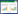扩展功能
文章信息
- 唐标, 张玲, 常江, 裘罕琦, 戴贤君, 夏效东, 杨华
- TANG Biao, ZHANG Ling, CHANG Jiang, QIU Han-Qi, DAI Xian-Jun, XIA Xiao-Dong, YANG Hua
- 两株分离自鸡粪便费格森埃希菌的耐药性
- Antimicrobial resistance of two Escherichia fergusonii strains isolated from chicken feces
- 微生物学通报, 2019, 46(11): 3022-3029
- Microbiology China, 2019, 46(11): 3022-3029
- DOI: 10.13344/j.microbiol.china.181032
-
文章历史
- 收稿日期: 2018-12-18
- 接受日期: 2019-03-25
- 网络首发日期: 2019-04-24
2. 西北农林科技大学食品科学与工程学院 陕西 杨凌 712100;
3. 中国计量大学现代科技学院 浙江 杭州 310018
2. College of Food Science and Engineering, Northwest Agriculture and Forestry University, Yangling, Shaanxi 712100, China;
3. College of Modern Science and Technology, China Jiliang University, Hangzhou, Zhejiang 310018, China
埃希氏菌属成员目前包括Escherichia albertii[1]、Escherichia blattae[2]、Escherichia fergusonii[3]、Escherichia hermanii[4]、Escherichia vulneris[5]、Escherichia marmotae[6]、Escherichia adecarboxylata[7]、Escherichia coli[8] 8个种。其中Escherichia fergusonii和Escherichia coli在表型和基因组上非常相似,分离过程中容易混淆[9-10]。目前从大肠杆菌类似菌中简单有效鉴定出费格森埃希菌(E. fergusonii)的方式有多重PCR[11]、荧光定量PCR[12]等方法,该菌对人和动物均具有致病性,最开始1985年从人的血液、尿液、粪便中分离到[3],后来又广泛地在动物上分离获得,比如山羊[13]、牛[14-15]、绵羊[14]、马[16]、火鸡[17]、鸡[18]和猪[19]等动物。国内也已经陆续报道从临床病人[20]、雏鸡[21]、羚牛[22]等宿主分离到费格森埃希菌。目前该菌的耐药性研究相对大肠埃希菌较少,但根据文献报道,费格森埃希菌和其他肠杆菌科内种属一样是存在多重耐药性的种[9, 19]。费格森埃希菌在动物和临床上均分离到多重耐药菌[9, 19, 23-24],包括超广谱β-内酰胺酶(Extended spectrum beta-lactamases,ESBL)携带菌株及粘菌素抗药菌株,例如Simmons等在肉鸡中发现ESBL携带的菌株[23-24],Glover等在非洲的非灵长类动物中分离的23株费格森埃希菌中发现14株对粘菌素耐药[10],北京大学人民医院团队在广东发现2株mcr-1携带费格森埃希菌[25],但是鲜有该菌对碳青霉烯类抗生素耐药的报道。
前期实验中在浙江省衢州蛋鸡场新鲜粪便中分离鉴定到2株费格森埃希菌[26],本研究中经过药敏检测、全基因组分析后,发现2株菌多重耐药且耐药基因丰富,其中一株菌对粘菌素耐药,并含有mcr-1耐药基因。此次研究为国内首次对粘菌素耐药的费格森埃希菌进行药敏实验验证及耐药基因分析的详细报道,为该菌的分布特征及耐药基因传播提供了思路,该菌的耐药性问题值得广泛重视。
1 材料与方法 1.1 主要试剂和仪器及培养基药敏检测试剂盒,上海星佰生物技术有限公司;利福霉素、粘菌素,生工生物工程(上海)股份有限公司;限制性内切酶、S1核酸酶、DNA聚合酶,宝生物工程(大连)有限公司。脉冲场电泳系统、凝胶成像系统、PCR扩增仪,Bio-Rad公司;LB培养基的配制参考文献[27]。
1.2 菌株来源与复苏两株费格森埃希菌EFCF053和EFCF056于2017年在浙江省衢州市某蛋鸡场的健康蛋鸡的粪便中分离获得。通过多重PCR、Viteck-2 Compact细菌鉴定系统和16S rRNA比对确认后,保藏至−80 ℃冰箱。划线于LB固体培养基上,37 ℃培养12 h复苏。E. coli EC600接合转移受体菌由中国农业大学动物医学院汪洋教授赠送。
