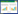扩展功能
文章信息
- 阳飞, 闵勇, 刘晓艳, 饶犇, 王开梅, 杨自文
- YANG Fei, MIN Yong, LIU Xiao-Yan, RAO Ben, WANG Kai-Mei, YANG Zi-Wen
- 甾类化合物微生物转化与分解代谢机制研究进展
- Advances in microbial transformation and catabolic mechanism of steroid compounds
- 微生物学通报, 2019, 46(10): 2743-2762
- Microbiology China, 2019, 46(10): 2743-2762
- DOI: 10.13344/j.microbiol.china.180811
-
文章历史
- 收稿日期: 2018-10-18
- 接受日期: 2019-04-02
- 网络首发日期: 2019-04-18
2. 湖北省农业科学院 湖北省生物农药工程研究中心 湖北 武汉 430064
2. National Biopesticide Engineering Research Center, Hubei Academy of Agricultural Sciences, Wuhan, Hubei 430064, China
甾类化合物在医药领域的市场需求仅次于抗生素,全球总产值已达400亿元,有400多种授权甾类药物在流通,而且未来的需求还会递增[1-3]。甾类化合物是重要的生命元素,是构成甾醇(Sterols)、胆汁酸(Bile acid)、维生素D (Vitamin D)、神经甾体(Neurosteroids)以及类固醇激素(Steroid hormones)的基本结构物质,具有重要的生理医药功能[3-4]。甾体药物大多是利用具有甾体骨架的天然产物进行结构改造及化学合成而成,甾体药物作为重要的激素药物对机体起着重要的调节作用,被开发成为麻醉药、抗心律失常药、抗细菌药、抗胆碱酯酶药、抗凝血剂、抗真菌药、抗肿瘤药、抗原生动物药、胆汁分泌剂、诊断剂、神经调节阻断剂、胆石分散剂、止血剂、钙调节剂、脂调节剂、神经病治疗剂、泻药与安定药等[5]。甾类药物最初是从动物肾上腺提取的,显然实际应用意义并不大。后来也有研究者尝试以有机小分子为原料出发通过化学方法全合成工艺的探索,虽然理论上可行,但是由于合成路线太长、收率低、反应性差且副产物处理困难,加之环境污染问题,严重缺乏工业生产应用价值[2, 6-7]。
甾体药物的工业化生产主要有化学合成法和微生物转化法两种。以植物体中提取的薯蓣皂素(Diosgenin)、剑麻皂素(Tigogenin)、蕃麻皂素(Hecogenin)等皂素为主导原料,进一步借助化学方法合成甾药关键中间体的工艺面临生产成本高、步骤较多、过程复杂以及环境污染严重等客观问题[8-11]。1970年薯蓣皂苷元及其它皂苷元正式被确立为甾醇药物合成的起始物质,但是由于供应量的减少和价格的攀升导致该工艺的利润有所下降,且1992年联合国卫生组织宣布禁止使用全化学合成法生产药物,已逐渐不适用甾醇工业化生产[12]。而微生物转化(Microbial transformation)是指利用微生物产生的酶作用于化合物的某一基团,使其转化为结构上类似且更有价值的新型产物,其反应本质为酶促反应,具有较强的专一性与手性选择性,可以弥补化学合成的不足。1908年Bondi研究了胆酸代谢途径以及各种分枝杆菌分解氧化甾醇类化合物,1913年Sohngen发现多种微生物能够以甾醇为唯一碳源生长,1952年Murray与Petersons首次利用黑根霉一步催化黄体酮为11α-羟基黄体酮,开创了微生物转化甾体化合物的先例。1944年第一次发现分枝杆菌可以降解胆固醇转化为4-雄烯二酮(4-Androstene-3, 17-dione,4-AD)与1, 4-雄烯二酮(Androsta-1, 4-diene-3, 17-dione,ADD),研究者相继发现更多不同微生物可以降解不同甾醇类化合物制备甾药关键中间体如4-AD或ADD,而4-AD或ADD是甾类激素药物不可替代的中间体,几乎可以用于合成所有的甾类激素药物,因此以植物甾醇(Phytosterols)为原料利用微生物降解而制备关键中间体工艺,相对于以皂素为原料的化学合成工艺因具有原料来源稳定、无污染以及成本低的优点而被采用。然而,微生物降解甾类化合物的分解代谢机制仍有待进一步深入探索,而且不同的微生物间还存在一定的差异性。本文介绍了微生物转化甾类化合物及其分解代谢机制方面的研究进展,为进一步研究代谢分子机制及生产应用提供一定参考。
1 甾类化合物的结构种类及主要来源甾体类化合物属于脂类,通常指一类特殊的多元环萜类化合物,包括核心环(Core ring)、C17位侧链(Side chain)以及不同官能团取代基(Functional groups)三部分,核心环(Core ring)由A、B、C三个完整封闭的六元环和一个五元环D环组成,C17位侧链通常为长烃类,包括8个碳链烷基、10个碳烷基或者烯基等,官能团取代基主要指核心环碳原子连接的相关的官能团,常见位点有C3、C9、C11、C17、C18、C19等。根据核心环特点通常包括五类基本结构单元(图 1A),即Cholest、Chol、Pregn、Androst、Estr等单元,其余的均为相应结构单元的衍生物结构。通常所说的甾醇类物质结构特点为C3位β-OH、C17位为8–10个碳链烷基或者烯基,C10位与C13位连有甲基,如胆固醇(Cholesterol) (图 1B)。甾类化合物拥有类似的结构且结构复杂数目繁多,在自然界中存在上千种甾类化合物,普遍存在于动植物和部分微生物中[1, 13-16],胆固醇(Cholesterol)、性激素(Sex hormones)、肾上腺皮质激素(Adrenocortical hormones)、胆汁酸(Bile acids)、维生素D (Vitamin D)及神经甾体(Neurosteroids)等存在于高等脊椎动物体中,蜕化类固醇(Ecdysteroids)存在于昆虫体内,麦角固醇(Ergosterol)存在于酵母细胞壁中,β-谷甾醇(β-Sitosterol)、豆甾醇(Stigmasterol)、菜油甾醇(Campesterol)、岩藻甾醇(Fucosterol)、菜籽甾醇(Brassicasterol)等(图 1B)存在于植物中。其中植物甾醇(Phytosterols,PSs)多达250种以上[1],由于植物甾醇来源广泛且价格相对便宜,因此用于生产重要的医药甾药化合物或中间体(图 1C)。
|
| 图 1 甾类化合物 Figure 1 Steroid compounds 注:A:甾类化合物常见基本结构单元;B:甾醇化合物;C:甾醇微生物降解关键中间体;D:甾体激素. Note: A: Common structure unit of steroid compounds; B: Sterols; C: Key intermediate in degradation of sterols by microorganism; D: Steroid hormones. |
|
|
