除中的应用[J]. 中国畜牧兽医, 2017, 44(7): 1934-1940. |
| [12] |
Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products[J]. Proceedings of the National Academy of Sciences of the United States of America, 2000, 97(12): 6640-6645. DOI:10.1073/pnas.120163297 |
| [13] |
Baba T, Ara T, Hasegawa M, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection[J]. Molecular Systems Biology, 2006, 2: 2006. |
| [14] |
Qian N, Stanley GA, Hahn-Hagerdal B, et al. Purification and characterization of two phosphoglucomutases from Lactococcus lactis subsp. lactis and their regulation in maltose- and glucose-utilizing cells[J]. Journal of Bacteriology, 1994, 176(17): 5304-5311. |
| [15] |
Bernstein RL, Robbins PW. Control aspects of uridine 5'-diphosphate glucose and thymidine 5'-diphosphate glucose synthesis by microbial enzymes[J]. The Journal of Biological Chemistry, 1965, 240: 391-397. |
| [16] |
Shinabarger D, Berry A, May TB, et al. Purification and characterization of phosphomannose isomerase-guanosine diphospho-D-mannose pyrophosphorylase. A bifunctional enzyme in the alginate biosynthetic pathway of Pseudomonas aeruginosa[J]. The Journal of Biological Chemistry, 1991, 266(4): 2080-2088. |
| [17] |
Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding[J]. Analytical Biochemistry, 1976, 72(1/2): 248-254. |
| [18] |
Sch gger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kD[J]. Analytical Biochemistry, 1987, 166(2): 368-379. DOI:10.1016/0003-2697(87)90587-2 |
| [19] |
Dixon M, Webb EC. Enzymes[M]. 3rd ed. New York: Academic Press, 1979.
|
| [20] |
Joshi JG, Handler P. Phosphoglucomutase. I. Purification and properties of phosphoglucomutase from Escherichia coli[J]. Journal of Biological Chemistry, 1964, 239: 2741-2751. |
| [21] |
Tang JY, Lan DM, Wang YH, et al. Cloning, expression, and characterization of a phosphoglucomutase from Paenibacillus mucilaginosus[J]. Modern Food Science and Technology, 2015, 31(3): 38-42. (in Chinese) 唐家毅, 蓝东明, 王永华, 等. 胶质芽孢杆菌葡萄糖磷酸变位酶基因的克隆、表达与酶学性质研究[J]. 现代食品科技, 2015, 31(3): 38-42. |
| [22] |
Huang HD, Li XY, Wu M, et al. Cloning, expression and characterization of a phosphoglucomutase/phosphomannomutase from sphingan-producing Sphingomonas sanxanigenens[J]. Biotechnology Letters, 2013, 35(8): 1265-1270. |
| [23] |
Kourtoglou E, Anasontzis GE, Mamma D, et al. Constitutive expression, purification and characterization of a phosphoglucomutase from Fusarium oxysporum[J]. Enzyme and Microbial Technology, 2011, 48(3): 217-224. DOI:10.1016/j.enzmictec.2010.10.007 |
| [24] |
Neves AR, Pool WA, Castro R, et al. The α-phosphoglucomutase of Lactococcus lactis is unrelated to the α-D-phosphohexomutase superfamily and is encoded by the essential gene pgmH[J]. Journal of Biological Chemistry, 2006, 281(48): 36864-36873. DOI:10.1074/jbc.M607044200 |
| [25] |
Thomas W, Baylac A, Alkim C, et al. The PGM3 gene encodes the major phosphoribomutase in the yeast Saccharomyces cerevisiae[J]. FEBS Letters, 2012, 586(23): 4114-4118. DOI:10.1016/j.febslet.2012.09.042 |
| [26] |
Mühlbach H, Schnarrenberger C. Properties and intracellular distribution of two phosphoglucomutases from spinach leaves[J]. Planta, 1978, 141(1): 65-70. DOI:10.1007/BF00387746 |
| [27] |
Takamiya S, Fukui T. Phosphoglucomutase from potato tubers: chemical and catalytic properties[J]. The Journal of Biochemistry, 1978, 84(3): 569-574. DOI:10.1093/oxfordjournals.jbchem.a132161 |
| [28] |
Popova TN, Matasova LV, Lapotko AA. Purification, separation and characterization of phosphoglucomutase and phosphomannomutase from maize leaves[J]. IUBMB Life, 1998, 46(3): 461-470. DOI:10.1080/15216549800203982 |
| [29] |
Matasova LV, Popova TN. Catalytic properties of phosphoglucomutase from pea chloroplasts[J]. Biochemistry, 1999, 64(7): 774-779. |
| [30] |