1.3 药敏检测采用肉汤微量稀释法测定以下抗菌药物对试验菌株的最低抑菌浓度(Minimal inhibition concentration,MIC),使用E. coli ATCC 25922 (American type culture collection,Manassas,VA)作为质控菌株。使用农业农村部动物源细菌耐药性监测网指定的商业化产品(上海星佰生物技术有限公司),抗菌药物包括氨苄西林(Ampicillin,AMP)、奥格门丁(Amoxicillin/Clavulanate,A/C)、庆大霉素(Gentamicin,GEM)、大观霉素(Spectinomycin,SPT)、四环素(Tetracycline,TET)、氟苯尼考(Florfenicol,FFC)、磺胺异恶唑(Sulfisoxazole,SF)、复方新诺明(Trimethoprim/Sulfamethoxazole,SXT)、头孢噻呋(Ceftiofur,CEF)、头孢他啶(Ceftazidime,CAZ)、恩诺沙星(Enrofloxacin,ENR)、氧氟沙星(Ofloxacin,OFL)、美罗培南(Meropenem,MEM)、安普霉素(Apramycin,APR)、粘杆菌素(Colistin,CL)。所有MIC数据参照美国临床实验室标准化委员会(Clinical and laboratory standards institute,CLSI) 2015版M100-S25文件解释[28]。
1.4 基因组DNA抽提参照上海捷瑞生物工程有限公司的细菌基因组DNA提取试剂盒说明书提取基因组DNA。抽取的DNA通过琼脂糖凝胶电泳和NanoDrop ONE进行检测,基因组条带清晰明亮,质量合格后保存于−20 ℃冰箱备用。
1.5 全基因组测序与分析使用Illumina HiSeq测序平台对分离获得的费格森埃希菌进行全基因组测序,序列使用Velvet软件进行组装。使用ResFinder 3.1数据库[29] (https://cge.cbs.dtu.dk/services/ResFinder)对耐药基因进行预测;利用PlasmidFinder 2.0数据库[30] (https://cge.cbs.dtu.dk/services/PlasmidFinder)进行质粒复制类型的预测;质粒序列比对使用Easyfig软件[31];平均核苷酸相似度(Average nucleotide identity,ANI)分析使用JSpeciesWS网站[32]。
1.6 S1-PFGE和Southern blotting杂交利用Xba I酶切标准菌株Salmonella enterica serovar Braenderup H9812作为分子标准,S1核酸酶处理包埋胶块,使用脉冲场电泳系统分离质粒。电泳条件为:最小分子量5 kb,最大分子量500 kb,电泳16 h,初始转换时间2.16 s,终末转换时间63.8 s,电压6 V/cm,电场夹角为120°。将地高辛标记的mcr-1特异性探针参考孙龙[33]的方法进行Southern blotting杂交。
1.7 接合转移实验以携带mcr-1的菌株EFCF056为供体菌,E. coli EC600为受体菌进行接合试验。分别用无菌生理盐水制备处在对数期2麦氏浊度单位的菌悬液,按供体菌与受体菌体积比1:1各吸取100 μL混合后,点在无抗LB培养基上的无菌滤膜上进行混合培养,37 ℃培养8 h后,1 mL冲洗混合细菌配成菌悬液。取100 μL涂布于含400 μg/mL利福霉素及4 μg/mL粘菌素的LB培养基上,同时取100 μL配制梯度浓度后分别在含400 μg/mL利福霉素的LB培养基上涂布,37 ℃培养过夜。双抗板上疑似菌落使用mcr-1引物进行菌落PCR验证。取相同浓度的供体菌和受体菌分别涂布在同时含400 μg/mL利福霉素及4 μg/mL粘菌素的双抗LB培养基上作为阴性对照。
2 结果与分析 2.1 药敏结果将2株菌进行了药物敏感试验,获得了对15种药物的MIC值并进行了比较(表 1)。两株菌均表现出多重耐药的特征,其中菌株EFCF053对氨苄西林、庆大霉素、四环素、氟苯尼考、磺胺异噁唑、复方新诺明表现出耐药,对恩诺沙星和氧氟沙星耐药则为中介值。菌株EFCF056耐药类型较多,对氨苄西林、头孢噻呋、大观霉素、庆大霉素、四环素、氟苯尼考、磺胺异噁唑、复方新诺明、粘菌素、恩诺沙星、氧氟沙星11个抗生素表现出耐药,同时对奥格门丁和头孢他啶则为中介值。EFCF056相对EFCF053耐药种类增加,比如对大观霉素、头孢噻呋、粘菌素、恩诺沙星、氧氟沙星耐药。同时判断为耐药的情况下,菌株EFCF056表现出较高的MIC,比如氨苄西林(>512 vs 512),庆大霉素(>512 vs 64)。
| 抗生素 Antimicrobial agents |
EFCF053 | EFCF056 |
| 氨苄西林Ampicillin (AMP) | 512* | >512* |
| 奥格门丁Amoxicillin/Clavulanate (A/C) | 4/2 | 16/8* |
| 头孢噻呋Ceftiofur (CEF) | 0.5 | >256* |
| 头孢他啶Ceftazidime (CAZ) | 0.25 | 8* |
| 美罗培南Meropenem (MEM) | ≤0.03 | ≤0.03 |
| 庆大霉素Gentamicin (GEM) | 64 | >512* |
| 大观霉素Spectinomycin (SPT) | 32 | 512* |
| 四环素Tetracycline (TET) | 128* | 128* |
| 氟苯尼考Florfenicol (FFC) | >256* | >256* |
| 磺胺异恶唑Sulfisoxazole (SF) | >512* | 512* |
| 复方新诺明Trimethoprim/Sulfamethoxazole (SXT) | >32/608* | >32/608* |
| 恩诺沙星Enrofloxacin (ENR) | 1 | 16* |
| 氧氟沙星Ofloxacin (OFL) | 4 | 64* |
| 安普霉素Apramycin (APR) | 8 | 8 |
| 粘杆菌素Colistin (CL) | 0.5 | 4* |