关于甾体药物的作用机理,大多比较认可的是“基因表达学说”,即由于甾体激素分子量小(300 Da左右)且脂溶性强,因此很容易透过细胞膜进入细胞内,首先与胞浆受体结合形成复合物,然后穿过细胞核膜与核内受体结合,进而启动或抑制DNA的复制转录,最终诱导或抑制新蛋白的生成,从而引起各种生理性状的改变[4, 7, 17]。甾体化合物的生理功能取决于核心环连接的官能团的种类、数目以及位置的构型[1, 18]。甾体化合物主要包括胆固醇、植物甾醇、胆汁酸、维生素D、神经甾体与类固醇激素等。胆固醇是构成细胞壁膜的成分之一,主要分布在磷脂双分子层之间,可以影响细胞膜的流动性以及调节细胞增殖与分化[19],主要生理功能体现在参与血浆蛋白的合成,在体内可转化为胆汁酸盐、维生素D3前体以及肾上腺皮质激素与性激素等(图 1D)。植物甾醇在食品中作为添加剂以及在化妆品中也有重要的应用[3, 13, 20-21]。尤其是植物甾醇中的豆甾醇可以阻碍胆固醇的吸收,从而降低血清中胆固醇的含量,具有抗氧化活性,预防和治疗高血压、冠心病等心血管疾病,能促进血纤维蛋白溶酶的激素因子,可以预防血栓症,此外还可以抑制癌细胞的形成。胆汁酸是构成胆汁的重要成分,在胆汁中主要以甘氨酸胆甾酸和牛磺酸胆甾酸的钠盐或钾盐的形式存在,减少水与脂肪的表面张力,促进脂溶性维生素的吸收与脂肪的消化,具有调节糖代谢与能量代谢、抗炎症、药物代谢与脱毒的功效[15, 22]。维生素D是一类具有抗佝偻病价值的维生素,对骨骼的形成有着重要的意义。神经甾体特指神经系统产生的甾体化合物,主要通过进入突触间隙调节GABAA (γ-氨基丁酸A型)受体的功能而影响神经细胞的兴奋性,对中枢神经系统有着广泛的生理作用,具有保护神经细胞、镇静催眠、麻醉、抗惊厥、抗焦虑、促进学习记忆和改善认知能力等作用,而没有神经毒性和传统甾体激素的副作用[23]。研究表明,去氢表雄酮(Dihydroepiandrosterone,DHEA)可以直接作用于神经元细胞表面,改变神经递质受体结构,从而调节神经刺激在细胞间的传递[1, 23-25]。
甾体激素又称类固醇激素,根据生理学功能分为肾上腺皮质激素和性激素,其中肾上腺皮质激素分为糖皮质激素(Glucocorticoids)和盐皮质激素(Mineralocorticoids),而性激素包括孕激素(Progestogens)、雌性激素(Estrogens)、雄性激素(Androgens)以及蛋白同化激素类(Anabolic steroids)。肾上腺皮质激素基本结构中甾体母核含有21个碳,C3位与C20位均为羰基,C4-5为双键。糖皮质激素在C17位有α-羟基以及C11位为羟基(如氢化可的松)或者羰基(如可的松),是机体应激反应最重要的调节激素,参与调节糖类、脂肪与蛋白质代谢,调节机体发育生长以及免疫功能等,具有抗炎、抗毒、抗过敏、抗休克、非特异性抑制免疫及退热等多种作用,也是临床上使用最广泛而有效的抗炎和免疫抑制剂,而盐皮质激素在C17位无α-羟基与C11位无氧(如去氧皮质酮)或者在C11位有氧但与C18位碳结合(如醛固酮),主要生理作用是促进肾小管重吸收钠而保留水并排泄钾,与下丘脑分泌的抗利尿激素相互协调,共同维持机体电解质平衡和体液容量[5, 7, 23]。孕激素基本结构中母核含有21个碳,C3位为羰基,C4-5为双键,C17位为β-甲基酮,主要生理作用是促进女性附性器官成熟及第二性征出现,并维持正常性欲及生殖功能的激素[26]。雌性激素基本结构中母核含有18个碳,A环为苯环结构,C3位为羟基,C17位为β-羟基,主要生理作用是促进雌性第二性征的发育和性器官的成熟,促进蛋白质合成,减少碳水化合物利用,调节水、盐分子与钙的平衡,与孕激素一起完成性周期、妊娠、哺乳等,用于治疗女性性功能疾病、更年期综合症、骨质疏松症等[4, 7]。雄性激素基本结构中母核含有19个碳,C3位为羰基,C4-5为双键,C17位为β-羟基,主要生理功能是促进男性器官及副性征的发育和成熟,对抗雌激素,抑制子宫内膜生长及垂体-性腺功能,增强免疫功能,促进蛋白质合成及骨质形成,刺激骨髓造血功能,使红细胞和血状红细胞增加[7]。蛋白同化激素是由雄性激素衍生出的一系列人工合成类固醇化合物,是一类外源性的以蛋白同化作用为主的甾体激素,可分为睾酮衍生物、雄烷衍生物、诺龙衍生物、杂环衍生物以及杂类合成类固醇,主要生理功能为雄性化作用显著减弱而蛋白同化作用增强,促进蛋白质合成和抑制蛋白质异化,促进组织钙化和生长,刺激骨髓造血功能,增加红血球量,促进组织新生和肉芽形成,降低胆固醇[5, 7, 18]。因此,开发甾类药物对改善人类健康体质显得尤为重要,具有非常重要的社会意义。
3 甾类化合物微生物转化与分解代谢机制 3.1 甾类化合物转化微生物甾体微生物转化是利用微生物细胞酶系对甾体化合物某一特定部位进行化学反应产生新的产物。微生物对甾类化合物很多位置的原子或基团都有可能进行转化[27-30],涉及的反应类型有氧化、还原、水解、酯化、缩合、异构化、重排以及侧链降解等,羟化反应主要位点有C9α、C11α、C11β、C15α、C16α、C16β、C17α、C19、C26,脱氢反应通常发生在A环的C1, 2和C4, 5位之间,芳香化反应主要发生在A环上,环氧化反应一般发生在C9, 11和C14, 15位之间,双键还原反应一般发生在A环C1, 2、C4, 5和B环C5, 6位之间,酮基还原主要体现在C3、C17、C20位上,羟基氧化为酮基常见位点C3α、C3β、C17α。甾类化合物转化研究较多的主要体现在羟基化反应、脱氢反应以及侧链降解反应等方面。甾体转化微生物包括细菌、放线菌、酵母菌以及霉菌等。化学法除了在甾体C17位比较容易引入羟基外,其它位点都很难引入,甾体微生物转化中羟化反应是最早用于工业化生产的,1952年Murray与Petersons首次利用黑根霉一步催化黄体酮为11α-羟基黄体酮,开创了微生物转化甾体化合物的先例。少根根霉(R. arrhizus)、黑曲霉(Aspergillus niger)、赭曲霉(A. orchraceus)、弯孢霉(Curvularia sp.)、小克银汉霉(Cunninghamella sp.)、金龟子绿僵菌(Metarhizium anisopliae)、球孢白僵菌(Beauveria bassiana)也可以催化甾类化合物C11位羟基化[31-32]。对甾体C16α位羟基化转化的微生物有玫瑰色链霉菌(Streptomyces roseochromogenes)、绿色链霉菌(S. argenteolus)、橄榄链霉菌(S. olivaceus)、银色链霉菌(S. argenteolus)等[5]。由于抗炎甾体激素药物在母核C1, 2位引入双键后能成倍地增加抗炎作用,微生物脱氢也成为甾体抗炎激素药物合成不可缺少的一步。研究发现,降解甾类化合物的微生物大多数为Arthrobacter spp.、Brevibacterium spp.、Pseudomonas spp.、Norcardia spp.、Comamonas spp.、Rhodococcus spp.以及Mycobacterium spp.属等[33-59]。大多数在生产甾类药物中间体时为了防止核心环降解通常需要加入抑制剂[1]。部分真菌经过诱变筛选后也可以降解甾醇转化关键甾类药物前体物质4-AD[60-61]、9-OHAD[62]和ADD[63-68]。在上述众多的微生物菌属中,其中红球菌属(Rhodococcus spp.)与分枝杆菌属(Mycobacterium spp.)能够有效积累甾类药物关键中间体4-AD、ADD或9-OHAD,因而引起研究者的广泛关注。红球菌是介于分枝杆菌与诺卡氏菌之间的一类微生物,与分枝杆菌很相似[69]。红球菌与分枝杆菌用于甾类转化生产主要基于几个方面特性:(1)多数不具有侵染致病性,且表现为快速生产、易于培养增殖[2];(2)拥有完善的甾类降解基因簇,含有丰富的氧化还原酶以及β-氧化酶系,有利于甾类化合物的快速分解代谢[70-74];(3)细胞壁中含有致密的分枝菌酸层,可以提高细胞的耐受力,适用于甾类的油水两相体系发酵工艺[75],促进细胞对疏水性甾类化合物的吸收与转化[3, 76]。
随着诱变育种技术的不断改进以及对微生物降解甾类机制的进一步揭示,研究者们已经选育出高效积累关键甾药前体物质的大量优良菌株[53, 77-81]。近些年来,研究的重心又转向分枝杆菌属的新金分枝杆菌(Mycolicibacterium neoaurum)[54, 56, 82-90]与耻垢分枝杆菌(Mycolicibacterium smegmatis)[91-95]。
3.2 甾类化合物转化微生物生物信息学甾类化合物分解代谢通路的推测源自对关键中间体的鉴定基础,因而也导致前期菌种选育方面遭遇到瓶颈。随着基因测序技术的发展,大多数降解甾醇、胆甾酸、睾酮等甾类的微生物也完成了基因测序[41, 46, 54, 56, 83, 88-90, 93, 96-101],其中多数为分枝杆菌(表 1)。
| Strain | GenBank No. | Size (bp) | CDS | kstR | kstR2 | igr |