Hirose M, Sugimoto E, Chiba H. Studies on crystalline yeast phosphoglucomutase: fundamental properties and chemical modification[J]. Biochimica et Biophysica Acta (BBA)-Enzymology, 1971, 250(3): 515-521. DOI:10.1016/0005-2744(71)90251-8 |
| [31] |
Fazi A, Piacentini MP, Piatti E, et al. Purification and partial characterization of the phosphoglucomutase isozyhes from human placenta[J]. Preparative Biochemistry, 1990, 20(3/4): 219-240. |
| [32] |
Weissborn AC, Liu Q, Rumley MK, et al. UTP: α-D-glucose-1-phosphate uridylyltransferase of Escherichia coli: isolation and DNA sequence of the galU gene and purification of the enzyme[J]. Journal of Bacteriology, 1994, 176(9): 2611-2618. DOI:10.1128/jb.176.9.2611-2618.1994 |
| [33] |
de Bruyn F, Beauprez J, Maertens J, et al. Unraveling the Leloir pathway of Bifidobacterium bifidum: significance of the uridylyltransferases[J]. Applied and Environmental Microbiology, 2013, 79(22): 7028-7035. DOI:10.1128/AEM.02460-13 |
| [34] |
Marques AR, Ferreira PB, Sá-Correia I, et al. Characterization of the ugpG gene encoding a UDP-glucose pyrophosphorylase from the gellan gum producer Sphingomonas paucimobilis ATCC 31461[J]. Molecular Genetics and Genomics, 2003, 268(6): 816-824. |
| [35] |
Koo HM, Yim SW, Lee CS, et al. Cloning, sequencing and expression of UDP-glucose pyrophosphorylase gene from Acetobacter xylinum BRC5[J]. Bioscience, Biotechnology, and Biochemistry, 2000, 64(3): 523-529. DOI:10.1271/bbb.64.523 |
| [36] |
Guranowski A, de Diego A, Sillero A, et al. Uridine 5'-polyphosphates (p4U and p5U) and uridine(5') polyphospho(5')nucleosides(UpnNs) can be synthesized by UTP: glucose-1-phosphate uridylyltransferase from Saccharomyces cerevisiae[J]. FEBS Letters, 2004, 561(1/2): 83-88. |
| [37] |
Chi S, Feng YJ, Liu T. Molecular cloning, characterization, and comparison of UDP-glucose pyrophosphorylase from Gracilaria chouae and Saccharina japonica[J]. Journal of Applied Phycology, 2016, 28(3): 2051-2059. DOI:10.1007/s10811-015-0738-7 |
| [38] |
Katsube T, Kazuta Y, Mori H, et al. UDP-glucose pyrophosphorylase from potato tuber: cDNA cloning and sequencing[J]. Journal of Biochemistry, 1990, 108(2): 321-326. DOI:10.1093/oxfordjournals.jbchem.a123200 |
| [39] |
Sowokinos JR, Spychalla JP, Desborough SL. Pyrophosphorylases in Solanum tuberosum. Ⅳ. Purification, tissue localization, and physicochemical properties of UDP-glucose pyrophosphorylase[J]. Plant Physiology, 1993, 101(3): 1073-1080. DOI:10.1104/pp.101.3.1073 |
| [40] |
Gustafson GL, Gander JE. Uridine diphosphate glucose pyrophosphorylase from Sorghum vulgare. Purification and kinetic properties[J]. The Journal of Biological Chemistry, 1972, 247(5): 1387-1397. |
| [41] |
Kawano Y, Sekine M, Ihara M. Identification and characterization of UDP-glucose pyrophosphorylase in cyanobacteria Anabaena sp. PCC 7120[J]. Journal of Bioscience and Bioengineering, 2014, 117(5): 531-538. DOI:10.1016/j.jbiosc.2013.10.015 |
| [42] |
Qi C. Purification and characterization of UDPase[J]. Journal of the Graduate School of the Chinese Academy of Sciences, 2004, 21(3): 345-351. (in Chinese) 祁超. Apocynaceae系细胞尿苷二磷酸葡萄糖焦磷酸化酶的纯化和表征[J]. 中国科学院研究生院学报, 2004, 21(3): 345-351. DOI:10.3969/j.issn.1002-1175.2004.03.010 |
| [43] |
Chacko CM, McCrone L, Nadler HL. Uridine diphosphoglucose pyrophosphorylase and uridine diphosphogalactose pyrophosphorylase in human skin fibroblasts derived from normal and galactosemic individuals[J]. Biochimica et Biophysica Acta (BBA)-Enzymology, 1972, 268(1): 113-120. DOI:10.1016/0005-2744(72)90204-5 |
| [44] |
Magee C, Nurminskaya M, Linsenmayer TF. UDP-glucose pyrophosphorylase: up-regulation in hypertrophic cartilage and role in hyaluronan synthesis[J]. Biochemical Journal, 2001, 360(3): 667-674. DOI:10.1042/bj3600667 |
| [45] |