| 注:*:判断为耐药. Note: *: Represent antimicrobial resistance. |
||
对2株费格森埃希菌EFCF053和EFCF056菌株进行了全基因组测序,基因组大小分别为4.77 Mb和5.02 Mb,序列已经提交至GenBank数据库,检索号分别为RWHR00000000和RWHS00000000。以上两株菌与典型菌株E. fergusonii ATCC 35469T (CU928158.2) ANI值分别为98.54%和98.43%,高于同种临界值95%[32],可从基因组水平确定为费格森埃希菌。
对获得性耐药基因进行了预测,结果显示2株菌的耐药表型都可以通过获得性耐药基因得到解释(表 2)。但是相同耐药表型的菌株耐药基因可能不同,例如对复方新诺明耐药的菌株EFCF053的耐药基因是dfrA14,而EFCF056为dfrA12;对磺胺异噁唑耐药的菌株EFCF053的耐药基因是sul2、sul3,而在EFCF056中是sul1、sul2;介导菌株EFCF053对氨苄西林耐药的是blaTEM-1A,而EFCF056中则是blaTEM-1B。多个基因共同介导耐药,具体耐药贡献不明确,例如菌株EFCF056对喹诺酮类耐药的基因有qnrS2、aac(6')-Ib-cr、oqxA、oqxB共4个。通过2株菌的耐药表型比较可以推测blaCTX-M-65、blaOXA-1、blaCTX-M-55基因介导了头孢他啶和头孢噻呋的耐药,基因rmtB、aadA2、aac(6')-Ib-cr更有可能参与大观霉素的耐药。预测到的fosA3、mph(A)、ARR-3耐药基因,分别可能介导菌株EFCF056对磷霉素、大环内脂类和利福霉素的耐药,但是缺少药敏实验验证。另外重要的是,在菌株EFCF056中发现了耐药基因mcr-1,造成了该菌对粘菌素的耐药;两株菌都含有编码超广谱β-内酰胺酶(ESBL)的基因,菌株EFCF053中的blaTEM-1A和EFCF056中的blaCTX-M-65、blaOXA-1、blaTEM-1B和blaCTX-M-55。最后对2株菌的耐药质粒进行了预测,发现多个复制子类型,菌株EFCF053中有IncFIB、IncFII、IncHI2、IncHI2A、IncX1复制子;而EFCF056中除了含有上述所有复制子外,还预测到IncQ1和p0111复制类型,这意味着2株菌都含有多个质粒。通过S1-PFGE可以进一步证明,菌株EFCF053和EFCF056中大于40 kb的质粒分别为4个和5个,且带型具有很大差异(图 1)。
| 抗生素类型 Antibiotic types |
EFCF053 | EFCF056 | ||||
| Susceptibility | Acquired resistance genes | Susceptibility | Acquired resistance genes | |||
| β-Lactam | AMP | R | blaTEM-1A | R | blaCTX-M-65, blaOXA-1, blaTEM-1B, blaCTX-M-55 | |
| A/C | S | I | ||||
| CEF | S | R | ||||
| CAZ | S | I | ||||
| MEM | S | S | ||||
| Aminoglycoside | GEM | R | aac(3)-IId, aph(3')-Ia, aph(3'')-Ib, aph(6)-Id | R | aac(3)-IId, aph(3')-Ia, aph(3'')-Ib, aph(6)-Id, rmtB, aac(6')-Ib-cr, aadA2 | |
| SPT | S | R | ||||
| Colistin | CL | S | R | mcr-1 | ||
| Fluoroquinolone | ENR | I | R | qnrS2, aac(6')-Ib-cr, oqxA, oqxB | ||
| OFL | I | R | ||||
| Fosfomycin | fosA3 | |||||
| MLS-Macrolide, Lincosamide and Streptogramin B | mph(A) | |||||
| Phenicol | FFC | R | floR | R | catA1, floR, catB3 | |
| Rifampicin | ARR-3 | |||||
| Sulfonamides | SF | R | sul2, sul3 | R | sul1, sul2 | |
| SXT | R | dfrA14 | R | dfrA12 | ||
| Tetracycline | TET | R | tet(A) | R | tet(A) | |
| 注:R:耐药;S:敏感. Note: R: Resistance;S: Sensitive. |
||||||
|