| Mycolicibacterium neoaurum ATCC 25795 | JMDW00000000.1 | 5 468 381 | 5 192 | RS0123370 | RS0123530 | Yes |
| Mycobacterium tuberculosis H37Rv | NC_000962.3 | 4 411 532 | 4 090 | Rv3574 | Rv3557c | Yes |
| Rhodococcus jostii RHA1 | NC_008268.1 | 7 804 765 | 9 189 | Ro04482 | Ro04598 | Yes |
| Mycolicibacterium neoaurum HGMS2 | CP031414.1 | 5 421 383 | 5 078 | RS00580 | RS00705 | Yes |
| Mycolicibacterium neoaurum MN2 | LQMX00000000.1 | 5 383 007 | 5 073 | RS22745 | RS22135 | Yes |
| Mycolicibacterium neoaurum MN4 | JXYZ00000000.1 | 5 390 529 | 5 068 | RS14335 | RS02720 | Yes |
| Mycolicibacterium neoaurum VKMAc-1815D | NC_023036.2 | 5 421 267 | 5 048 | RS24245 | RS24120 | Yes |
| Mycobacterium sp. VKMAc-1816D | AOHQ00000000.1 | 5 413 599 | 5 091 | RS6380 | RS2135 | Yes |
| Mycolicibacterium neoaurum VKMAc-1817D | CP009914.1 | 6 324 222 | 6 024 | RS26515 | RS26275 | Yes |
| Mycolicibacterium neoaurum DSM 440744 | CCDR00000000.1 | 5 504 703 | 5 272 | RS24985 | RS24790 | Yes |
| Mycobacterium sp. NRRL B-3805 | CP011022 | 5 421 338 | 5 049 | RS24255 | RS24130 | Yes |
| Mycolicibacterium smegmatis MC2155 | NC_008596.1 | 6 988 209 | 6 947 | MSMEG_6042 | MSMEG_6009 | Yes |
| Pimelobacter simplex VKM Ac-2033D | CP009896.1 | 5 637 360 | 5 463 | KR76_12270 | KR76_25115 | Lost chsE1 |
| Note: kstR/2: TetR-type transcriptional regulator/2; igr: Intracellular growth. | ||||||
研究显示降解甾类化合物的相关基因通常成簇排列且多数基因会由于甾类化合物诱导显著上调,通过比较红球菌Rhodococcus jostii RHA1与结核分枝杆菌M. tuberculosis H37Rv甾醇代谢基因簇显示相似性比较高,且研究显示该基因簇受到甾类等底物诱导的水平也非常相似[91, 95, 102-104]。基于对红球菌、分枝杆菌等微生物甾类分子降解机制的研究,学者们逐步揭示了甾类降解关键基因簇并阐明了部分关键基因及其功能[52, 72, 95, 104-111] (图 2)。Griffin通过高密度转座子突变及高深度测序证明,结核分枝杆菌M. tuberculosis H37Rv与甾醇代谢的基因还有约60%位于上述基因簇之外的其它位置且呈分散状态分布,一些快速生长型分枝杆菌还因同工酶的存在也给甾类化合物降解机制与工程菌改造等研究增加了困难[2, 82, 84, 111-129]。随着转录组学与蛋白组学的发展,部分微生物也进行了甾类化合物诱导下的转录组、蛋白组以及基因微阵列的测定[74, 91-92, 102-103, 130-132]。
3.3 甾类化合物微生物分解代谢机制解析通常甾类化合物如甾醇等在微生物体内分解代谢过程大致包括4个阶段:(1)甾类化合物的摄取与转运(Uptake and transportation);(2)底物活化(Substrate activation);(3)侧链降解(Side-chain degradation);(4)核心环降解(Core ring cleavage)。甾类化合物在微生物体内分解代谢过程属于典型的有氧代谢,前期的研究主要是基于某些代谢中间体推测可能的代谢途径[58, 133-154]。但随着基因测序以及甾类化合物降解基因簇的揭示,结合核磁共振、高效液相色谱、高效液相-质谱联用、气相色谱以及气相色谱-质谱联用技术,借助基因工程手段,甾类化合物分解代谢的关键酶及其功能结构被逐步鉴定,实现大部分基因与代谢途径的准确匹配关联,多步反应机制得以准确阐明,从而进一步明确甾类化合物降解机制[2, 27, 45-46, 71-73, 82, 85, 94-95, 102, 105-106, 109-111, 115, 122, 155-175]。根据代谢过程中一些中间体,将代谢路径及方向大致拟定为4-AD途径、ADD途径、9-OHAD途径、HMP途径、HMPD途径、9-DHMP途径、4-BNC途径、1, 4-BNC途径、Ts途径、BD途径[2, 63, 82, 86, 119, 123, 176-179]。以β-Sitosterol和Cholesterol为例,结合基因功能从分子水平探讨甾醇分解代谢机制(图 3)。

|
| 图 3 甾醇微生物分解代谢机制 Figure 3 Speculated catabolic mechanism of sterols in microorganism Note: A: Sterol substrate activation; B: Side-chain degradation; C: Core ring cleavage. 1, 4-BNC: 3-Oxo-23, 24-bisnorchola-1, 4-dien-22-oic acid; 1, 4-BNC-CoA: 3-Oxo-23, 24-bisnorchola-1, 4-diene-22-oyl-CoA; 3, 4-DHSA: 3, 4-Dihydroxy-9, 10-secoandrost-1, 3, 5(10)-triene-9, 17-dione; 3-HSA: 3-Hydroxy-9, 10-secoandrost-1, 3, 5(10)-triene-9, 17-dione; 3-HSBNC-CoA: 3-Hydroxy-9-oxo-9, 10-seco-23, 24-bisnorchola-1, 3, 5(10)-trien-22-oyl-CoA; 3-OBNCT-CoA: 3-Oxo-23, 24-bisnorchola-1, 4, 17-trien-22-oyl-CoA; 3-OP20CA: 3-Oxo-pregn-4-en-20-carbaldehyde; 3-OCO-CoA: 3-Oxo-chol-4-en-24-oyl-CoA; 3-OCDO-CoA: 3-Oxo-chola-4, 22-dien-24-oyl-CoA; 3-OPC-CoA: 3-Oxo-4-pregnene-20-carboxyl-CoA; 3-OPDC-CoA: 3-Oxo-4, 17-pregnadiene-20-carboxyl-CoA; 4, 9-DSHA: 4, 5-9, 10-Diseco-3-hydroxy-5, 9, 17-trioxo-androsta-1(10), 2-diene-4-oic acid; 4-AD: 4-Androstene-3, 17-dione; 4-BNC: 3-Oxo-23, 