扩展功能
文章信息
- 吴霖, 葛洋, 张海坤, 李岩, 胡晓珂
- WU Lin, GE Yang, ZHANG Hai-Kun, LI Yan, HU Xiao-Ke
- 一株高ACE抑制活性乳杆菌的筛选鉴定、培养条件优化及其基因组分析
- Screening, identification, optimization of culture condition and genome analysis of a bacterium with high ACE inhibitory activity
- 微生物学通报, 2019, 46(11): 2830-2847
- Microbiology China, 2019, 46(11): 2830-2847
- DOI: 10.13344/j.microbiol.china.180927
-
文章历史
- 收稿日期: 2018-11-19
- 接受日期: 2019-04-10
- 网络首发日期: 2019-06-11
2. 中国科学院烟台海岸带研究所 海岸带生物学与生物资源利用重点实验室 山东 烟台 264003;
3. 福建农林大学 福建 福州 350002;
4. 青岛海洋科学与技术试点国家实验室 海洋生物学与生物技术功能实验室 山东 青岛 266237
2. Key Laboratory of Coastal Biology and Bio-resource Utilization, Yantai Institute of Coastal Zone Research, Chinese Academy of Sciences, Yantai, Shandong 264003, China;
3. Fujian Agriculture and Forestry University, Fuzhou, Fujian 350002, China;
4. Laboratory for Marine Biology and Biotechnology, Qingdao National Laboratory for Marine Science and Technology, Qingdao, Shandong 266237, China
高血压是我国最常见的慢性疾病之一。现代人生活压力大、饮食作息不规律等使得它的发病率呈现逐年上升的趋势,且发病人群趋于年轻化[1]。高血压主要依靠长期服用降压药物(血管紧张素转换酶活性抑制剂)进行治疗,目前市售的降压药物多为人工合成,抑制效果较强[2]。但是,长期服用人工合成的血管紧张素转化酶抑制剂不仅会对人体某些脏器如肾脏产生副作用,还会伴有疼痛、发烧、恶心等症状,使人体正常的生理功能受到不同程度的损伤[3-4]。天然食物中分离出的ACE抑制剂药效温和、对人体生理机能毒副作用小,具有广阔的应用前景[5]。因此,越来越多的天然产品,包括鱼[6-7]、山羊奶[8]、鸡蛋[9]和植物来源[10-15]中具有ACE抑制活性的物质陆续被分离并评估其潜在的抗高血压效果。
作为调节血压的关键酶,ACE可以催化ANG-I转化为ANG-Ⅱ,从而使血管收缩导致血压升高[16]。因此,大量报道研究了如何通过抑制ACE活性来缓解高血压[13, 17-19]。ACE抑制肽是一类具有抑制ACE活性的小分子多肽物质,分子量一般低于1 000 Da,这些多肽的氨基酸序列和肽链长度各不相同,但其功能类似。微生物发酵或蛋白酶水解是生产ACE抑制肽的最常用方法,目前已从牛奶蛋白、鱼类蛋白、大豆蛋白等食物中分离出具有ACE抑制活性的多肽(ACE抑制肽)[20-21]。乳杆菌作为公认的安全食品微生物,是一类动植物益生菌,被广泛应用于食品及医疗保健业。以乳杆菌为主要发酵菌种的传统发酵食品中,其多肽具有相对更强的ACE抑制活性,同时所含的乳杆菌本身还具有免疫促进、改善肠胃等益生功能[22-24]。瑞士乳杆菌是乳杆菌的一种,广泛分布于自然界,大部分属于人类肠道的正常菌群,几乎无致病性报道[25]。瑞士乳杆菌营养条件相对苛刻,需要多种游离氨基酸,这些氨基酸在牛奶中的含量较少,但由于枯草杆菌蛋白酶样丝氨酸蛋白酶的作用,活性细胞包膜蛋白酶(Cell-envelope proteinase,CEP)可为瑞士乳杆菌在牛奶中的快速生长提供支持。因此,瑞士乳杆菌在牛乳中的生长取决于复合蛋白水解系统的存在,该系统可水解占牛乳中存在的所有蛋白质约80%的乳酪蛋白。该蛋白水解系统通常由3部分构成:首先,CEP将酪蛋白水解成多肽;其次,转运系统将多肽转移进入细菌;最后,许多特异性不同或部分重叠的细胞内肽酶作用,将多肽在细胞内水解为氨基酸[26-27]。CEP不仅对发酵乳中细菌的生长存在关键作用,还有助于改善发酵产品的风味和质地[28-29],发酵过程中产生的具有不同性质的生物活性肽,有益人类健康,可抗高血压、抑制细菌致病活性、降低结肠癌的风险、调节免疫活性等[30-33]。乳酸菌中,瑞士乳杆菌产生的ACE抑制肽活性最高[34]。
海洋作为巨大的生物资源宝库,蕴含着丰富的微生物资源,各领域研究者从海洋沉积物中筛选出了具有不同实用价值的菌株。例如,林学政等[35]在北极地区海洋沉积物中分离筛选出了可在以石油为唯一碳源的无机盐培养基中生长的石油烃降解菌。夏玉林等[36]在南海海洋沉积物中筛选出具有抗肿瘤活性的菌株。针对海洋来源的乳杆菌研究主要集中在发酵、水产养殖和疾病防护等方面,如青贮饲料发酵[37]、水产养殖抗病增产[38-39]、食品防腐、降低胆固醇[40-41]、增强人体免疫力、抑制肿瘤发生发展[42]等方面的应用。此外,黄君阳[43]还从海带中筛选到一株乳杆菌,其发酵液具有α-葡萄糖苷酶抑制活性,可降血糖。但是,关于海洋源ACE抑制活性的乳杆菌研究相对较少,涉及海洋源瑞士乳杆菌的研究更少。基于此,本文以挖掘海洋源ACE抑制活性的乳杆菌为目的,以未培养和纯培养结合的方式从海洋沉积物中筛选出乳杆菌,并对其中一株高ACE抑制活性的瑞士乳杆菌进行发酵工艺优化,显著提高了其ACE抑制活性。同时本研究还针对该菌株基因组信息进行分析,为安全高效的降压药物开发提供更广阔的天然ACE抑制剂来源和研究基础。
1 材料与方法 1.1 实验材料本实验使用的沉积物于2016年4月22日采自渤海(119.999°E,39.092°N),使用箱式采泥器采集并取表层1-2 cm的沉积物,混匀后用5 mL的冻存管分装并保存在-20 ℃用于DNA的提取,另保存300-500 g样品在4 ℃用于分离筛选菌株。
1.2 培养基乳酸细菌培养基(MRS)肉汤培养基参考文献[44];MRS琼脂培养基:在液体培养基的基础上加琼脂15.0 g/L;MRS碳酸钙琼脂培养基:配制好的MRS琼脂培养基,1×105 Pa灭菌15 min后加入已灭菌的碳酸钙溶液适量(15.0-20.0 g/L);MRS溴甲酚绿琼脂培养基:在MRS固体培养基的基础上,加0.02%的溴甲酚绿酒精溶液。蛋白胨水培养基(g/L):蛋白胨10.0,NaCl 5.0,pH 7.2-7.4。石蕊牛奶培养基(g/L):脱脂奶粉100.000,石蕊试剂0.075,pH 6.8。脱脂乳培养基(g/L):脱脂奶粉100.0,pH 6.3±0.2,0.3×105 Pa灭菌10-15 min。
1.3 主要试剂和仪器溴甲酚绿,天津光复精细化工研究所;GenColour,北京金博益生物技术有限公司;Loading buffer、DNA buffer,TRANS公司;琼脂、石蕊,国药集团化学试剂有限公司;其他生化试剂,生工生物工程(上海)股份有限公司;PowerSoilTM DNA Isolation Kit、UltraClean Microbial DNA Isolation Kit,MOBIO公司;Phusion High-Fidelity PCR Master Mix with GC Buffer,New England Biolabs公司;10×PCR Rxn Buffer-MgCl2、Taq DNA Polymerase,Invitrogen公司。
水平电泳仪,北京六一生物科技有限公司;涡旋仪,上海智诚分析仪器制造有限公司;紫外分光光度计,PerkinElmer股份有限公司。
1.4 沉积物中细菌多样性分析前处理取贮存于-20 ℃的渤海表层沉积物20 g于50 mL离心管,在-80 ℃预冷后,用真空冷冻干燥机进行冷冻干燥。
1.5 细菌总DNA提取与PCR扩增参照提取说明书,使用MOBIO公司生产的PowerSoilTM DNA Isolation Kit提取沉积物样品中的全部基因组。将样品送至北京诺禾致源科技股份有限公司进行测序,以提取的基因组DNA为模板,使用带Barcode的特异引物扩增16S rRNA基因V4–V5区(392 bp),正向引物F:5′-CCTACGGGNB GCASCAG-3′,反向引物R:5′-GACTACNVGGGTA TCTAATCC-3′。PCR反应体系:2×Phusion Master Mix 15 μL,Primer F (2 μmol/L) 1.5 μL,Primer R (2 μmol/L) 1.5 μL,gDNA (1 ng/μL) 10 μL,ddH2O 2 μL。PCR反应条件:98 ℃ 1 min;98 ℃ 10 s,50 ℃ 30 s,72 ℃ 30 s,30个循环;72 ℃ 5 min。
1.6 测序结果处理使用QIIME软件对双末端测序结果进行拼接,并使用默认参数对拼接后的序列进行质控清洗聚类分析[45],主要流程包括:根据质量分数质控、移除嵌合体、97%相似性水平聚类及分类水平鉴定[46-47]。分类水平鉴定使用的数据库为GreenGene (http://www.greengene.com/)。使用相对丰度标准化测序结果,并在属水平上汇总。
1.7 高ACE抑制活性乳杆菌的筛选 1.7.1 选择性富集分离乳杆菌取沉积物样品0.5 g,置于含20 mL MRS液体培养基的锥形瓶中(1×105 Pa灭菌15 min),37 ℃、160 r/min培养2-3 h,待沉积物与培养基混合均匀,将锥形瓶放置37 ℃静置培养,隔夜取出,采用稀释涂布平板法取梯度稀释液均匀涂布于MRS培养基上,37 ℃恒温厌氧培养24 h,挑取生长状态良好的菌株,反复划线分离,获得的纯培养菌株放置在4 ℃备用。
1.7.2 乳杆菌的筛选及鉴定采用溴甲酚绿指示剂法[48]和CaCO3溶钙圈法进行筛选,将经过纯化的疑似乳杆菌菌落进行革兰氏染色,并在显微镜下进行观察,进行生理生化鉴定[49],包括过氧化氢酶测定、明胶液化实验、H2S产生实验、吲哚实验和石蕊牛奶实验等。
1.7.3 16S rRNA基因扩增及系统发育分析参照提取说明书,使用UItraClean Microbial DNA Isolation Kit提取样品基因组。扩增引物采用通用引物[50]27f (5′-AGAGTTTGATCCTGGCTCA G-3′)和1492r (5′-CTACGGCTACCTTGTTACGA-3′)。PCR反应体系:10×PCR Rxn Buffer-MgCl2 2.50 μL,dNTPs (2.5 mmol/μL) 2.00 μL,引物27f (10 μmol/L) 0.50 μL,引物1492r (10 μmol/L) 0.50 μL,Taq DNA Polymrease (5 U/μL) 0.50 μL,模板1.5 μL,ddH2O 17.5 μL。PCR反应条件:94 ℃ 5 min;92 ℃ 30 s,48 ℃ 2 min,72 ℃ 1.5 min,25个循环;72 ℃ 10 min。样品由英潍捷基(上海)贸易有限公司测序。将测得的16S rRNA基因序列提交GenBank,并在NCBI应用BLAST程序在线与数据库中相似性高的相关菌株的16S rRNA基因序列进行比对。应用MEGA 6.0软件将测序获得的Lactobacillus helveticus GY-3菌株16S rRNA基因序列进行多序列完全比对,去掉两端差异序列后,采用Kimura-2参数,进行邻接法(Neighbor-Joining method)分析各类群间的核苷酸差异值,设置自展值(Bootstrap)为1 000,构建进化树。
1.7.4 发酵乳的制备选取具有典型乳杆菌特征的菌株,将活化好的瑞士乳杆菌按3% (体积比)接入已灭菌的脱脂乳培养基中,在37 ℃恒温静置发酵18-22 h,待发酵乳pH降至4.5左右,取发酵乳800 mL,6 000×g离心10 min,取上清,用5 mol/L NaOH调整pH至8.3,10 000×g离心10 min,取上清,用于检测ACE抑制活性。
1.7.5 发酵乳指标的检测(1) pH值的测定:采用精密pH计测定pH值。
(2) 总酸度(Total acidity,TA)的测定[51]:在5 g发酵乳中加入40 mL去CO2水,再加入0.5 mL 0.5%酚酞乙醇溶液,用浓度为0.1 mol/L的NaOH溶液滴定至红色,在一分钟内不褪色即为滴定终点。计算公式如下:
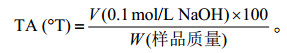
|
(3) OD600值的测定[52]:将0.5 mL发酵乳加入冰EDTA (0.2%,pH 12.0) 4.5 mL,振荡均匀,使用可见光分光光度计在600 nm处测定浊度。