| 图 1 菌株EFCF053、EFCF056的S1-PFGE与mcr-1基因的Southern blotting分析 Figure 1 S1-PFGE and Southern blotting with mcr-1 gene analysis of strains EFCF053 and EFCF056 注:M:H9812 marker;1、2:菌株EFCF053和EFCF056的S1-PFGE图,显示了各自的质粒条带;3、4:菌株EFCF053和EFCF056的中的质粒与mcr-1基因探针的Southern blotting图;箭头:含有mcr-1基因的质粒. Note: M: H9812 marker;1, 2: S1-PFGE of strains EFCF053 and EFCF056, showing the sizes of the respective plasmids;3, 4: The plasmids of strains EFCF053 and EFCF056 with mcr-1 gene probe in the Southern blotting;The arrows: The plasmid harboring the mcr-1 gene. |
|
|
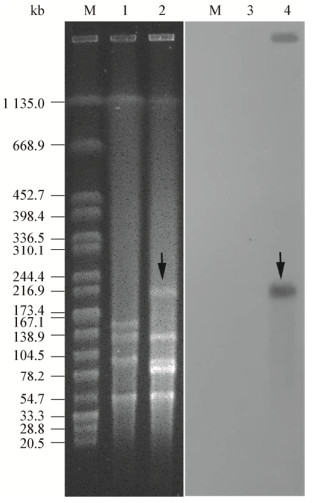
从表 1可以发现菌株EFCF056对粘菌素耐药,MIC为4 μg/mL,进一步在其基因组序列中预测到mcr-1基因。该基因位于组装序列的Contig101上,片段大小为47 800 bp。将该序列进行BLAST,与比对到的2个最同源的质粒进行了比较(图 2)。两个质粒均来源于大肠杆菌,分别为pHS13-1-IncHI2和pMCR1_025943,前者在深圳腹泻病人中分离到,后者从美国临床病人的直肠拭子中分离获得,2个质粒均大于260 kb。在mcr-1耐药基因下游发现重组酶,意味着该基因具有可移动元件,具有发生水平转移的能力。针对菌株ECEF056中的该质粒进行了接合转移实验,在含有利福霉素和粘菌素的双抗板上生长的疑似接合子进行了菌落PCR鉴定,结果发现均能扩增出1 626 bp全长的mcr-1基因(图 3),并通过测序验证。该实验可证实菌株ECEF056中携带mcr-1基因的质粒可以发生接合转移,通过计算,接合转移效率为0.56×10−6。
|
| 图 2 mcr-1基因所在片段与同源质粒的比较 Figure 2 Comparison of the fragment harboring mcr-1 gene with homologous plasmids |
|
|

|
| 图 3 接合子的mcr-1基因验证 Figure 3 Verification of mcr-1 gene in transconjugants 注:M: DL5000 DNA marker;+:阳性对照;−:阴性对照;1−5:疑似接合子的菌落PCR产物. Note: M: DL5000 DNA marker;+: Positive control;−: Negative control;1−5: PCR products of suspected transconjugant colonies. |
|
|
在本次研究中,对分离获得的2株费格森埃希菌进行了全基因组测序和药敏实验。结果显示出2株菌耐药谱广、耐药基因丰富,同时检出ESBL和mcr-1基因,这是国内第一次报道该菌同时含有这两类基因。此次研究中,mcr-1基因携带质粒可证实具有接合转移的能力,有传播风险。两株菌所有的耐药表型都可以通过相应的获得性耐药基因得到解释。但是部分耐药表型同时包含多个耐药基因,目前无法确认哪一个占主要因素,可能是共同作用的结果。另外,由于药敏实验的限制,没有穷尽所有的抗生素,但是预测到了磷霉素耐药基因fosA3、大环内脂类耐药基因mph(A)和利福霉素耐药基因ARR-3。这意味着菌株EFCF056的耐药谱比药敏实验获得的结果更加广泛。另外,两株菌质粒较多,可以确定的2株菌具有大质粒4−5个。另外,2株菌没有获得基因组完成图,所以质粒的数目和耐药基因的存在位置无法确认,未来会通过全基因组完成图进一步探究。
费格森埃希菌的宿主非常广泛,在健康或疾病动物、人体中都被分离得到。费格森埃希菌和大肠杆菌不容易区分,难以判断,所以该菌的研究可能容易被忽略。通过本次研究,显示出费格森埃希菌是重要的耐药基因存储库,可能与家禽养殖兽用抗生素的不规范使用相关,其耐药性应引起广泛重视。未来应该对该菌进行动物源及临床上筛查和耐药性监测,深入研究其MIC频率分布,更全面深入地研究该菌的耐药现状和流行规律。
| [1] |
Huys G, Cnockaert M, Janda JM, et al. Escherichia albertii sp. nov., a diarrhoeagenic species isolated from stool specimens of Bangladeshi children[J]. International Journal of Systematic and Evolutionary Microbiology, 2003, 53(3): 807-810. DOI:10.1099/ijs.0.02475-0 |
| [2] |
Priest FG, Barker M. Gram-negative bacteria associated with brewery yeasts: reclassification of Obesumbacterium proteus biogroup 2 as Shimwellia pseudoproteus gen. nov., sp. nov., and transfer of Escherichia blattae to Shimwellia blattae comb. nov.[J]. International Journal of Systematic and Evolutionary Microbiology, 2010, 60(4): 828-833. DOI:10.1099/ijs.0.013458-0 |