24-bisnorchol-4-en-20-oic acid; 4-CSO: Cholest-5-en-3-one; 4-ECSO: 24-Ethyl-cholest-4-en-3-one; 4-CSO: Cholest-4-en-3-one; 5-COHNCA-CoA: 5-Carboxy-9-oxo-1, 2, 3, 4, 10, 19-hexanor-cholan-22-oyl-CoA; 5-DOHNAA-CoA: 9, 17-Dioxo-1, 2, 3, 4, 10, 19-hexanorandrostan-5-oyl-CoA; 5-ECSO: 24-Ethyl-cholest-5-en-3-one; 6-HOHNAE-CoA: 9-Hydroxy- 17-oxo-1, 2, 3, 4, 10, 19-hexanor-androst-6-en-oyl-CoA; 7-HDONAA-CoA: 7-Hydroxy-9, 17-dioxo-1, 2, 3, 4, 5, 6, 10, 19-octanorandrostan-7-oyl-CoA; 9, 17-DHOPC-CoA: 9α, 17β-Dihydroxy-3-oxo-pregn-4-ene-20-carboxyl-CoA; 9-DHMPD: 9, 22-Dihydroxy-20-methylpregna-1, 4-diene-3-one; 9-DHMP: 9, 21-Dihydroxy-20-methylpregn-4-en-3-one; 9-DHMPD: 9, 22-Dihydroxy-20-methylpregna-1, 4-diene-3-one; 9-HDOHNAA-CoA: 9-Hydroxy-7, 17-dioxo-1, 2, 3, 4, 10, 19-hexanorandrostan-5-oyl-CoA; 9-HBNCD: 9-Hydroxy-3-oxo-23, 24-bisnorchola-1, 4-diene-20-oic acid; 9-HOHNAA-CoA: 9-Hydroxy-17-oxo-1, 2, 3, 4, 10, 19-hexanor-androstan-5-oyl-CoA; 9-HOONAA-CoA: 9-Hydroxy-17-oxo-1, 2, 3, 4, 5, 6, 10, 19- otanorandrostan-7-oyl-CoA; 9-HOPDC-CoA: 9-Hydroxy-3-oxo-pregna-1, 4-diene-20-carboxyl-CoA; 9-HOPC-CoA: 9α-Hydroxy-3-oxo-4- pregnene-20-carboxyl-CoA; 9-HOPCD-CoA: 9α-Hydroxy-3-Oxo-4, 17-pregnadiene-20-carboxyl-CoA; 9-OHAD: 9α-Hydroxy-4-androstene- 3, 17-dione; 9OH-ADD: 9α-Hydroxy-androsta-1, 4-diene-3, 17-dione; 9-OHTs: 9α-Hydroxy-testosterone; 17-BHBNCD-CoA: 17β-Hydroxy-3- oxo-23, 24-bisnorchola-1, 4-dien-22-oyl-CoA; 17-BHOPC-CoA: 17β-Hydroxy-3-oxo-4-pregnene-20-carboxyl-CoA; 17-COHNCE-CoA: 5-Carboxy-9-oxo-1, 2, 3, 4, 10, 19-hexanor-chol-17-en-22-oyl-CoA; 22-DOCO-CoA: 3, 22-Dioxo-chol-4-en-24-oyl-CoA; 22-HOCO-CoA: 22-Hydroxy-3-oxo-chol-4-en-24-oyl-CoA; 17-HCOHNCA-CoA: 17-Hydroxy-5-carboxy-9-oxo-1, 2, 3, 4, 10, 19-hexanor-cholan-22-oyl-CoA; 24-HOCS27CoA: 24-Hydroxy-3-oxo-4-cholesten-27-oyl-CoA; 24-HCEOCS27CoA: 24-Hydroxy-24-(1-carboxyethyl)-3-oxo-4-cholesten- 27-oyl-CoA; 26-DOCS: 3, 24-Dioxo-4-cholesten-26-oic acid; 26-DOCS-CoA: 3, 24-Dioxo-4-cholesten-26-oyl-CoA; 27-CEOCSD-CoA: 24-(1-Carboxyethyl)-3-oxo-4, 24-cholestadien-27-oyl-CoA; 27-DOCS-CoA: 3, 24-Dioxo-4-cholesten-27-oyl-CoA; 27-EOCS-CoA: 24-Ethyl- 3-oxo-4-cholestene-27-oyl-CoA; 27-HCSO: 27-Hydroxycholest-4-en-3-one; 27-HECSO: 24-Ethyl-27-hydroxycholest-4-en-3-one; 27-EOCS: 24-Ethyl-3-oxo-4-cholestene-27-oic acid; 27-OCS: 3-Oxo-4-cholestene-27-oic acid; 27-OCS-CoA: 3-Oxo-4-cholestene-27-oyl-CoA; 27-OCSD-CoA: 3-Oxo-4, 24-cholestadien-27-oyl-CoA; ADD: Androsta-1, 4-diene-3, 17-dione; BD: Boldenone; COCHEA-CoA: (R)-2-(2- Carboxyethyl)-3-methyl-6-oxocyclohex-1-ene-1-carboxyl-CoA; DHOHNAA-CoA: 7, 9-Dihydroxy-17-oxo-1, 2, 3, 4, 10, 19-hexanorandrostan- 5-oyl-CoA; DHSBNC-CoA: 3, 4-Dihydroxy-9-oxo-9, 10-seco-23, 24-bisnorchola-1, 3, 5(10)-trien-22-oyl-CoA; DSHDA-CoA: 4, 5-9, 10-Diseco- 3-hydroxy-5, 9-dioxo-androsta-1(10), 2-dien-4-oyl-CoA; DOHNAA: 9, 17-Dioxo-1, 2, 3, 4, 10, 19-hexanorandrostan-5-oic acid; EOCS25CA: 24-Ethyl-3-oxo-4-cholestene-25-carbaldehyde; HHD: 2-Hydroxy-cis-hex-2, 4-dienoate; HIEC-CoA: (7aS)-7a-Methyl-1, 5-dioxo-2, 3, 5, 6, 7, 7a- hexahydro-1H-indene-4-carboxyl-CoA; HMP: 21-Hydroxy-20-methylpregn-4-en-3-one; HMPD: 20-Hydroxymethylpregna-1, 4-dien-3-one; HOHA: 4-Hydroxy-2-oxohexanoic acid; MDOODA-CoA: 4-Methyl-5, 7-dioxo-octanedioyl-CoA; MHOODA-CoA: 4-Methyl-7-hydroxy-5- Oxo-octanedioyl-CoA; MeDODA-CoA: 6-Methyl-3, 7-dioxodecanedioyl-CoA; MOHDA-CoA: 4-Methyl-5-oxo-hexanedioyl-CoA; MOODA-CoA: 4-Methyl-5-oxo-octanedioyl-CoA; MOODE-CoA: 4-Methyl-5-oxo-7-octenedioyl-CoA; OCS25CA: (20S, 25S)-3-Oxo-4- cholestene-25-carbaldehyde; Ts: Testosterone. |