(4) 游离氨基氮含量的测定[53]:将100 mmol/L的硼砂50 mL、20% (质量比)的十二烷基硫酸钠5 mL、80 mg的邻苯二甲醛(溶解于2 mL的甲醇)和200 μL β-巯基乙醇混合后用去离子水定容至100 mL配制成反应试剂后,取50 μL样品混合于2 mL反应试剂中,在室温下反应2 min后用紫外分光光度计在340 nm处进行检测。以胰蛋白胨为标准品确定抑制成分含量。
(5) ACE抑制活性的测定[54]:取2 mL离心管,加入5 mmol/L HHL为底物的50 mmol/L硼酸钠缓冲液50 μL,再加入20 μL待测乳清,混合,在37 ℃水浴中预热3 min,加入10 μL ACE酶液,在37 ℃反应30 min,再加入100 μL 1 mol/L盐酸终止反应。加入1.7 mL乙酸乙酯,振荡15 s,3 000×g离心10 min,取上层的1 mL乙酸乙酯,加热挥发去除有机溶剂,再加入0.3 mL去离子水,并振荡使马尿酸完全溶解,用微量紫外分光光度计在228 nm测定样品吸光值。ACE抑制率(ACEI)计算公式如下:
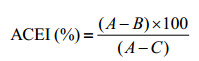
|
其中,A表示ACE与HHL反应完全生成马尿酸的吸光值(对照);B表示乳清与ACE和HHL反应后生成马尿酸的吸光值(样品);C表示先加入盐酸ACE和HHL反应后生成马尿酸的吸光值。
1.7.6 发酵培养基的优化分别对不同温度、接种量、碳源、氮源和盐离子条件下发酵乳ACE抑制活性进行测定,并确定ACE抑制活性最高的条件,同时设置3因素4水平的正交实验,分析其发酵的最适条件。
1.7.7 全基因组DNA提取、数据处理及生物信息学分析使用UltraClean Microbial DNA Isolation Kit提取Lactobacillus helveticus GY-3菌株全基因组,交由北京诺禾致源公司测序。将所获得的序列在QIIME (V.1.8.0)[46]和Mothur (V.1.36)[25]完成数据处理和分析,数据经过严格质控,仅保留高质量Reads,进一步使用SOAPdenovo (V2.04)[55]对所获得全基因组序列进行组装,并对结果进行评价,采用GeneMarkS (V4.17)[56]对基因组进行编码基因预测,通过RepeatMasker (Vopen-4.0.5)[57]和TRF (Tandem repeats finder,V4.07 b)[58]进行散在重复序列预测与搜寻串联重复序列等,同时采用多种数据库对编码基因序列进行功能注释。
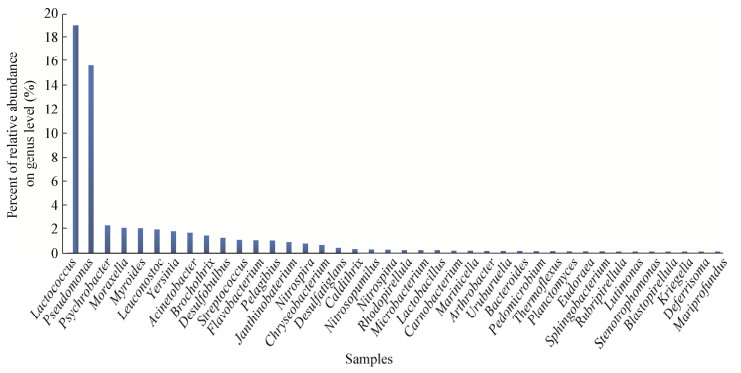
2 结果与分析 2.1 沉积物中细菌多样性分析如图 1所示,本次采样所获得渤海表层沉积物中的优势菌主要是乳球菌属(Lactococcus,18.77%)及假单胞菌属(Pseudomonas,15.6%)。其中乳杆菌属(Lactobacillus)所占相对丰度为0.24%。这表明渤海沉积物中存在比较丰富(相对丰度 > 0.1%)的乳杆菌菌种资源。
|
| 图 1 渤海沉积物中属水平上细菌组成 Figure 1 Bacterial composition on genus level in sediments of Bohai Sea 注:只显示相对丰度 > 0.1%的可鉴定属. Note: Species with relative abundance (> 1%) were chosen for the statistics. |
|
|
利用MRS溴甲酚绿固体培养基筛选法分离出14株疑似乳杆菌菌株,经过形态学观察和革兰氏染色后发现,共有9株潜在的乳杆菌菌株,对这9株菌进行过氧化氢酶测定、明胶液化实验、H2S产生实验、吲哚实验和石蕊牛奶实验,各项测定结果见表 1。
| 菌株编号 Strain No. |
过氧化氢酶测定 H2O2 enzyme |
明胶液化 Gelatin liquefaction |
H2S产生 Production of H2S |
吲哚实验 Indole experiment |
石蕊牛奶 Litmus milk |
| 1 | - | - | - | - | + |
| 2 | - | - | + | - | + |
| 3 | - | - | - | - | + |
| 4 | - | - | + | - | - |
| 5 | - | - | - | - | + |
| 6 | - | - | + | - | + |
| 7 | - | - | - | - | + |
| 8 | - | - | - | - | + |
| 9 | - | - | - | - | - |
| 注:+:反应呈阳性,即石蕊变红,有H2S产生;-:反应呈阴性,即明胶未液化,未产生吲哚,无H2S产生. Note: +: Positive results, such as the litmus turns red and H2S is produced; -: Negative results, such as gelatin is not liquefied and no indole or H2S is produced. |
|||||
结果显示,9株菌中有5株具有典型的乳杆菌菌落特征,其他4株菌本文将不做研究。
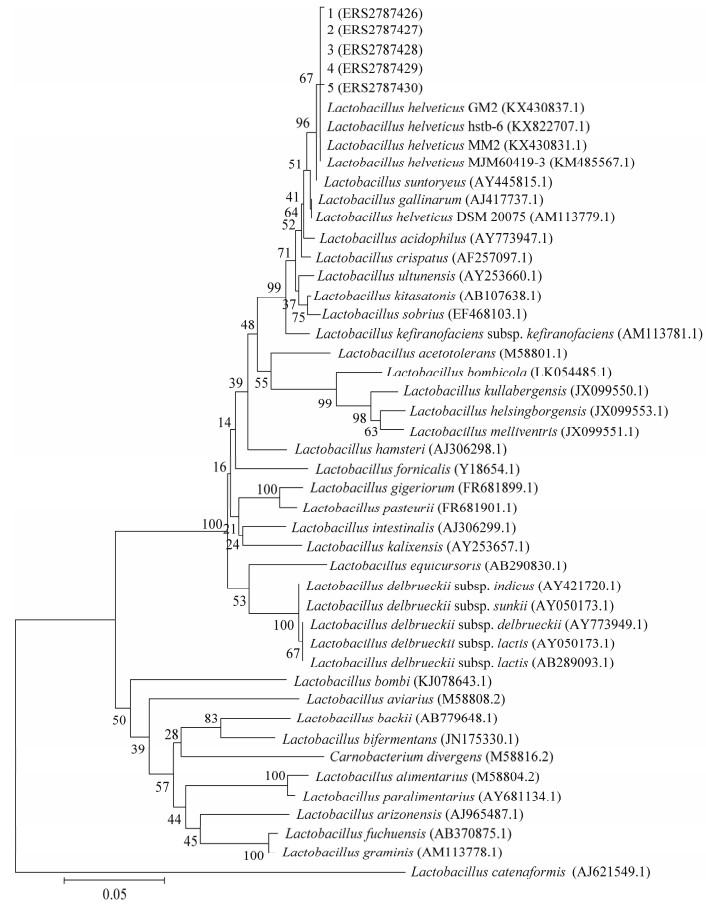
2.2.2 菌株的纯化及16S rRNA基因系统发育分析对上述5株菌进行生化实验鉴定,确定5株菌均为乳酸杆菌。为进一步确定菌种类型和名称,对5株乳杆菌分别编号,进行16S rRNA基因扩增及测序(已上传至ENA数据库,登录号ERS2787426- ERS2787430)。系统发育树分析如图 2所示。
|
| 图 2 通过邻接法构建的基于5株乳杆菌及相关菌株16S rRNA基因序列的系统发育树 Figure 2 The phylogenetic tree based on 16S rRNA gene sequences of five strains of Lactobacillus and other related strains, constructed by Neighbor-Joining method 注:括号中序号:相关菌株登录号;分支点上的数字:Bootstrap支持率;标尺刻度0.05:序列差异的分支长度. Note: Numbers in parentheses: The sequence accession numbers; Numbers in each branch points: The percentages supported by bootstrap; Bar=0.05: Nucleotide divergence. |
|
|
结果表明,上述5株乳杆菌具有革兰氏染色呈阳性、不液化明胶、能分解乳糖、能产酸、不产生H2S和H2O2等典型的乳杆菌特征,1-5号菌株均属于Lactobacillus,各菌株间相似性在99.8%-100%,与Lactobacillus helveticus (GenBank登录号为AM113779.1)相似性为99.3%,因此上述5株菌均属于瑞士乳杆菌(Lactobacillus helveticus)。
2.3 5株瑞士乳杆菌发酵乳ACE抑制活性检测将上述5株瑞士乳杆菌分别按3% (体积比)的接种量接种于已灭菌的脱脂乳培养基上,37 ℃恒温静置发酵,待发酵乳pH下降为4.5时终止发酵。对发酵乳的各项生化指标进行检测,以ACE抑制活性为标准,筛选出发酵乳ACE活性较高的菌株。5株瑞士乳杆菌ACE抑制活性及其他生化检测结果见表 2。
| 菌株编号 Strain No. |
发酵乳吸光值 OD600 |
滴定酸度TA (°T) |
游离氨基氮含量 Free amino nitrogen (mmol/L) |
ACE抑制活性 ACE inhibitory activity (%) |
pH | 发酵时间 Time (h) |
| 1 | 0.350±0.020 | 106.58±1.96 | 9.89±0.36 | 38.57±2.55 | 4.48±0.04 | 20.0 |
| 2 | 0.143±0.040 | 100.63±1.26 | 8.65±0.03 | 5.25±2.79 | 4.56±0.01 | 20.5 |
| 3 | 0.292±0.010 | 95.42±0.74 | 9.06±0.10 | 58.56±0.48 | 4.48±0.02 | 19.0 |
| 4 | 0.085±0.020 | 88.34±2.12 | 7.96±0.05 | 26.60±1.35 | 4.39±0.07 | 22.0 |
| 5 | 0.137±0.040 | 98.20±2.86 | 9.14±0.13 | 45.27±2.01 | 4.50±0.02 | 20.0 |
根据以上结果,3号菌株,即Lactobacillus helveticus GY-3的ACE抑制活性最高。
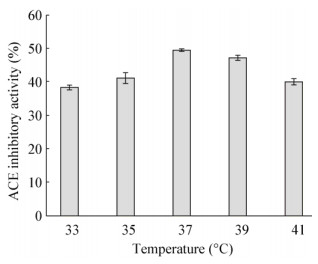
2.4 Lactobacillus helveticus GY-3发酵条件优化 2.4.1 不同发酵温度对ACE抑制活性的影响检测Lactobacillus helveticus GY-3在33、35、37、39、41℃不同温度下的发酵乳ACE抑制活性,结果见图 3。
|
| 图 3 不同温度对ACE抑制活性的影响 Figure 3 Effect of temperature on ACE inhibitory activity in fermented milk |
|
|
以上结果显示,温度≤37 ℃,随着温度的升高Lactobacillus helveticus GY-3发酵乳ACE抑制活性不断上升,37 ℃时活性达到最大值。而在超过37 ℃以后,随着温度的升高,ACE抑制活性并不再继续增大,而是开始减小。
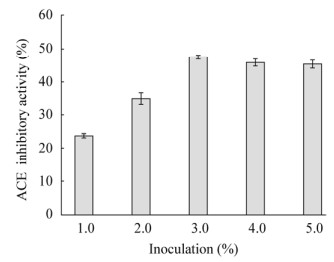
2.4.2 不同接种量对ACE抑制活性的影响在Lactobacillus helveticus GY-3接种量分别为1.0%、2.0%、3.0%、4.0%、5.0% (体积比)时进行发酵乳ACE活性检测,结果见图 4。
|
| 图 4 不同接种量对发酵乳ACE抑制活性的影响 Figure 4 Effect of inoculation on ACE inhibitory activity in fermented milk |
|
|
ACE抑制活性检测结果显示,Lactobacillus helveticus GY-3接种量为3.0%时,菌体和脱脂乳之间的配比较为适当,Lactobacillus helveticus GY-3发酵乳ACE抑制活性达到最大值。
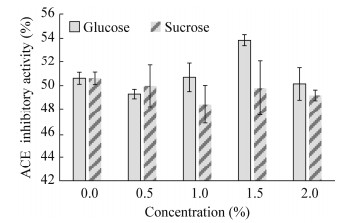
2.4.3 不同碳源对ACE抑制活性的影响研究不同碳源对Lactobacillus helveticus GY-3发酵乳ACE抑制活性的影响,结果见图 5。Lactobacillus helveticus GY-3在葡萄糖添加量为0.5%、1.0%、2.0% (质量体积比)时,ACE抑制活性没有明显增强,添加量为1.5% (质量体积比)时,ACE抑制活性最高。碳源为蔗糖时,添加量为0.5%、1.0%、1.5%、2.0% (质量体积比)时,ACE抑制活性均无明显的增加。
|
| 图 5 不同碳源对ACE抑制活性的影响 Figure 5 Effect of different concentrations of glucose and sucrose on ACE inhibitory activity |
|
|
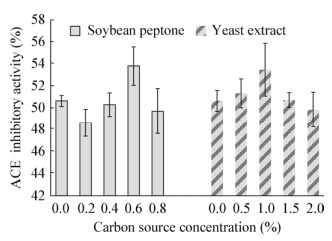
研究不同的氮源对Lactobacillus helveticus GY-3发酵乳ACE抑制活性的影响,结果见图 6。结果显示,在脱脂乳培养基中大豆蛋白胨为氮源时,添加0.6% (质量体积比)的大豆蛋白胨,Lactobacillus helveticus GY-3发酵乳ACE抑制活性最高。当以酵母浸粉为氮源时,添加0.5%-1.0%的酵母浸粉能提高ACE抑制活性,在添加1.0% (质量体积比)酵母浸粉时,发酵乳ACE抑制活性最高。
|
| 图 6 不同浓度氮源对ACE抑制活性的影响 Figure 6 Effect of different concentrations of soybean peptone and yeast extract on ACE inhibitory activity |
|
|
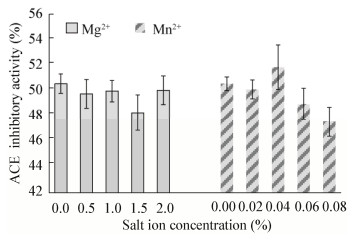
不同的盐离子对Lactobacillus helveticus GY-3发酵乳ACE抑制活性的影响结果见图 7。在脱脂乳培养基中添加不同浓度的MgSO4·7H2O,均未增强Lactobacillus helveticus GY-3发酵乳ACE抑制活性,表明MgSO4·7H2O对该菌发酵乳ACE抑制活性无明显影响。添加0.04%的MnSO4·4H2O时Lactobacillus helveticus GY-3发酵乳ACE抑制活性最高。
|