| [3] |
Farmer Ⅲ JJ, Fanning GR, Davis BR, et al. Escherichia fergusonii and Enterobacter taylorae, two new species of Enterobacteriaceae isolated from clinical specimens[J]. Journal of Clinical Microbiology, 1985, 21(1): 77-81. |
| [4] |
Brenner DJ, Davis BR, Steigerwalt AG, et al. Atypical biogroups of Escherichia coli found in clinical specimens and description of Escherichia hermannii sp. nov.[J]. Journal of Clinical Microbiology, 1982, 15(4): 703-713. |
| [5] |
Brenner DJ, McWhorter AC, Knutson JKL, et al. Escherichia vulneris: a new species of Enterobacteriaceae associated with human wounds[J]. Journal of Clinical Microbiology, 1982, 15(6): 1133-1140. |
| [6] |
Liu S, Jin D, Lan RT, et al. Escherichia marmotae sp. nov., isolated from faeces of Marmota himalayana[J]. International Journal of Systematic and Evolutionary Microbiology, 2015, 65(7): 2130-2134. DOI:10.1099/ijs.0.000228 |
| [7] |
Izard D, Mergaert J, Gavini F, et al. Separation of Escherichia adecarboxylata from the Erwinia herbicola-Enterobacter agglomerans complex and from the other Enterobacteriaceae by nucleic acid and protein electrophoretic techniques[J]. Annales de l'Institut Pasteur/Microbiologie, 1985, 136(2SB): 151-159, 161-168. |
| [8] |
Skerman VBD, McGowan V, Sneath PHA. Approved lists of bacterial names[J]. International Journal of Systematic and Evolutionary Microbiology, 1980, 30(1): 225-420. DOI:10.1099/00207713-30-1-225 |
| [9] |
Gaastra W, Kusters JG, van Duijkeren E, et al. Escherichia fergusonii[J]. Veterinary Microbiology, 2014, 172(1/2): 7-12. |
| [10] |
Glover B, Wentzel J, Jenkins A, et al. The first report of Escherichia fergusonii isolated from non-human primates, in Africa[J]. One Health, 2017, 3: 70-75. DOI:10.1016/j.onehlt.2017.05.001 |
| [11] |
Lindsey RL, Garcia-Toledo L, Fasulo D, et al. Multiplex polymerase chain reaction for identification of Escherichia coli, Escherichia albertii and Escherichia fergusonii[J]. Journal of Microbiological Methods, 2017, 140: 1-4. DOI:10.1016/j.mimet.2017.06.005 |
| [12] |
Simmons K, Rempel H, Block G, et al. Duplex PCR methods for the molecular detection of Escherichia fergusonii isolates from broiler chickens[J]. Applied and Environmental Microbiology, 2014, 80(6): 1941-1948. DOI:10.1128/AEM.04169-13 |
| [13] |
Hariharan H, López A, Conboy G, et al. Isolation of Escherichia fergusonii from the feces and internal organs of a goat with diarrhea[J]. The Canadian Veterinary Journal, 2007, 48(6): 630-631. |