|
|
研究表明在M. tuberculosis中Mce4操纵子编码的功能蛋白与甾醇转运密切相关,是M. tuberculosis摄取宿主细胞胆固醇的关键系统[180-181]。Mce (Mammalian cell entry)操纵子存在于Mycobacteriaceae、Nocardiaceae、Intrasporangiaceae、Norcardioidaceae、Pseudonocardiaceae、Streptomycetaceae中,通常包含2种yrbE基因(yrbEA、yrbEB)与6种mce基因(mceA、mceB、mceC、mceD、mceE、mceF)[182-183]。在M. tuberculosis中存在4套Mce操纵子[184],即Mce1、Mce2、Mce3、Mce4,均可编码与ABC转运体(ATP-binding cassette transporters)类似的跨膜蛋白以及其它一些必需的膜表蛋白[2, 185]。微生物对甾体底物的主动摄取主要跟Mce4密切相关,这也是部分致病菌侵染哺乳动物细胞后依靠降解细胞膜内胆固醇而长期寄生的关键[186]。研究显示在M. smegmatis中Mce4操纵子与胆固醇的转运也密切相关[187-188]。研究发现在Rhodococcus jostii RHA1也存在类似M. tuberculosis的Mce4功能蛋白家族,受Cholesterol显著诱导,当部分基因或全部基因缺失时菌株无法利用Cholesterol、β-Sitosterol、5α-Cholesterol、5α-Cholestanone等物质而生长,但对4-AD、Progesterone、Cholic acid等甾类物质的利用几乎无影响,Mce4编码的转运子仅能识别C17取代有至少8个碳原子的非极性类基团如胆固醇等物质[2, 104, 185]。
3.3.2 底物活化(Substrate activation)甾醇底物活化主要包括3个阶段(图 3A):
第一阶段:甾醇首先氧化为胆甾-5-烯-3-酮(5-Cholesten-3-one),然后异构化为胆甾-4-烯-3-酮(4-Cholesten-3-one),这一步主要由胆固醇氧化酶(Cholesterol oxidase,ChO)催化完成。ChO为寡聚黄素氧化酶,以FAD为辅基,可同时催化甾醇C3位的羟基氧化脱氢与C5位向C4位的异构化,包括GMC (Glucose-methanol-choline,与FAD非共价结合)和BOC2 (与FAD共价结合)两大类。不同来源的胆固醇氧化酶均可以催化多种3β-羟基底物,上述两大类ChO对底物动力学和氧化还原能力差异性比较显著。研究显示ChO除上述功能外还可以催化胆固醇生成6β-羟基-胆甾-4-烯-3-酮[2, 189-191]。在M. neoaurum ATCC 25795基因组中存在两种类型的ChoM1与ChoM2,ChoM2虽然位于kstR (TetR-type transcriptional regulator)调控区外但属于胞外分泌型,是参与氧化脱氢与异构化这一阶段的关键酶,而ChoM1位于kstR调控区内,与膜结合起辅助作用[2]。然而,在M. tuberculosis与M. smegmatis等微生物中催化这一阶段的关键酶并不是ChO,而是由另一类甾类脱氢酶3β-Hydroxysteroid dehydrogenase (3β-HSD),以类似NAD或者NADP为辅基,该类蛋白家族普遍存在一段甘氨酸密集区(GX2GX2G)与活性中心(YX2K),虽然位于降解甾醇基因簇之外,却受到kstR转录因子的调控[111, 192-193]。姚抗发现在M. neoaurum ATCC 25795基因组中也存在一种3β-HSD,但证实不是甾醇代谢所必需的酶[2]。除了上述的ChO与3β-HSD具有C5位向C4位异构化功能外,在Mycobacterium、Pseudomonas putida、Comamonas testosteroni等菌中还存在另一类甾酮异构酶Δ5-3-Ketosteroid isomerase (KSI),也可以直接催化胆甾-5-烯-3-酮向胆甾-4-烯-3-酮转化[194-204],部分分枝杆菌还存在多种同工酶,分析推测可能在异构化这一步起协助催化作用。
第二阶段:胆甾-4-烯-3-酮C27羟基化再氧化成甾醛(3-Oxo-4-cholestene-25-carbaldehyde),最后氧化为甾酸(3-Oxo-4-cholestene-27-oic acid),这是启动侧链降解的关键第一步。这一阶段主要由Cytochrome P450催化完成,主要包括Cyp125与Cyp142,存在于M. tuberculosis、M. smegmatis,两者催化位点在C27位,但Cyp125催化产生25S羟化构型产物,而Cyp142产生25R羟化构型产物[74, 107, 126, 205-208]。然而Rhodococcus jostii RHA1[54, 106]与Mycobacterium bovis BCG[209]的Cyp125却主要催化C26羟基化。Su等研究M. neoaurum TCCC 11978发现存在3种Cyp125,其中Cyp125-1位于kstR调节区igr因子,过表达引起ADD(D)产率提升[84],通过同源性以及注释比对分析M. neoaurum ATCC 25795基因组也发现存在3种Cyp125。
第三阶段:甾酸在乙酰辅酶A合成酶(Acyl-CoA synthetase)下发生转乙酰作用而被活化,侧链进一步进行类似脂肪酸的β-氧化(β-Oxidation)过程,与这一步催化反应相关的酶为FadD19 (Steroid-26-oyl-CoA synthetase),位于kstR调控区[102]。Yang等[166]证实FadD19可以催化3-Oxo-cholest-4-ene-26-oic acid转化为3-Oxo-cholest-4-ene-26-oyl CoA,但Wilbrink等[210]在RG32研究中发现敲除fadD19基因后不影响胆固醇的侧链降解,但可以阻断含有C24支链的菜油甾醇与β-谷甾醇的侧链降解,与Wrońska等研究M. smegmatis MC2 155结论基本一致[171]。姚抗研究M. neoaurum ATCC 25795敲除kshA-fadD19-echA19后发现对植物甾醇转化效果不佳,但不影响胆固醇代谢[2]。虽然Cyp125产生25S构型甾酸产物,Cyp142产生25R构型甾酸产物,研究显示M. tuberculosis H37Rv的FadD19对构型没有选择性,可以同时催化这两种构型产物[128, 166]。可推断FadD19是植物甾醇降解的关键酶,而不是胆固醇代谢的限速因子,因此可能还存在其它催化该步骤的同工酶FadD36[111],植物甾醇和胆固醇差异性体现在C24存在支链,可知FadD19应该是一个多功能酶。
3.3.3 侧链降解(Side-chain degradation)胆固醇的侧链降解类似于脂肪酸的β-氧化(β-Oxidation)过程,共进行三轮氧化过程,前两轮基本上与β-氧化一致,最后一轮脱丙酰辅酶A (Propionyl CoA)属于反醇醛反应,此轮反应不同于脂肪酸的β-氧化而有显著差异性,经过三轮氧化胆固醇依次释放出各1分子丙酰辅酶A (Propionyl CoA)、乙酰辅酶A (Acetyl CoA)和丙酰辅酶A (Propionyl CoA),而β-谷甾醇发生氧化作用则依次释放出2分子丙酰辅酶A (Propionyl CoA)、1分子乙酰辅酶A (Acetyl CoA)、1分子丙酰辅酶A (Propionyl CoA) (图 3B)。脂肪酸的β-氧化酶系包括FadD family (Fatty-acid-CoA ligase)、FadE family (Acyl-CoA dehydrogenase)、EchA family (Enoyl-CoA hydratase)、FadB family (β-hydroxyacyl-CoA dehydrogenase)、FadA family (Acetoscetyl-CoA thiolase)。由3.3.1所述可知FadD19参与了第一步活化反应,因而推动了后续催化反应。