| 图 7 不同浓度盐离子对ACE抑制活性的影响 Figure 7 Effect of different concentrations of MgSO4·7H2O and MnSO4·4H2O on ACE inhibitory activity |
|
|
在脱脂乳培养基中添加不同的碳源、氮源、无机盐离子对Lactobacillus helveticus GY-3发酵乳ACE抑制活性有不同程度的影响,为了得出4种影响因素对Lactobacillus helveticus GY-3发酵乳ACE抑制活性影响程度的大小,及ACE抑制活性最高的发酵条件,以葡萄糖、大豆蛋白胨、酵母浸粉和MnSO4·4H2O及其浓度差异设计4因素3水平的正交试验。
从表 3分析结果可知,对Lactobacillus helveticus GY-3发酵乳ACE抑制活性具有促进作用的4项因素效应值大小为大豆蛋白胨 > 酵母浸粉 > 葡萄糖 > MnSO4·4H2O。其最佳发酵组合为在脱脂乳培养基中添加1.0%葡萄糖,0.6%大豆蛋白胨,1.0%酵母浸粉,0.04% MnSO4·4H2O,ACE抑制活性可达79.52%,提高28.84%。
| 序号 No. |
葡萄糖 Glucose (g/100 mL) |
大豆蛋白胨 Soy peptone (g/100 mL) |
酵母浸粉 Yeast extract (g/100 mL) |
MnSO4·4H2O (g/100 mL) |
ACE抑制活性 ACE inhibitory activity (%) |
| 1 | 1 (1.0%) | 1 (0.4%) | 1 (0.5%) | 1 (0.02%) | 65.03±2.01 |
| 2 | 1 (1.0%) | 2 (0.6%) | 2 (1.0%) | 2 (0.04%) | 79.52±1.24 |
| 3 | 1 (1.0%) | 3 (0.8%) | 3 (1.5%) | 3 (0.06%) | 61.14±1.64 |
| 4 | 2 (1.5%) | 1 (0.4%) | 2 (1.0%) | 3 (0.06%) | 76.73±0.78 |
| 5 | 2 (1.5%) | 2 (0.6%) | 3 (1.5%) | 1 (0.02%) | 66.37±1.58 |
| 6 | 2 (1.5%) | 3 (0.8%) | 1 (0.5%) | 2 (0.04%) | 53.31±1.35 |
| 7 | 3 (2.0%) | 1 (0.4%) | 3 (1.5%) | 2 (0.04%) | 63.67±2.20 |
| 8 | 3 (2.0%) | 2 (0.6%) | 1 (0.5%) | 3 (0.06%) | 62.89±3.12 |
| 9 | 3 (2.0%) | 3 (0.8%) | 2 (1.0%) | 1 (0.02%) | 52.75±1.03 |
| Average 1 | 68.563 | 68.477 | 60.410 | 61.383 | |
| Average 2 | 65.470 | 69.593 | 69.667 | 65.500 | |
| Average 3 | 59.770 | 55.733 | 63.727 | 66.920 | |
| Range | 8.793 | 13.860 | 9.257 | 5.537 |
将文献中乳杆菌发酵乳ACEI与Lactobacillus helveticus GY-3进行比较。由表 4中可以看出,优化条件下菌株GY-3的ACE抑制活性(79.52%)比大部分乳杆菌菌株高,仅有菌株Lactobacillus casei 17与Lactobacillus delbrueckii QS701的ACEI稍高,但二者发酵时间均在24 h以上,长于GY-3的18 h,因此发酵优化后的Lactobacillus helveticus GY-3具有较为优越的ACE抑制活性。
| Name | Fermentation conditions | ACE inhibitory activity (%) |
||||
| Inoculum (%) | Time (h) | Temperature (℃) | Supplementation | pH | ||
| Lb. helveticus GY-3 | 3 | 18 | 37 | Glucose 1.0% (V/V) | - | 79.52±1.24 |
| Soy peptone 0.6% (V/V) | ||||||
| Yeast extract 1.0% (V/V) | ||||||
| MnSO4·4H2O 0.04% (V/V) | ||||||
| Lb. acidophilus ATCC 4356 | 3 | 24 | - | 3% Caseinate | 6.0 | 74.00 |
| Lb. raffinolactis ATCC 43920 | - | 44 | - | - | - | 19.00 |
| Lb. casei 17 | 3 | 24 | - | 1% Lactose+Lactase | 4.2 | 80.00 |
| L. casei 34143S | 1 | 24 | 30 | - | 3.8 | 34.80±1.80 |
| L. casei S-3-1 | 1 | 24 | 30 | - | 3.8 | 33.90±0.50 |
| L. casei 6-2-05 | 1 | 24 | 30 | - | 3.7 | 30.40±3.00 |
| Lb. curvatus ATCC 25601 | - | 44 | - | - | - | 5.00 |
| Lb. helveticus 89 | - | 44 | - | - | - | 31.00 |
| Lb. helveticus ATCC 15009 | - | 44 | - | - | - | 32.00 |
| Lb. helveticus CNRZ32 | 3 | 24 | - | - | 3.9 | 37.00 |
| Lb. jensenii ATCC 25258 | 3 | 24 | - | - | 5.4 | 72.00 |
| Lb. rhamnosus ATCC 7469 | - | 44 | - | - | - | 14.00 |
| Lb. reuteri ATCC 55730 | - | 44 | - | - | - | 19.00 |
| Lb. reuteri ATCC 23272 | - | 44 | - | - | - | 19.00 |
| Lb. delbrueckii QS701 | 3 | 72 | 43 | - | - | 81.58 |
| Lb. plantarum 6214 | 1 | 24 | 30 | - | 3.6 | 40.60±3.40 |
| Lb. helveticus ND01 | 3 | 24 | 37 | - | 3.5 | 69.51±2.32 |
| Lb. helveticus H-1 | 4 | 18 | 30 | - | - | 33.07 |
将获得的Lactobacillus helveticus GY-3全基因组测序信息进行组装,组装前采用基于K-mer统计的分析方法来估计基因组大小,得到样品的基因组大小约为2.38 Mb。组装后将Reads比对到组装好的基因组序列上,通过统计组装序列(G+C)mol%含量和Reads覆盖深度,总结基因组的(G+C)mol%偏向性和重复序列情况。Lactobacillus helveticus GY-3的全基因组长度为2 022 469 bp,(G+C)mol%含量为37.22%,包括2 159个编码基因,序列总长度为1 730 880 bp,占基因总长度的85.58%。
2.5.2 基因组组分分析Lactobacillus helveticus GY-3的基因组中共含有散在重复序列151个,tRNA 61个,5S rRNA 3个,16S rRNA和23S rRNA各1个,sRNA 31个。
2.5.3 基因注释从细胞学组件、分子功能及生物学途径采用不同数据库对基因组信息进行注释。GO数据库[59]对1 423个基因进行了注释;KEGG数据库[60-61]从细胞过程、遗传信息处理及代谢相关基因进行注释,得到577个功能基因;而在蛋白数据库COG[62]中1 278个基因被注释。NR数据库[63]物种注释统计显示,Lactobacillus helveticus GY-3与瑞士乳杆菌Lactobacillus helveticus基因最为匹配。
2.5.4 ACE抑制肽相关基因分析将所测序的基因组信息进行注释,Lactobacillus helveticus GY-3与ACE抑制肽产生有关的基因及信息见表 4。该菌株含有与ACE抑制肽产生相关的全部重要功能基因,包括胞壁结合蛋白(PrtH4)、多肽转运系统(OppB1)及肽酶系统(PepT、PepD、PepO3、PepE、PepO、PepF、PepI、PepCE等)。Lactobacillus helveticus GY-3具有强大的水解功能及有利于ACE抑制肽保留的肽酶系统组成。已将所有ACE抑制肽相关基因原始序列上传至NCBI数据库,获得的序列号为MK675035- MK675051,具体信息见表 5。
| 注释编号 Annotation No. |
序列号 Accession No. |
参考序列 Reference sequence |
相似性 Identity (%) |
描述 Description |
系统 System |
| LAC_GM001726 | MK675035 | gi|504731207|ref| WP_014918309.1 |
100.0 | Lactocepin H4 proteinase PrtH4 [Lactobacillus helveticus R0052] | Cell wall binding protein |
| LAC_GM000899 | MK675036 | gi|504731495|ref| WP_014918597.1 |
100.0 | ATP-binding cassette domain-containing protein [Lactobacillus helveticus R0052] | Polypeptide transport system |
| LAC_GM000900 | MK675037 | gi|501168412|ref| WP_012212411.1 |
100.0 | Oligopeptide ABC transport protein ATP-binding component OppB1 [Lactobacillus helveticus CNRZ32] | Polypeptide transport system |
| LAC_GM000901 | MK675038 | gi|504731496|ref| WP_014918598.1 |
100.0 | Oligopeptide ABC transporter permease [Lactobacillus helveticus R0052] | Polypeptide transport system |
| LAC_GM000902 | MK675039 | gi|504731497|ref| WP_014918599.1 |
100.0 | Oligopeptide ABC transporter substrate-binding protein [Lactobacillus helveticus] | Polypeptide transport system |
| LAC_GM000898 | MK675040 | gi|504731494|ref| WP_014918596.1 |
99.7 | Peptide ABC transporter ATPase [Lactobacillus helveticus] | Polypeptide transport system |
| LAC_GM001735 | MK675041 | gi|504730965|ref| WP_014918067.1 |
100.0 | Pro-Xaa aminopeptidase I PepI [Lactobacillus helveticus R0052] | Peptide enzyme system |
| LAC_GM001872 | MK675042 | gi|504377167|ref| WP_014564269.1 |
100.0 | Peptidase M1, membrane alanine aminopeptidase-like protein [Lactobacillus helveticus H10] | Peptide enzyme system |
| LAC_GM000032 | MK675043 | gi|578023951|gb| AHI12348.1| |
100.0 | Aminopeptidase N [Lactobacillus helveticus H9] | Peptide enzyme system |
| LAC_GM000420 | MK675044 | gi|489723717|ref| WP_003627833.1 |