| [14] |
Bain MS, Green CC. Isolation of Escherichia fergusonii in cases clinically suggestive of salmonellosis[J]. The Veterinary Record, 1999, 144(18): 511. |
| [15] |
Rimoldi GM, Moeller RB Jr. Escherichia fergusonii associated with pneumonia in a beef cow[J]. Journal of Veterinary Medicine, 2013, 2013: 829532. |
| [16] |
Weiss ATA, Lübke-Becker A, Krenz M, et al. Enteritis and septicemia in a horse associated with infection by Escherichia fergusonii[J]. Journal of Equine Veterinary Science, 2011, 31(7): 361-364. DOI:10.1016/j.jevs.2011.01.005 |
| [17] |
Herráez P, Rodríguez AF, Espinosa De Los Monteros A, et al. Fibrino-necrotic typhlitis caused by Escherichia fergusonii in ostriches (Struthio camelus)[J]. Avian Diseases, 2005, 49(1): 167-169. DOI:10.1637/7221-061104r |
| [18] |
Oh JY, Kang MS, An BK, et al. Isolation and epidemiological characterization of heat-labile enterotoxin-producing Escherichia fergusonii from healthy chickens[J]. Veterinary Microbiology, 2012, 160(1/2): 170-175. |
| [19] |
Rayamajhi N, Cha SB, Shin SW, et al. Plasmid typing and resistance profiling of Escherichia fergusonii and other Enterobacteriaceae isolates from South Korean farm animals[J]. Applied and Environmental Microbiology, 2011, 77(9): 3163-3166. DOI:10.1128/AEM.02188-10 |
| [20] |
Song Y, Ma SQ. A case of multi-drug resistant Escherichia fergusonii isolated from blood stream infection induced by ascending urinary tract infection[J]. Chinese Journal of Microecology, 2015, 27(4): 475-476. (in Chinese) 宋宇, 马淑青. 尿路感染上行所致血流感染中分离出多重耐药弗格森埃希菌一例[J]. 中国微生态学杂志, 2015, 27(4): 475-476. |
| [21] |
Lu Y, Zhao HY, Qi XL, et al. Identification of Escherichia fergusonii and analysis of its drug resistance[J]. Progress in Veterinary Medicine, 2013, 34(5): 48-51. (in Chinese) 陆彦, 赵红玉, 齐晓丽, 等. 弗格森埃希菌的鉴定及药敏试验[J]. 动物医学进展, 2013, 34(5): 48-51. DOI:10.3969/j.issn.1007-5038.2013.05.010 |
| [22] |
Ye N, Quan ZF, Wang B, et al. Isolation, idenfication and drug sensitivity test of Escherichia fergusonii from Takin[J]. Progress in Veterinary Medicine, 2015, 36(2): 68-71. (in Chinese) 叶泥, 权自芳, 王彬, 等. 一株扭角羚弗格森埃希菌的分离鉴定及药敏试验[J]. 动物医学进展, 2015, 36(2): 68-71. |
| [23] |
Simmons K, Islam MR, Rempel H, et al. Antimicrobial resistance of Escherichia fergusonii isolated from broiler chickens[J]. Journal of Food Protection, 2016, 79(6): 929-938. DOI:10.4315/0362-028X.JFP-15-575 |