第一轮脱氢酶为FadE26-FadE27 (ChsE4-ChsE5),第二轮脱氢酶为FadE28 (ChsE3),第三轮脱氢酶为FadE28-FadE29 (ChsE1-ChsE2),全部位于kstR调控区[2, 46, 71-72, 102, 110, 166, 175, 211]。Yang等研究M. tuberculosis H37Rv表明ChsE4-ChsE5对第一步26-OCS-CoA的脱氢反应活力最强,ChsE3专一性催化第二步3-OCO-CoA的脱氢反应,而ChsE1-ChsE2专一性催化3-OPC-CoA的脱氢反应,此外ChsE4-ChsE5也可以催化3-OCO-CoA与3-OPDC-CoA的脱氢反应,对ChsE3与ChsE1-ChsE2起到一定的协助作用[166]。而Wilbrink等研究发现RG32敲除fadE26-fadE27后对菜油甾醇失去降解能力,而对β-谷甾醇与胆固醇降解几乎没有影响[210]。同样情况也发生在M. neoaurum ATCC 25795中,敲除fadE26-fadE27后对主产物4-AD与ADD影响比较少,只是不再积累宝丹酮(Boldenone,BD)与睾酮(Testerone,Ts)[2]。虽然M. tuberculosis H37Rv的FadD19对C27甾酸构型没有选择性,但是ChsE4-ChsE5却只专一性催化25S构型产物,而25R构型产物需要在α-Methyl acyl CoA (MCR)异构酶转化为25S构型才能被进一步降解[128]。FadE26-FadE27并非显著作用于β-谷甾醇与胆固醇侧链降解,也可能还存在其它同工酶FadE5或FadE25发挥相关作用[111]。ChsE1-ChsE2催化3-OPC-CoA为3-OPDC-CoA也得到了证实[166, 170, 175]。
Yang等证实ChsH1-ChsH2可以催化3-OPDC-CoA为17-BHOPC-CoA[175]。Wrońska等研究发现M. smegmatis MC2 155敲除echA19基因后对β-谷甾醇与胆固醇降解几乎无影响[171],这与姚抗研究M. neoaurum ATCC 25795的结论一致[2]。综上可知,对于甾醇侧链降解前两步催化Enoyl-CoA hydratase相关酶如EchA9[111]还有待进一步深入研究。
在M. tuberculosis H37Rv基因组中β-Hydroxyacyl-CoA dehydrogenase共有5种(FadB1、FadB2、FadB3、FadB4、FadB5)[101, 110]。Taylor等研究发现当敲除fadB2基因后几乎无影响[212]。截至目前,分枝杆菌第一个氧化过程催化β-羟基转化为酮基的β-Hydroxyacyl-CoA dehydrogenase尚未报道。姚抗研究M. neoaurum ATCC 25795时证实Hsd4A是第二轮氧化中催化22-HOCO-CoA为22-DOCO-CoA的关键酶,敲除该基因后C22中间体β-氧化代谢完全被抑制,转向新的旁路代谢途径HMP途径而生成HMPD,此外该酶还具有C17-β-羟基氧化作用,为双功能酶[2, 165]。
Nesbitt等研究发现M. tuberculosis H37Rv的FadA5具有Thiolase功能,它是转化胆固醇为4-AD与ADD产物的关键酶[172]。姚抗研究发现当敲除M. neoaurum ATCC 25795的fadA5基因后植物甾醇代谢过程不再积累4-AD与ADD产物,反而大量积累HMPD,应该是转向旁路代谢途径HMP途径所致,说明发生β-氧化的4-AD途径与ADD途径完全被抑制[2]。Schaefer等证实M. tuberculosis H37Rv的FadA5具有催化22-DOCO-CoA为3-OPC-CoA的功能,推测可能也具有催化27-DOCS-CoA为3-OCO-CoA的功能,也就是第一轮β-氧化的最后一步[173]。第三轮β-氧化最后一步则由醛缩酶Ltp2完成,ltp2属于igr操纵子,姚抗研究发现当敲除M. neoaurum ATCC 25795的ltp2基因后代谢产物发生显著变化,不再积累4-AD与ADD产物,而出现未见报道的6种复杂物质,但由于没有分离出单体物质而无法表征[2]。Gilbert等证实Ltp2可以转化17-BHOPC-CoA为4-AD产物,证实M. tuberculosis H37Rv的Ltp2协同ChsH2形成复合体发挥最大催化作用,在生物体内可能形成Ltp2-ChsH2-ChsH1复合体发生作用[170]。
β-谷甾醇发生氧化作用与胆固醇氧化作用差异性源于C24存在乙基支链,在第一轮脱氢后并非直接发生水合作用,而是C28位首先发生羧化作用,这一步由Methyl-crotonyl-CoA carboxylase催化完成,该酶受到β-Sitosterol严格诱导,而Cholesterol则不能诱导[148]。接着由Ltp3-Ltp4发生反醛醇反应脱去1分子丙酰辅酶A,Wilbrink等在研究RG32时证实Ltp3-Ltp4是催化含有C24支链β-Sitosterol与Campesterol等降解的关键酶系,具有醛缩酶功能[129],而姚抗研究发现当敲除M. neoaurum ATCC 25795的ltp3-ltp4基因后植物甾醇代谢仍旧积累4-AD与ADD产物,与野生型基本一致,说明在该菌中Ltp3-Ltp4并非是植物甾醇降解的关键酶系,可能还存在其它同工酶作用,可能是侧链降解的关键酶系[2]。
综上所述,经过侧链降解后,1分子胆固醇可以产生2分子丙酰辅酶A和1分子乙酰辅酶A,而1分子β-谷甾醇则可以产生3分子丙酰辅酶A和1分子乙酰辅酶A,丙酰辅酶A与乙酰辅酶A最后会进入TCA (Tricarboxylic acid cycle)循环进一步氧化分解。有研究者估算1分子的胆固醇经过氧化最后可得7分子的FADH2和18分子的NADH,至少产生80单位的ATP[148]。
3.3.4 核心环降解(Core ring cleavage)核心环降解代谢(图 3C)是3-甾酮-Δ1-脱氢酶(3-Ketosteroid-Δ1-dehydrogenase,KstD)与3-甾酮-9α-羟化酶(3-Ketosteroid-9α-hydroxylase,KSH)共同作用引起的,KstD使A环的C1(2)发生脱氢作用,而KSH则是使B环C9发生羟基化作用,从而产生9α-羟基-1, 4-二烯甾体的不稳定结构,由于过高的分子势能自发引发B环C9(10)开环裂解,通常称为9, 10-开环降解途径(9, 10-sec-pathway)。因此KstD与KSH成为重要甾药化合物筛选需要考虑的关键酶,KstD的失活或缺失可以通过发酵富集制备9-OHAD、9-DHMP、4-BNC以及Ts等,而KSH的失活或缺失可以通过发酵富集制备ADD、HMPD、1, 4-BNC以及BD等,当KstD与KSH两者同时缺失或失活时可以制备4-AD、HMP等。然而不同微生物中KstD与KSH的种类与功能存在较大的差异性,对底物的选择性有显著性差异,在生物体内是否受甾类物质诱导也存在明显不同。不同微生物的KstD与KSH可能存在多种同工酶基因。研究表明,KstD多数为寡聚膜蛋白,结构中存在穿膜螺旋区,以黄素蛋白FAD为辅酶,在N端存在高度保守的GSG(A/G)(A/G)(A/G)X17E结合区,该酶经纯化后未检测到活性,可能原因在于该酶的功能与细胞膜的生理状态密切相关[2, 169]。Rohman等从结构推测出KstD的催化机制为Tyr487的羟基和Gly491的酰胺键与底物C3的羰基作用导致底物C3酮式-烯醇式互变异构,Tyr318与Tyr119通过氢键作用夺取C2的β-氢质子而C2变成碳负离子,当负电荷转移到C1时,FAD的异咯嗪环N5原子夺取C1的α-氢,此时在C1与C2之间形成双键[169]。KSH属于Class Ⅳ型单加氧酶,由末端加氧酶(KshA)与铁流还原酶(KshB)双组分组成,KshB存在3处参与电子传递的高度保守功能区,即RCYSL为FAD结合区,GSGITP为NAD结合区,CX4CX2CX29C为电子传递性铁流蛋白[2, 213]。KSH羟基化反应源于NADH的氧化,自由电子通过KshB辅酶FAD传递到铁流还原中心,过渡到KshA的Rieske结构域,最后在KshA活性中心Fe2+处结合氧分子,从而完成C9-α位的羟基化反应[2, 122, 162]。