100.0 | MULTISPECIES: prolyl aminopeptidase [Lactobacillus|Lactobacillus helveticus] | Peptide enzyme system |
| LAC_GM000946 | MK675045 | gi|504731058|ref| WP_014918160.1 |
100.0 | Oligoendopeptidase E PepE [Lactobacillus helveticus R0052] | Peptide enzyme system |
| LAC_GM001534 | MK675046 | gi|504732346|ref| WP_014919448.1 |
100.0 | Oligoendopeptidase F PepF [Lactobacillus helveticus] | Peptide enzyme system |
| LAC_GM000490 | MK675047 | gi|504731022|ref| WP_014918124.1 |
100.0 | Endopeptidase O3 PepO3 [Lactobacillus helveticus R0052] | Peptide enzyme system |
| LAC_GM001839 | MK675048 | gi|504731848|ref| WP_014918950.1 |
100.0 | Peptidase CE PepCE [Lactobacillus helveticus R0052] | Peptide enzyme system |
| LAC_GM001046 | MK675049 | gi|504731458|ref| WP_014918560.1 |
100.0 | x-prolyl-dipeptidyl aminopeptidase [Lactobacillus helveticus] | Peptide enzyme system |
| LAC_GM000966 | MK675050 | gi|489729178|ref| WP_003633294.1 |
99.6 | MULTISPECIES: methionine aminopeptidase [Lactobacillus|Lactobacillus helveticus] | Peptide enzyme system |
| LAC_GM000834 | MK675051 | gi|578023095|gb| AHI11492.1| |
98.9 | Aminopeptidase I zinc metalloprotease [Lactobacillus helveticus H9] | Peptide enzyme system |
瑞士乳杆菌对乳蛋白的水解作用可概括为以下三步:(1)在细菌细胞外进行,细菌通过分泌蛋白酶将酪蛋白剪切成多肽;(2)将第一步产物,即多肽经转运系统运输进入细胞内;(3)在细菌细胞内进行,在胞内酶的作用下多肽被进一步水解成氨基酸。
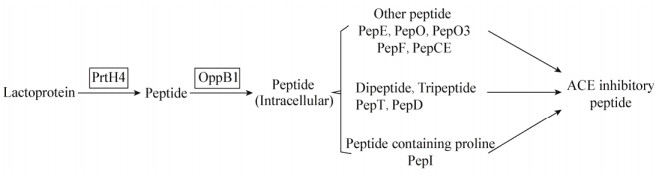
由此可知,乳蛋白在Lactobacillus helveticus GY-3 CEP系统(PrtH4)作用下水解为一系列多肽,再由多肽转运系统(OppB1)运输至细胞内,在细胞内由肽酶系统(PepT、PepD、PepO3、PepE、PepO、PepF、PepI、PepCE等)的作用下进一步水解为具有降血压活性的ACE抑制肽,其基因通路如图 8所示。
|
| 图 8 Lactobacillus helveticus GY-3产ACE抑制肽基因通路 Figure 8 The gene pathway of biosynthesis of ACE inhibitory peptide in Lactobacillus helveticus GY-3 |
|
|
本文对来自渤海M5站位的沉积物细菌群落组成进行探索,通过未培养方法对沉积物中乳杆菌的多样性及相对丰度进行分析,结果显示渤海沉积物中有较为丰富的乳杆菌菌种资源。基于此,我们从沉积物中富集并选择性分离出5株典型乳杆菌,对其进行发酵乳ACE抑制活性检测,筛选出其中发酵乳ACE抑制活性最高的菌株Lactobacillus helveticus GY-3。对该菌发酵条件进行优化并进行正交实验,得到其ACE抑制活性最高的发酵乳条件为:接种量3.0% (体积比),发酵温度37 ℃,培养基中添加1.0% (体积比)的葡萄糖、0.6% (体积比)的大豆蛋白胨、1.0% (体积比)的酵母浸粉和0.04% (体积比)的MnSO4·4H2O,恒温发酵18-22 h,ACE抑制率最高,可达79.52%,比未优化提高了28.84%。Donkor等[64]曾对益生菌发酵酸奶中的ACE活性抑制率进行检测,发现大部分菌株发酵乳在28 d的发酵期内ACEI在70%-90%之间,且在7 d内随发酵时间增长,ACE抑制率逐渐上升,该研究还针对相应酶的体外实验进行了ACE抑制率测定,结果约在39%-75%之间。Pihlanto等[65]对25株乳酸菌进行体外发酵实验,经过44 h的发酵产生ACE抑制活性最高的为Lactobacillus casei 17,可达74%,而瑞士乳杆菌属的几株菌的ACEI均在30%-40%间,经过优化后,菌株Lactobacillus casei在添加1%乳糖和乳糖酶的牛奶中产ACE抑制肽效率高达80%。Luo等[66]对沙丁鱼肽(Sardine peptides,SP)和槲皮素(Quer cetin,QC)及其复合物对发酵乳中ACEI的影响进行了研究,发现二者的复合物(体积比=8:2)能有效提高ACEI至93%,比单独添加SP或QC分别提高了19.56%和44.44%。Kim等[67]对发酵时间影响ACEI进行了研究,结果表明发酵时间在18-24个月间的发酵乳抑制活性最高,这与Park等[68]的研究结果是一致的。这说明菌株Lactobacillus helveticus GY-3的ACEI在所有已报道菌株中处于中等偏上,在已报道海洋细菌中处于较高水平。
本研究还对菌株Lactobacillus helveticus GY-3进行了全基因组测序分析,结果显示其拥有完整的产ACE抑制肽所需的基因,包括胞壁结合蛋白基因(PrtH4)、多肽转运系统基因(OppB1)及肽酶系统基因(PepT、PepD、PepO3、PepE、PepO、PepF、PepI、PepCE等)。
已有研究表明,不同乳酸菌ACE抑制活性有显著地差异。Yamamoto等[34]和王宇[69]的研究中对不同种属的乳酸菌发酵乳进行ACE抑制活性研究,结果表明并不是所有的乳酸菌发酵乳都具有降血压的功能,乳杆菌属具有高ACE抑制活性。本研究表明海洋可以作为新的乳杆菌菌种资源库,为开发高效产ACE抑制肽的乳杆菌提供了更为广阔的来源。随着研究的深入,高产ACE抑制肽的乳杆菌的遗传信息以及底物促进乳杆菌ACE抑制活性的机理逐步被揭示。其中,Lactobacillus helveticus GY-3将乳蛋白水解并产生ACE抑制肽的过程如下:(1)通过CEP (主要为PrtH4,最早作为额外推定基因被报道)实现水解乳蛋白的初步水解[70-72]。研究表明,乳杆菌属ACE抑制活性相对较高,其中瑞士乳杆菌和干酪乳杆菌的ACE抑制活性最为突出[34, 73],大多数乳酸球菌自身蛋白水解能力较弱,在乳中生长能力缓慢,所以释放ACE抑制肽的能力较差[69]。Yamamoto等[34]认为这可能与不同种属乳酸菌产生胞外蛋白酶的能力有关。Fung等[74]的实验也同样显示乳酸菌蛋白水解活性与ACE抑制活性高度相关。(2)经过CEP系统水解的多肽由多肽转运系统运送至细胞内。研究表明,不能表达编码Opp系统的基因突变菌株不能实现多肽的转运,也不能在乳中生长,Opp寡肽转运系统对于乳酸菌的蛋白水解体系是最重要的。Lactobacillus helveticus GY-3中的多肽转运系统为OppB1,可将多肽运输到细胞内进行进一步水解。(3)在细胞内,Lactobacillus helveticus GY-3的肽酶系统(PepT、PepD、PepO3、PepE、PepO、PepF、PepI、PepCE等)将多肽进一步水解为具有降血压活性的ACE抑制肽。在肽酶系统中,一些二肽酶属于脯氨酰氨基酸二肽酶或氨酰基脯氨酸二肽酶,主要作用于N-端或C-端具有脯氨酸的肽[75-76]。研究表明,脯氨酸含量和含脯氨酸的肽的数量与ACE抑制活性的大小具有一定关系。在已被发现的降血压肽中,大多C-端含有脯氨酸[77]。Cheung等[78]的研究也表明ACE抑制肽的活性大小主要取决于C-端氨基酸,C-端氨基酸为脯氨酸和芳香族氨基酸(包括色氨酸、酪氨酸、苯丙氨酸)时其ACE抑制活性较高。Lactobacillus helveticus GY-3基因测序结果显示,其编码二肽和三肽基因含量较少,在细胞内的水解作用中保留了较多N-端或C-端含有脯氨酸的肽。Kilpi等[79]分别将Lactobacillus helveticus CNRZ32中总氨基肽酶(PepN)和x-脯氨酰基-二肽基-氨肽酶(PepX)去除,获得两株基因缺陷菌株,经过基因改造后的菌株ACE抑制活性明显高于原始菌株,由此表明PepN和PepX与ACE抑制肽的进一步水解密切相关。而在Lactobacillus helveticus GY-3与ACE抑制肽产生有关的基因中,并未检测到PepN和PepX两种基因。因此,Lactobacillus helveticus GY-3肽酶系统的编码基因组成及由此编码的蛋白更有利于ACE抑制肽的保留。
此外,大多数瑞士乳杆菌CEPs的表观大小在170-180 kD范围内,且所研究的所有瑞士乳杆菌CEP几乎都属于丝氨酸蛋白酶家族,但其中CEPs的特异性不同,这使CEP的活性位点如何与这些各种氨基酸结构相互作用成为疑问。自催化过程移除的前结构域(PP,约150 aa)起伴侣的作用,保证CEP正确折叠。CEP的特征在于细胞表面位置,这是由于催化结构域序列下游的长延伸。在PrtH和PrtH2中,C-末端延伸由A、B、H和W四个结构域构成。Leenhouts等[80]描述了乳酸菌细胞表面的5种蛋白质锚定模式:跨膜锚、脂蛋白锚、LPxTG基序锚、AcmA型蛋白锚和表层蛋白锚,因此在CEP锚定中可能涉及两种类型的机制:CEP LPxTG基序与细菌壁肽聚糖的肽键之间的共价结合或CEP C-末端的正电荷与细菌壁的带负电的磷壁酸之间的静电相互作用。来自瑞士乳杆菌的CEP难以提取和纯化,并且迅速失活[81]。实验测定了3种状态的CEP活性,即嵌入细菌包膜中的CEP[82-87]、从细胞包膜中提取的CEP[83, 88]和纯化的CEP[81, 89-92]。瑞士乳杆菌中CEP按全细胞、包膜提取物到纯化的CEP的顺序活性状态依次下降[93-94],但Hebert等则发现细胞包膜组分中CEP的活性比全细胞中更高[88]。通常,CEP活性在指数生长期最大,然后逐渐降低,这可能是由于CEP释放到培养基中或已降解[88, 95-97]。同时,CEP对发酵乳的风味也有至关重要的作用[98]。本实验中所筛选到的瑞士乳杆菌中含有较为少见的PrtH4,今后可进行更多相关分子机制方面的研究,相信能更进一步了解瑞士乳杆菌发酵产生ACE抑制肽的机制。
在基于本研究基础上,结合生物信息学更加深入的对基因调控机制进行分析,从而达到利用代谢调控手段进一步提高Lactobacillus helveticus GY-3产ACE抑制肽的水平。在体内外安全试验的基础上,开发基于Lactobacillus helveticus GY-3的发酵乳产品,达到研产结合。另外,有大量研究表明,菌种混合发酵可能产生更高的ACE抑制活性或更好的发酵乳品质,今后可以对Lactobacillus helveticus GY-3进行复合菌种发酵,以期获得更好的发酵乳品质以及更高的产ACE抑制肽的水平,提升发酵乳产品品质。关于Lactobacillus acidophilus L10发酵豆乳的研究结果显示,延长保藏时间同样可以提高发酵乳对ACE抑制率[99]。因此能否合理延长保藏时间,能否适应低温发酵,是否具有抗氧化作用,对提升发酵乳品质、延长货架期等方面均有重要的意义,这也是对Lactobacillus helveticus GY-3提出的新的要求,对于研发以该菌为主要发酵菌种的发酵乳有着重要的意义。今后可据此展开进一步的研究,使该类发酵乳产品更具有市场竞争力,更好地实现研产结合的效益最大化。
| [1] |
Sang SD, Wang J. Current status and prevention and treatment of basic hypertension in China[J]. Women's Health Research, 2017(23): 33-34. (in Chinese) 桑树东, 王锦. 我国基层高血压病的现状与防治进展[J]. 中外女性健康研究, 2017(23): 33-34. DOI:10.3969/j.issn.2096-0417.2017.23.014 |
| [2] |