| [24] |
Lagacé-Wiens PRS, Baudry PJ, Pang P, et al. First description of an extended-spectrum-β-lactamase-producing multidrug-resistant Escherichia fergusonii strain in a patient with cystitis[J]. Journal of Clinical Microbiology, 2010, 48(6): 2301-2302. DOI:10.1128/JCM.00364-10 |
| [25] |
Wang RB, van Dorp L, Shaw LP, et al. The global distribution and spread of the mobilized colistin resistance gene mcr-1[J]. Nature Communications, 2018, 9(1): 1179. DOI:10.1038/s41467-018-03205-z |
| [26] |
Zhang L, Chang J, Cao LJ, et al. Rapid identification and clustering analysis on E. coli isolates from chicken fecal by MALDI-TOF MS[J]. China Poultry, 2018, 40(20): 23-27. (in Chinese) 张玲, 常江, 曹刘杰, 等. 鸡粪便来源大肠杆菌的质谱法快速鉴定及聚类分析[J]. 中国家禽, 2018, 40(20): 23-27. |
| [27] |
Rédei GP. LB (Luria-Bertani) bacterial medium[A]//Rédei GP. Encyclopedia of Genetics, Genomics, Proteomics and Informatics[M]. Dordrecht: Springer, 2008: 1087
|
| [28] |
Clinical and Laboratory Standards Institute. CLSI Document M100-S25 Performance standards for antimicrobial susceptibility testing;25th Informational Supplement[S]. Wayne, PA: Clinical and Laboratory Standards Institute, 2015
|
| [29] |
Zankari E, Hasman H, Cosentino S, et al. Identification of acquired antimicrobial resistance genes[J]. Journal of Antimicrobial Chemotherapy, 2012, 67(11): 2640-2644. DOI:10.1093/jac/dks261 |
| [30] |
Carattoli A, Zankari E, García-Fernández A, et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing[J]. Antimicrobial Agents and Chemotherapy, 2014, 58(7): 3895-3903. DOI:10.1128/AAC.02412-14 |
| [31] |
Sullivan MJ, Petty NK, Beatson SA. Easyfig: a genome comparison visualizer[J]. Bioinformatics, 2011, 27(7): 1009-1010. DOI:10.1093/bioinformatics/btr039 |
| [32] |
Richter M, Rosselló-Móra R, Oliver Glckner F, et al. JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison[J]. Bioinformatics, 2016, 32(6): 929-931. DOI:10.1093/bioinformatics/btv681 |
| [33] |
Sun L. Study on the biological characters, molecular typing and carbapenem resistant mechanisms of Proteus mirabilis deficient in swarming motility[D]. Hangzhou: Master's Thesis of Zhejiang University, 2016 (in Chinese) 孙龙.无迁徙生长碳青霉烯耐药奇异变形杆菌生物学特性、耐药机制及分子分型研究[D].杭州: 浙江大学硕士学位论文, 2016 http://cdmd.cnki.com.cn/Article/CDMD-10335-1017046069.htm |
 2019, Vol. 46
2019, Vol. 46