在Rhodococcus erythropolis SQ1中发现分别存在两种KstD[214-215]与KSH[117, 122]。大多数微生物一般只含有一种KshB组分,部分微生物可能存在多种KshB组分[54, 105],目前大多数研究主要集中在KshA组分及其功能与结构的研究,而对KshB组分、功能与结构研究较少,此外也有研究表明KshB组分甾体C26-羟基化密切相关[122, 216]。魏巍在M. neoaurum NwIB-01中发现各存在1种KstD、KshA及KshB,KstD对4-AD催化效率较高,敲除或者使KSH失活会导致ADD产量增加[7, 217-219]。而在M. tuberculosis H37Rv基因组中也仅存在1种KstD、KshA及KshB,虽然KstD不能催化4-AD转化为ADD[123],但KstD与KSH催化关键中间4-BNC与4-BNC-CoA表现出较高酶活性[176, 220],同样在HGMS2中也发现仅存在1种KstD[221]。在M. neoaurum ATCC 25795中存在3种KstD与2种KshA及1种KshB[2, 179],MN-KstD1显著受甾醇与9OHAD底物诱导,而MN-KstD3受4-AD显著诱导,MN-KstD2则诱导作用不明显,推测MN-KstD1是9-OHAD途径关键脱氢酶,MN-KstD3则是参与4-AD途径起到协助降解甾醇作用,MN-KstD1对9-OHAD体现出高催化活性,而MN-KstD3对4-AD具有高催化能力;MN-KshA1受到甾醇底物的显著诱导,MN-KshA2B催化能力总体较弱,而MN-KshA1B对4-BNC与1, 4-BNC表现出高催化能力,其次为ADD,而对4-AD催化作用较弱。在M. neoaurum DSM1381中存在3种KstD,其中KstD1受植物甾醇的显著诱导对HMP催化作用强,而KstD2对4-AD催化能力较HMP强[82]。在Rhodococcus ruber Chol-4中存在3种KstD[119]与3种KshA及1种KshB[114],KstD1对9OHAD与Ts具有高催化能力,KstD2则对Ts与Deoxycorticosterone (DOC)显示高活性,而KstD3仅仅体现在5α-Ts;KshA2对含有短支链的甾类底物催化能力强,KshA3则对长支链甾醇底物体现出高活力,KshA1可能与胆甾酸代谢有关。在Rhodococcus rhodochrous DSM 43269中存在3种KstD[120]与5种KshA及2种KshB[116],KstD1是导致9-OHAD降解的主要原因,其次是KstD2,而KstD3无催化作用;5种KshA同工酶显示出截然不同的底物特性,各自具有特定的底物诱导机制。9-OHAD高产菌种M. neoaurum VKM Ac-1817D中也存在5种KstD与5种KshA组分及2种KshB组分,可以看出5种同工酶KstD对4-AD与9OHAD催化能力很弱,KSH催化4-AD的能力可能相对较强,可以催化Ts为9OH-Ts,也可以催化HMP为9-DHMP[54-55, 87, 222-223]。在4-AD高产菌种M. neoaurum VKM Ac-1815D中仅存在1种KstD与2种KshA及2种KshB组分,表明KstD对4-AD、9-OHAD、HMP催化能力很弱,KSH催化4-AD的能力也可能比较弱[53-54, 90, 222, 224]。在ADD高产菌种M. neoaurum VKM Ac-1816D中仅存在1种KstD与2种KshA及2种KshB组分,表明KstD对4-AD催化能力很强,且同时也可以催化HMP为HMPD,而KSH催化4-AD的能力也可能比较弱[54, 222]。
KstD与KSH是核心环降解的两个关键酶,现在一致认同的是9-OHADD自发发生B环C9(10)位断链为3-HSA,然后在M. tuberculosis的Hsa家族蛋白HsaA/B、HsaC、HsaD或者是C. testosteroni的Tes家族蛋白TesA1/2、TesB、TesD催化降解为HHD (原A环结构)与DOHNAA,HHD在M. tuberculosis的HsaE、HsaF、HsaG或者是C. testosteroni的TesE、TesG、TesF进一步降解为丙酰辅酶A与丙酮酸,而DOHNAA在FadD3、IpdF、FadE30、FadE33、FadE31-32、IpdC、IpdA/B、FadA6、EchA20等酶系进一步转化为丙酰辅酶A和琥珀酰辅酶A (Succinyl-CoA)[45, 73, 95, 104, 157-161, 225-228]。Capyk等研究发现胆固醇环降解酶系对含有支链的侧链代谢产物(如9-HBNCD,9-Hydroxy-3-oxo- 23, 24-bisnorchola-1, 4-diene-20-oic acid;9-HOPDC- CoA,9-Hroxy-3-oxo-pregna-1, 4-diene-20-carboxyl-CoA)的活力要高于侧链完全降解产物(如4-AD)[176]。另外,Rhodococcus spp.在胆甾酸降解中存在类似中间体降解开环途径[229-230]。此外,也有研究表明甾醇侧链降解与核心环部分降解几乎同时进行[176],这也是导致部分关键甾药中间体摩尔收率下降的主要原因。
在Steroidobacter denitrificans研究中发现还存在2, 3-开环降解途径(2, 3-sec-pathway),Yang等[155]研究该菌降解雄性激素(睾酮)发生2, 3-开环降解途径(图 4A),Wang等[156]在研究该菌降解胆固醇代谢时也发现发生了2, 3-开环降解途径(图 4B)。

|
| 图 4 2, 3-开环降解途径 Figure 4 2, 3-sec-pathway of degradation |
|
|
综上所述,1分子的胆固醇经过完全降解后将产生4分子丙酰辅酶A、4分子乙酰辅酶A、1分子丙酮酸和1分子琥珀酰辅酶A,而1分子的β-谷甾醇经过完全降解后将产生5分子丙酰辅酶A、4分子乙酰辅酶A、1分子丙酮酸和1分子琥珀酰辅酶A,丙酰辅酶A、乙酰辅酶A与琥珀酰辅酶A最后进入TCA循环发生彻底氧化,产生大量的FADH2、NADH以及ATP。
4 展望微生物在甾类化合物转化过程中发挥了重要的作用,将先进的生物制造理念引进甾体药物生产工业,从而提高资源利用率,降低能耗,实现绿色制造可持续发展。因此,基于基因组学、蛋白组学以及转录组学进一步揭示关键酶系及其催化机理不仅具有重要的理论意义,而且具有重要的产业化价值。一方面,在已有的研究基础上进一步研究并探明甾类降解微生物分解代谢HMP途径、4-AD途径、ADD途径、9-OHAD途径、HMP途径等关键酶系及分流机制,借助基因工程手段,通过基因敲除构建新型甾药中间体生产工程菌,实现4-AD、ADD、9-OHAD、DHEA、HMP等甾药关键中间体单一产物的生产,提高转化率的同时实现破解产物分离纯化瓶颈难题,几乎所有的甾类激素药物都可以基于4-AD或ADD为原料合成,开发酶法或化学法新型制备工艺。另一方面,基于对已有转化微生物或者酶系结构功能的研究,通过转录组学与蛋白组学测定分析获得微生物转化酶基因序列并克隆构建新型工程菌表达体系,进一步采用基因突变技术筛选高催化活性酶,借助酶工程技术实现酶法转化甾体药物,完成化学合成法不能完成的反应,以及克服化学合成法的区域与立体选择性不足,拓展KstD、KSH、C11/16/17位羟化酶等甾药转化酶系的应用研究,用于生产甾药活性成分(Active pharmaceutical ingredients,APIs)如Testosterone、Boldenone、Dexamethasone等,克服微生物细胞水平转化副产物的生成,提高甾药活性的同时避免化学法有害试剂残留对人体的危害。