Lei Y, Fan ZC. Current status and treatment progress of hypertension in China[J]. Journal of Modern Medicine & Health, 2015, 31(13): 1959-1962. (in Chinese) 雷宇, 范忠才. 我国高血压的现况与治疗进展[J]. 现代医药卫生, 2015, 31(13): 1959-1962. DOI:10.3969/j.issn.1009-5519.2015.13.012 |
| [3] |
Du HX. Therapeutic effect of valsartan and enalapril on QT interval dispersion and premature ventricular contraction in patients with hypertension[J]. Chinese Journal of Cardiovascular Rehabilitation Medicine, 2017, 26(3): 295-299. (in Chinese) 杜海霞. 缬沙坦与依那普利对高血压患者QT间期离散度与室性早搏的疗效[J]. 心血管康复医学杂志, 2017, 26(3): 295-299. DOI:10.3969/j.issn.1008-0074.2017.03.018 |
| [4] |
Cai T, Huang XL, Zeng YB, et al. Meta-analysis on risk factors of hypertension among Chinese[J]. Modern Preventive Medicine, 2015, 42(5): 831-836. (in Chinese) 蔡婷, 黄晓玲, 曾雁冰, 等. 中国居民高血压的危险因素Meta分析[J]. 现代预防医学, 2015, 42(5): 831-836. |
| [5] |
Ondetti MA, Williams NJ, Sabo EF, et al. Angiotensin-converting enzyme inhibitors from the venom of Bothrops jararaca. Isolation, elucidation of structure, and synthesis[J]. Biochemistry, 1971, 10(22): 4033-4039. DOI:10.1021/bi00798a004 |
| [6] |
Lee JK, Hong S, Jeon JK, et al. Purification and characterization of angiotensin I converting enzyme inhibitory peptides from the rotifer, Brachionus rotundiformis[J]. Bioresource Technology, 2009, 100(21): 5255-5259. DOI:10.1016/j.biortech.2009.05.057 |
| [7] |
Song HZ, Bi L, Lv S, et al. Antihypertensive effects of angiotensin-converting enzyme (ACE) inhibitory peptides from channel catfish skin gelatin[J]. Modern Food Science and Technology, 2014, 30(2): 78-83. (in Chinese) 宋华曾, 毕琳, 吕顺, 等. 鮰鱼皮明胶ACE抑制肽降血压活性的研究[J]. 现代食品科技, 2014, 30(2): 78-83. |
| [8] |
Ibrahim HR, Ahmed AS, Miyata T. Novel angiotensin-converting enzyme inhibitory peptides from caseins and whey proteins of goat milk[J]. Journal of Advanced Research, 2017, 8(1): 63-71. |
| [9] |
Miguel M, Aleixandre A. Antihypertensive peptides derived from egg proteins[J]. The Journal of Nutrition, 2006, 136(6): 1457-1460. DOI:10.1093/jn/136.6.1457 |
| [10] |
Daskaya-Dikmen C, Yucetepe A, Karbancioglu-Guler F, et al. Angiotensin-I-converting enzyme (ACE)-inhibitory peptides from plants[J]. Nutrients, 2017, 9(4): E316. DOI:10.3390/nu9040316 |
| [11] |
Persson IA, Persson K, Hägg S, et al. Effects of cocoa extract and dark chocolate on angiotensin-converting enzyme and nitric oxide in human endothelial cells and healthy volunteers—a nutrigenomics perspective[J]. Journal of Cardiovascular Pharmacology, 2011, 57(1): 44-50. DOI:10.1097/FJC.0b013e3181fe62e3 |
| [12] |
Akinyemi AJ, Ademiluyi AO, Oboh G. Aqueous extracts of two varieties of ginger (Zingiber officinale) inhibit angiotensin I-converting enzyme, iron (Ⅱ), and sodium nitroprusside-induced lipid peroxidation in the rat heart in vitro[J]. Journal of Medicinal Food, 2013, 16(7): 641-646. DOI:10.1089/jmf.2012.0022 |
| [13] |
Dong JJ, Xu XQ, Liang YR, et al. Inhibition of angiotensin converting enzyme (ACE) activity by polyphenols from tea (Camellia sinensis) and links to processing method[J]. Food & Function, 2011, 2(6): 310-319. |
| [14] |
Ademiluyi AO, Oboh G. Phenolic-rich extracts from selected tropical underutilized legumes inhibit α-amylase, α-glucosidase, and angiotensin I converting enzyme in vitro[J]. Journal of Basic and Clinical Physiology and Pharmacology, 2012, 23(1): 17-25. |
| [15] |
Persson IAL, Persson K, Andersson RGG. Effect of Vaccinium myrtillus and its polyphenols on angiotensin-converting enzyme activity in human endothelial cells[J]. Journal of Agricultural and Food Chemistry, 2009, 57(11): 4626-4629. DOI:10.1021/jf900128s |
| [16] |
Lee DH, Kim JH, Park JS, et al. Isolation and characterization of a novel angiotensin I-converting enzyme inhibitory peptide derived from the edible mushroom Tricholoma giganteum[J]. Peptides, 2004, 25(4): 621-627. DOI:10.1016/j.peptides.2004.01.015 |
| [17] |
Actis-Goretta L, Ottaviani JI, Fraga CG. Inhibition of angiotensin converting enzyme activity by flavanol-rich foods[J]. Journal of Agricultural and Food Chemistry, 2006, 54(1): 229-234. DOI:10.1021/jf052263o |
| [18] |
Al Shukor N, van Camp J, Gonzales GB, et al. Angiotensin-converting enzyme inhibitory effects by plant phenolic compounds: a study of structure activity relationships[J]. Journal of Agricultural and Food Chemistry, 2013, 61(48): 11832-11839. DOI:10.1021/jf404641v |
| [19] |
Wang W, Chen W, Yang YS, et al. New phenolic compounds from Coreopsis tinctoria Nutt. and their antioxidant and angiotensin I-converting enzyme inhibitory activities[J]. Journal of Agricultural and Food Chemistry, 2015, 63(1): 200-207. |
| [20] |
Zhao HZ, Lu ZX, Liu ZM. Research progress of angiotensin-I converting enzyme inhibitory peptides[J]. Chinese Journal of Biochemical Pharmaceutics, 2004, 25(5): 315-317. (in Chinese) 赵海珍, 陆兆新, 刘战民. 天然食品来源的血管紧张素转换酶抑制肽的研究进展[J]. 中国生化药物杂志, 2004, 25(5): 315-317. DOI:10.3969/j.issn.1005-1678.2004.05.021 |
| [21] |
You X. Functional Fermented Products[M]. Beijing: China Light Industry Press, 2000: 215-224. (in Chinese) 尤新. 功能性发酵制品[M]. 北京: 中国轻工业出版社, 2000: 215-224. |
| [22] |
Chen YF, Liu WJ, Xue JG, et al. Angiotensin-converting enzyme inhibitory activity of Lactobacillus helveticus strains from traditional fermented dairy foods and antihypertensive effect of fermented milk of strain H9[J]. Journal of Dairy Science, 2014, 97(11): 6680-6692. DOI:10.3168/jds.2014-7962 |
| [23] |
Ramchandran L, Shah NP. Influence of addition of Raftiline HP® on the growth, proteolytic, ACE-and α-glucosidase inhibitory activities of selected lactic acid bacteria and Bifidobacterium[J]. LWT-Food Science and Technology, 2010, 43(1): 146-152. |
| [24] |