Gao H, Chen Y, Leary JA. Kinetic measurements of phosphoglucose isomerase and phosphomannose isomerase by direct analysis of phosphorylated aldose–ketose isomers using tandem mass spectrometry[J]. International Journal of Mass Spectrometry, 2005, 240(3): 291-299. DOI:10.1016/j.ijms.2004.09.017 |
| [46] |
Desvergnes S, Courtiol-Legourd S, Daher R, et al. Synthesis and evaluation of malonate-based inhibitors of phosphosugar-metabolizing enzymes: class Ⅱ fructose-1, 6-bis-phosphate aldolases, type Ⅰ phosphomannose isomerase, and phosphoglucose isomerase[J]. Bioorganic & Medicinal Chemistry, 2012, 20(4): 1511-1520. |
| [47] |
Sousa SA, Moreira LM, Wopperer J, et al. The Burkholderia cepacia bceA gene encodes a protein with phosphomannose isomerase and GDP-D-mannose pyrophosphorylase activities[J]. Biochemical and Biophysical Research Communications, 2007, 353(1): 200-206. DOI:10.1016/j.bbrc.2006.12.016 |
| [48] |
Lee HJ, Chang HY, Venkatesan N, et al. Identification of amino acid residues important for the phosphomannose isomerase activity of PslB in Pseudomonas aeruginosa PAO1[J]. FEBS Letters, 2008, 582(23/24): 3479-3483. |
| [49] |
Zhang Y, Cui TB, Song Y. Gene cloning and expression of a phosphomannose isomerase from Bacillus cereus CZ and characterization of the recombinant enzyme[J]. Science and Technology of Food Industry, 2017, 38(6): 195-200. (in Chinese) 张瑶, 崔堂兵, 宋妍. 蜡状芽孢杆菌CZ磷酸甘露糖异构酶基因的克隆表达及酶学性质研究[J]. 食品工业科技, 2017, 38(6): 195-200. |
| [50] |
Tran ST, Le DT, Kim YC, et al. Cloning and characterization of phosphomannose isomerase from Sphingomonas chungbukensis DJ77[J]. BMB Reports, 2009, 42(8): 523-528. DOI:10.5483/BMBRep.2009.42.8.523 |
| [51] |
Bernard AR, Wells TNC, Cleasby A, et al. Selenomethionine labelling of phosphomannose isomerase changes its kinetic properties[J]. European Journal of Biochemistry, 1995, 230(1): 111-118. |
| [52] |
Roux C, Lee JH, Jeffery CJ, et al. Inhibition of type Ⅰ and type Ⅱ phosphomannose isomerases by the reaction intermediate analogue 5-phospho-D-arabinonohydroxamic acid supports a catalytic role for the metal cofactor[J]. Biochemistry, 2004, 43(10): 2926-2934. DOI:10.1021/bi035688h |
| [53] |
Fang WX, Yu XY, Wang B, et al. Characterization of the Aspergillus fumigatus phosphomannose isomerase Pmi1 and its impact on cell wall synthesis and morphogenesis[J]. Microbiology, 2009, 155(10): 3281-3293. DOI:10.1099/mic.0.029975-0 |
| [54] |
Maruta T, Yonemitsu M, Yabuta Y, et al. Arabidopsis phosphomannose isomerase 1, but not phosphomannose isomerase 2, is essential for ascorbic acid biosynthesis[J]. Journal of Biological Chemistry, 2008, 283(43): 28842-28851. DOI:10.1074/jbc.M805538200 |
| [55] |
Murata T. Studies on the phosphomannose isomerase of Amorphophallus konjac C. koch. I. Its isolation and some enzymic properties[J]. Plant and Cell Physiology, 1975, 16(6): 953-961. DOI:10.1093/oxfordjournals.pcp.a075241 |
| [56] |
Zhang YL. Cloning, analysis and expression of phosphomannose isomerase gene (PMI) from kelp[D]. Qingdao: Master's Thesis of Ocean University of China, 2013 (in Chinese) 张亚兰.海带磷酸甘露糖异构酶基因(PMI)的克隆、分析及表达[D].青岛: 中国海洋大学硕士学位论文, 2013 http://cdmd.cnki.com.cn/Article/CDMD-10423-1013369229.htm |
| [57] |
Pichaud N, Briatte S, Desrosiers V, et al. Metabolic capacities and immunocompetence of sea scallops (Placopecten magellanicus, gmelin) at different ages and life stages[J]. Journal of Shellfish Research, 1943, 28(4): 865-876. |
| [58] |
Proudfoot AEI, Turcatti G, Wells TNC, et al. Purification, cDNA cloning and heterologous expression of human phosphomannose isomerase[J]. European Journal of Biochemistry, 1994, 219(1/2): 415-423. |
| [59] |
Turnquist RL, Hansen RG. 2 Uridine diphosphoryl glucose pyrophosphorylase[J]. The Enzymes, 1973, 8: 51-71. DOI:10.1016/S1874-6047(08)60062-1 |
 2019, Vol. 46
2019, Vol. 46