Ge ST, Zhou LP, Ding X. Screening and identification of a lactic acid bacterium for γ-aminobutyric acid production[J]. Modern Food Science and Technology, 2009, 25(8): 940-943. (in Chinese) 葛松涛, 周立平, 丁兴. 一株产GABA的植物乳酸菌的筛选及鉴定[J]. 现代食品科技, 2009, 25(8): 940-943. |
| [25] |
Hamme V, Sannier F, Piot JM, et al. Crude goat whey fermentation by Kluyveromyces marxianus and Lactobacillus rhamnosus: contribution to proteolysis and ACE inhibitory activity[J]. Journal of Dairy Research, 2009, 76(2): 152-157. DOI:10.1017/S0022029908003877 |
| [26] |
Kunji ERS, Mierau I, Hagting A, et al. The proteotytic systems of lactic acid bacteria[J]. Antonie van Leeuwenhoek, 1996, 70(2/4): 187-221. |
| [27] |
Savijoki K, Ingmer H, Varmanen P. Proteolytic systems of lactic acid bacteria[J]. Applied Microbiology and Biotechnology, 2006, 71(4): 394-406. |
| [28] |
Fox PF, Wallace JM. Formation of flavor compounds in cheese[J]. Advances in Applied Microbiology, 1997, 45: 17-85. DOI:10.1016/S0065-2164(08)70261-2 |
| [29] |
Law J, Haandrikman A. Proteolytic enzymes of lactic acid bacteria[J]. International Dairy Journal, 1997, 7(1): 1-11. DOI:10.1016/0958-6946(95)00073-9 |
| [30] |
Hartmann R, Meisel H. Food-derived peptides with biological activity: from research to food applications[J]. Current Opinion in Biotechnology, 2007, 18(2): 163-169. DOI:10.1016/j.copbio.2007.01.013 |
| [31] |
Hata Y, Yamamoto M, Ohni M, et al. A placebo-controlled study of the effect of sour milk on blood pressure in hypertensive subjects[J]. The American Journal of Clinical Nutrition, 1996, 64(5): 767-771. DOI:10.1093/ajcn/64.5.767 |
| [32] |
Seppo L, Jauhiainen T, Poussa T, et al. A fermented milk high in bioactive peptides has a blood pressure-lowering effect in hypertensive subjects[J]. The American Journal of Clinical Nutrition, 2003, 77(2): 326-330. DOI:10.1093/ajcn/77.2.326 |
| [33] |
Takano T. Anti-hypertensive activity of fermented dairy products containing biogenic peptides[J]. Antonie van Leeuwenhoek, 2002, 82(1/4): 333-340. DOI:10.1023/A:1020600119907 |
| [34] |
Yamamoto N, Akino A, Takano T. Antihypertensive effects of different kinds of fermented milk in spontaneously hypertensive rats[J]. Bioscience, Biotechnology, and Biochemistry, 1994, 58(4): 776-778. DOI:10.1271/bbb.58.776 |
| [35] |
Lin XZ, Shen JH, Du N, et al. Screening and phylogenetic analysis of petroleum-degrading bacteria in sediments from the Arctic Ocean[J]. Acta Scientiae Circumstantiae, 2009, 29(3): 536-541. (in Chinese) 林学政, 沈继红, 杜宁, 等. 北极海洋沉积物石油降解菌的筛选及系统发育分析[J]. 环境科学学报, 2009, 29(3): 536-541. DOI:10.3321/j.issn:0253-2468.2009.03.011 |
| [36] |
Xia YL, Cheng LH, Li H. Screening and identification of the marine bacteria with antitumor activity from marine sediment of the South China Sea[J]. Journal of Guangdong Pharmaceutical University, 2014, 30(1): 76-80. (in Chinese) 夏玉林, 程丽慧, 李荷. 南海海洋沉积物中抗肿瘤活性菌株的初步筛选与鉴定[J]. 广东药学院学报, 2014, 30(1): 76-80. DOI:10.3969/j.issn.1006-8783.2014.01.017 |
| [37] |
Reyes-Becerril M, Ascencio-Valle F, Macias ME, et al. Effects of marine silages enriched with Lactobacillus sakei 5-4 on haemato-immunological and growth response in Pacific red snapper (Lutjanus peru) exposed to Aeromonas veronii[J]. Fish & Shellfish Immunology, 2012, 33(4): 984-992. |
| [38] |
Chen G, Yu LJ, Li ZX, et al. Beneficial effect of Lactobacillus helveticus and its combination with other probiotics on the growth and survival of juvenile turbots (Scophthalmus maximus)[J]. Chinese Journal of Microecology, 2016, 28(6): 639-644. (in Chinese) 陈刚, 于乐军, 李振兴, 等. 瑞士乳杆菌和几种有益微生物对大菱鲆幼鱼生长及成活率的影响[J]. 中国微生态学杂志, 2016, 28(6): 639-644. |
| [39] |
Liu D, Dong XH, Tan BP, et al. Effects of Lactobacillus acidophilus on growth performance, immunity and disease resistance in Pacific white leg shrimp Litopenaeus vannamei[J]. Fisheries Science, 2016, 35(1): 37-42. (in Chinese) 刘迪, 董晓慧, 谭北平, 等. 嗜酸乳杆菌对凡纳滨对虾生长、免疫力和抗病力的影响[J]. 水产科学, 2016, 35(1): 37-42. |
| [40] |
Ajitha S. Role of Bacillus and Lactobacillus from marine environment for sustainable aquaculture practices[D]. India: Doctoral Dissertation of Central Marine Fisheries Research Institute, 2004
|
| [41] |
Ao XL, Pu B, Cai YM, et al. Research progress of Lactobacillus fermentum and its probiotic characteristics[J]. Journal of Food Science and Biotechnology, 2015, 34(2): 121-127. (in Chinese) 敖晓琳, 蒲彪, 蔡义民, 等. 发酵乳杆菌及其益生特性研究进展[J]. 食品与生物技术学报, 2015, 34(2): 121-127. |
| [42] |
Gao Y. Antioxidant activity of marine lactic acid bacteria[D]. Dalian: Master's Thesis of Dalian Polytechnic University, 2016 (in Chinese) 高原.海洋乳酸菌抗氧化活性研究[D].大连: 大连工业大学硕士学位论文, 2016 |
| [43] |
Huang JY. Oriental acetic acid bacteria YZD-09 and its application: CN, 106635916[P]. 2017-05-10 (in Chinese) 黄君阳.东方醋酸菌YZD-09及其应用: 中国, 106635916[P]. 2017-05-10 |
| [44] |
de Man JC, Rogosa M, Sharpe ME. A medium for the cultivation of lactobacilli[J]. Journal of Applied Microbiology, 1960, 23(1): 130-135. |
| [45] |
Caporaso JG, Kuczynski J, Stombaugh J, et al. QⅡME allows analysis of high-throughput community sequencing data[J]. Nature Methods, 2010, 7(5): 335-336. DOI:10.1038/nmeth.f.303 |
| [46] |
Edgar RC, Haas BJ, Clemente JC, et al. UCHIME improves sensitivity and speed of chimera detection[J]. Bioinformatics, 2011, 27(16): 2194-2200. DOI:10.1093/bioinformatics/btr381 |
| [47] |
Edgar RC. Search and clustering orders of magnitude faster than BLAST[J]. Bioinformatics, 2010, 26(19): 2460-2461. DOI:10.1093/bioinformatics/btq461 |
| [48] |
Shen P, Fan XR, Li GW. Microbiology Experiment[M]. 3rd ed. Beijing: Higher Education Press, 1996: 119-120. (in Chinese) 沈萍, 范秀容, 李广武. 微生物学实验[M]. 3版. 北京: 高等教育出版社, 1996: 119-120. |
| [49] |
Huang XL, Xia LQ. Microbiology Experiment Guide[M]. Beijing: Higher Education Press; Germany: Springer Press, 1999: 39-60. (in Chinese) 黄秀梨, 夏立秋. 微生物学实验指导[M]. 北京: 高等教育出版社; 德国:施普林格出版社, 1999: 39-60. |
| [50] |
Scarpellini M, Mora D, Colombo S, et al. Development of genus/species-specific PCR analysis for identification of Carnobacterium strains[J]. Current Microbiology, 2002, 45(1): 24-29. DOI:10.1007/s00284-001-0043-3 |
| [51] |
Jia YT, Guo YM, Cai YA, et al. Research of quality and antioxidant capacity for fermented milk with different strains[J]. China Dairy Industry, 2017, 45(9): 22-25. (in Chinese) 贾亚婷, 郭艳梅, 蔡逸安, 等. 不同菌种发酵乳品质与抗氧化能力研究[J]. 中国乳品工业, 2017, 45(9): 22-25. DOI:10.3969/j.issn.1001-2230.2017.09.005 |
| [52] |
Hughes DB, Hoover DG. Viability and enzymatic activity of Bifidobacteria in milk[J]. Journal of Dairy Science, 1995, 78(2): 268-276. DOI:10.3168/jds.S0022-0302(95)76634-6 |
| [53] |
Church FC, Swaisgood HE, Porter DH, et al. Spectrophotometric assay using o-phthaldialdehyde for determination of proteolysis in milk and isolated milk proteins[J]. Journal of Dairy Science, 1983, 66(6): 1219-1227. DOI:10.3168/jds.S0022-0302(83)81926-2 |
| [54] |
Cushman DW, Cheung HS. Spectrophotometric assay and properties of the angiotensin-converting enzyme of rabbit lung[J]. Biochemical Pharmacology, 1971, 20(7): 1637-1648. DOI:10.1016/0006-2952(71)90292-9 |
| [55] |
Li RQ, Li YR, Kristiansen K, et al. SOAP: short oligonucleotide alignment program[J]. Bioinformatics, 2008, 24(5): 713-714. DOI:10.1093/bioinformatics/btn025 |
| [56] |
Stanke M, Diekhans M, Baertsch R, et al. Using native and syntenically mapped cDNA alignments to improve de novo gene finding[J]. Bioinformatics, 2008, 24(5): 637-644. DOI:10.1093/bioinformatics/btn013 |
| [57] |
Saha S, Bridges S, Magbanua ZV, et al. Empirical comparison of ab initio repeat finding programs[J]. Nucleic Acids Research, 2008, 36(7): 2284-2294. DOI:10.1093/nar/gkn064 |
| [58] |
Benson G. Tandem repeats finder: a program to analyze DNA sequences[J]. Nucleic Acids Research, 1999, 27(2): 573-580. |
| [59] |
Ashburner M, Ball CA, Blake JA, et al. Gene Ontology: tool for the unification of biology[J]. Nature Genetics, 2000, 25(1): 25-29. DOI:10.1038/75556 |
| [60] |
Kanehisa M, Goto S, Kawashima S, et al. The KEGG resource for deciphering the genome[J]. Nucleic Acids Research, 2004, 32(Database issue): D277-D280. |
| [61] |
Kanehisa M, Goto S, Hattori M, et al. From genomics to chemical genomics: new developments in KEGG[J]. Nucleic Acids Research, 2006, 34(Database issue): D354-D357. |
| [62] |
Tatusov RL, Fedorova ND, Jackson JD, et al. The COG database: an updated version includes eukaryotes[J]. BMC Bioinformatics, 2003, 4: 41. DOI:10.1186/1471-2105-4-41 |
| [63] |
Li WZ, Jaroszewski L, Godzik A. Tolerating some redundancy significantly speeds up clustering of large protein databases[J]. Bioinformatics, 2002, 18(1): 77-82. |
| [64] |
Donkor ON, Henriksson A, Singh TK, et al. ACE-inhibitory activity of probiotic yoghurt[J]. International Dairy Journal, 2007, 17(11): 1321-1331. DOI:10.1016/j.idairyj.2007.02.009 |
| [65] |
Pihlanto A, Virtanen T, Korhonen H. Angiotensin I converting enzyme (ACE) inhibitory activity and antihypertensive effect of fermented milk[J]. International Dairy Journal, 2010, 20(1): 3-10. DOI:10.1016/j.idairyj.2009.07.003 |
| [66] |
Luo JM, Zhang C, Liu QY, et al. Combinative effect of sardine peptides and quercetin alleviates hypertension through inhibition of angiotensin I converting enzyme activity and inflammation[J]. Food Research International, 2017, 100: 579-585. DOI:10.1016/j.foodres.2017.07.019 |
| [67] |
Kim HJ, Kang SG, Jaiswal L, et al. Identification of four new angiotensin I-converting enzyme inhibitory peptides from fermented anchovy sauce[J]. Applied Biological Chemistry, 2016, 59(1): 25-31. |
| [68] |
Park JH, Kim SM. Biofunctionality of peptides purified from naturally fermented anchovy sauce[J]. Journal of the Korean Society of Food Science and Nutrition, 2003, 32(7): 1120-1125. DOI:10.3746/jkfn.2003.32.7.1120 |
| [69] |
Wang Y. Study on the lactic acid bacteria fermented milk with angiotensin converting enzyme inhibitory effect[D]. Wuxi: Master's Thesis of Jiangnan University, 2008 (in Chinese) 王宇.乳酸菌发酵乳抑制血管紧张素转化酶活性的研究[D].无锡: 江南大学硕士学位论文, 2008 |
| [70] |
Broadbent JR, Thompson RL, Hughes JE, et al. Comparative genome analysis of the obligately homofermentative lactic acid bacterium Lactobacillus helveticus[A]//Proceedings of the 9th Symposium on Lactic Acid Bacteria: Health, Evolution, and Systems Biology[C]. The Netherlands: Utah State University Libraries, 2008
|
| [71] |
Pederson JA, Mileski GJ, Weimer BC, et al. Genetic characterization of a cell envelope-associated proteinase from Lactobacillus helveticus CNRZ32[J]. Journal of Bacteriology, 1999, 181(15): 4592-4597. |
| [72] |
Smeianov VV, Wechter P, Broadbent JR, et al. Comparative high-density microarray analysis of gene expression during growth of Lactobacillus helveticus in milk versus rich culture medium[J]. Applied and Environmental Microbiology, 2007, 73(8): 2661-2672. DOI:10.1128/AEM.00005-07 |
| [73] |
Nakamura Y. Studies on anti-hypertensive peptides in milk fermented with Lactobacillus helveticus[J]. Bioscience and Microflora, 2004, 23(4): 131-138. DOI:10.12938/bifidus.23.131 |
| [74] |
Fung WY, Liong MT. Evaluation of proteolytic and ACE-inhibitory activity of Lactobacillus acidophilus in soy whey growth medium via response surface methodology[J]. LWT-Food Science and Technology, 2010, 43(3): 563-567. |
| [75] |
Christensen JE, Dudley EG, Pederson JA, et al. Peptidases and amino acid catabolism in lactic acid bacteria[J]. Antonie van Leeuwenhoek, 1999, 76(1/4): 217-246. DOI:10.1023/A:1002001919720 |
| [76] |
Savijoki K, Palva A. Purification and molecular characterization of a tripeptidase (PepT) from Lactobacillus helveticus[J]. Applied and Environmental Microbiology, 2000, 66(2): 794-800. DOI:10.1128/AEM.66.2.794-800.2000 |
| [77] |
Zhang JC, Wang HY, Wang ZP, et al. The properties of lactic acid bacteria with ACE inhibitory activity[J]. China Dairy Industry, 2003, 31(2): 9-11. (in Chinese) 张佳程, 王海燕, 王忠萍, 等. 具有ACE抑制活性乳酸菌的特性研究[J]. 中国乳品工业, 2003, 31(2): 9-11. DOI:10.3969/j.issn.1001-2230.2003.02.003 |
| [78] |
Cheung HS, Wang FL, Ondetti MA, et al. Binding of peptide substrates and inhibitors of angiotensin-converting enzyme. Importance of the COOH-terminal dipeptide sequence[J]. The Journal of Biological Chemistry, 1980, 255(2): 401-407. |
| [79] |
Kilpi EER, Kahala MM, Steele JL, et al. Angiotensin I-converting enzyme inhibitory activity in milk fermented by wild-type and peptidase-deletion derivatives of Lactobacillus helveticus CNRZ32[J]. International Dairy Journal, 2007, 17(8): 976-984. DOI:10.1016/j.idairyj.2006.12.001 |
| [80] |
Leenhouts K, Buist G, Kok J. Anchoring of proteins to lactic acid bacteria[J]. Antonie van Leeuwenhoek, 1999, 76(1/4): 367-376. DOI:10.1023/A:1002095802571 |
| [81] |
Zevaco C, Gripon JC. Properties and specificity of a cell-wall proteinase from Lactobacillus helveticus[J]. Le Lait, 1988, 68(4): 393-407. DOI:10.1051/lait:1988425 |
| [82] |
Gilbert C, Blanc B, Frot-Coutaz J, et al. Comparison of cell surface proteinase activities within the Lactobacillus genus[J]. Journal of Dairy Research, 1997, 64(4): 561-571. DOI:10.1017/S0022029997002355 |
| [83] |
Hébert EM, Raya RR, de Giori GS. Characterisation of a cell-envelope proteinase from Lactobacillus helveticus[J]. Biotechnology Letters, 1999, 21(9): 831-834. DOI:10.1023/A:1005590731382 |
| [84] |
Hebert EM, Raya RR, de Giori GS. Nutritional requirements and nitrogen-dependent regulation of proteinase activity of Lactobacillus helveticus CRL 1062[J]. Applied and Environmental Microbiology, 2000, 66(12): 5316-5321. DOI:10.1128/AEM.66.12.5316-5321.2000 |
| [85] |
Hebert EM, Raya RR, de Giori GS. Modulation of the cell-surface proteinase activity of thermophilic lactobacilli by the peptide supply[J]. Current Microbiology, 2002, 45(6): 385-389. DOI:10.1007/s00284-002-3780-z |
| [86] |
Jensen MP, Vogensen FK, Ardö Y. Variation in caseinolytic properties of six cheese related Lactobacillus helveticus strains[J]. International Dairy Journal, 2009, 19(11): 661-668. DOI:10.1016/j.idairyj.2009.04.001 |
| [87] |
Sadat-Mekmene L, Jardin J, Corre C, et al. Simultaneous presence of PrtH and PrtH2 proteinases in Lactobacillus helveticus strains improves breakdown of the pure αS1-casein[J]. Applied and Environmental Microbiology, 2011, 77(1): 179-186. DOI:10.1128/AEM.01466-10 |
| [88] |
Hebert EM, Raya R, de Giori GS. Characterization of a cell membrane-associated proteinase from Lactobacillus helveticus CRL 581[J]. Current Microbiology, 1997, 35(3): 161-164. DOI:10.1007/s002849900231 |
| [89] |
Martín-Hernández MC, Alting AC, Exterkate FA. Purification and characterization of the mature, membrane-associated cell-envelope proteinase of Lactobacillus helveticus L89[J]. Applied Microbiology and Biotechnology, 1994, 40(6): 828-834. DOI:10.1007/BF00173983 |
| [90] |
Ono H, Yamamoto N, Maeno M, et al. Purification and characterization of a cell-wall associated proteinase of Lactobacillus helveticus CP53[J]. Milchwissenschaft, 1997, 52: 373-377. |
| [91] |
Scolari G, Vescovo M, Zacconi C, et al. Extraction and partial characterization of proteolytic activities from the cell surface of Lactobacillus helveticus Zuc2[J]. Journal of Dairy Science, 2006, 89(10): 3800-3809. DOI:10.3168/jds.S0022-0302(06)72421-3 |
| [92] |
Yamamoto N, Akino A, Takano T. Purification and specificity of a cell-wall-associated proteinase from Lactobacillus helveticus CP790[J]. The Journal of Biochemistry, 1993, 114(5): 740-745. DOI:10.1093/oxfordjournals.jbchem.a124247 |
| [93] |
Fira D, Kojic M, Banina A, et al. Characterization of cell envelope-associated proteinases of thermophilic lactobacilli[J]. Journal of Applied Microbiology, 2001, 90(1): 123-130. DOI:10.1046/j.1365-2672.2001.01226.x |
| [94] |
Kojic M, Fira D, Bojovic B, et al. Comparative study on cell envelope-associated proteinases in natural isolates of mesophilic lactobacilli[J]. Journal of Applied Bacteriology, 1995, 79(1): 61-66. DOI:10.1111/j.1365-2672.1995.tb03124.x |
| [95] |
Ezzat N, El-Soda M, Bouillance C, et al. Cell wall associated proteinases in Lactobacillus helveticus, Lactobacillus bulgaricus and Lactobacillus lactis[J]. Milchwissenschaft, 1985, 40: 140-143. |
| [96] |
Hebert EM, Raya R, Oliver G, et al. Characterization of proteolytic activity of Lactobacillus helveticus strains in milk[J]. Microbiologie, Aliments, Nutrition: the Journal of the International College Alexandre Tessier, 1996, 14(1): 65-72. |
| [97] |
Hebert EM, Mamone G, Picariello G, et al. Characterization of the pattern of αs1- and β-casein breakdown and release of a bioactive peptide by a cell envelope proteinase from Lactobacillus delbrueckii subsp. lactis CRL 581[J]. Applied and Environmental Microbiology, 2008, 74(12): 3682-3689. DOI:10.1128/AEM.00247-08 |
| [98] |
Exterkate FA. The lactococcal cell envelope proteinases: differences, calcium-binding effects and role in cheese ripening[J]. International Dairy Journal, 1995, 5(8): 995-1018. DOI:10.1016/0958-6946(95)00042-9 |
| [99] |
Donkor ON, Henriksson A, Vasiljevic T, et al. Probiotic strains as starter cultures improve angiotensin-converting enzyme inhibitory activity in soy yogurt[J]. Journal of Food Science, 2005, 70(8): m375-m381. DOI:10.1111/j.1365-2621.2005.tb11522.x |
 2019, Vol. 46
2019, Vol. 46