| [1] |
Donova MV, Egorova OV. Microbial steroid transformations: current state and prospects[J]. Applied Microbiology and Biotechnology, 2012, 94(6): 1423-1447. DOI:10.1007/s00253-012-4078-0 |
| [2] |
Yao K. Investigation into the molecular mechanism of microbial sterol degradation and its metabolic engineering for the production of steroid pharmaceutical precursors[D]. Shanghai: Doctoral Dissertation of East China University of Science and Technology, 2014 (in Chinese) 姚抗.分枝杆菌甾醇代谢机制的解析以及其代谢工程改造应用于制备重要甾药中间体的研究[D].上海: 华东理工大学博士学位论文, 2014 http://cdmd.cnki.com.cn/Article/CDMD-10251-1014251212.htm |
| [3] |
Wang FQ, Yao K, Wei DZ. From soybean phytosterols to steroid hormones[A]//El-Shemy H. Soybean and Health[M]. Rijeka, Croatia: InTech, 2011: 230-254
|
| [4] |
Baker ME. Origin and diversification of steroids: co-evolution of enzymes and nuclear receptors[J]. Molecular and Cellular Endocrinology, 2011, 334(1/2): 14-20. |
| [5] |
Lin YL. Studies on the microbial transformation of steroids[D]. Jinan: Doctoral Dissertation of Shandong University, 2009 (in Chinese) 林彦良.甾体化合物微生物转化的研究[D].济南: 山东大学博士学位论文, 2009 http://cdmd.cnki.com.cn/Article/CDMD-10422-2009248538.htm |
| [6] |
Blickenstaff RT. Total Synthesis of Steroids: Organic Chemistry: A Series of Monographs[M]. New York: Academic Press, 1974: 1-337.
|
| [7] |
Wei W. Identification and gene modification of key genes about 3-ketosteroid Δ1-dehydrogenase and 3-ketosteroid 9α-hydroxylase involved in steroid nucleus degradation in Mycobacterium neoaurum NwIB-01 and strain development via genetic manipulation[D]. Shanghai: Doctoral Dissertation of East China University of Science and Technology, 2010 (in Chinese) 魏巍. Mycobacterium neoaurum NwIB-01降解甾醇母核关键酶3-甾酮-△1-脱氢酶和3-甾酮-9α-羟化酶基因的鉴定及其基因工程改造[D].上海: 华东理工大学博士学位论文, 2010 http://cdmd.cnki.com.cn/Article/CDMD-10251-2010138215.htm |
| [8] |
Marker RE, Krueger J. CXⅡ. Sapogenins. XLI. The preparation of Trillin and its conversion to progesterone[J]. Journal of the American Chemical Society, 1940, 62(12): 3349-3350. DOI:10.1021/ja01869a023 |
| [9] |
Marker RE. Sterols. CXⅢ. Sapogenins. XLⅡ. The conversion of the sapogenins to the pregnenolones[J]. Journal of the American Chemical Society, 1940, 62(12): 3350-3352. DOI:10.1021/ja01869a024 |
| [10] |
Djerassi C, Rosenkranz G, Pataki J, et al. Steroids, XXVⅡ. Synthesis of allopregnane-3beta, 11beta, 17alpha-, 20beta, 21-pentol from cortisone and diosgenin[J]. The Journal of Biological Chemistry, 1952, 194(1): 115-118. |
| [11] |
Hanson JR. Steroids: reactions and partial synthesis[J]. Natural Product Reports, 2005, 22(1): 104-110. |
| [12] |
Zhang WQ. Expression of 3-ketosteroid-Δ1-dehydrogenase from Mycobacterium neoaurum in Bacillus subtilis and its application[D]. Wuxi: Master's Thesis of Jiangnan University, 2013 (in Chinese) 张温清.分枝杆菌3-甾酮-Δ1-脱氢酶在枯草芽孢杆菌中的表达及其应用研究[D].无锡: 江南大学硕士学位论文, 2013 http://cdmd.cnki.com.cn/Article/CDMD-10295-1013309495.htm |
| [13] |
Fernandes P, Cabral JMS. Phytosterols: applications and recovery methods[J]. Bioresource Technology, 2007, 98(12): 2335-2350. DOI:10.1016/j.biortech.2006.10.006 |
| [14] |
Funder JW. Minireview: Aldosterone and mineralocorticoid receptors: past, present, and future[J]. Endocrinology, 2010, 151(11): 5098-5102. DOI:10.1210/en.2010-0465 |
| [15] |
Li TG, Chiang JY. Bile acid signaling in metabolic disease and drug therapy[J]. Pharmacological Reviews, 2014, 66(4): 948-983. DOI:10.1124/pr.113.008201 |
| [16] |
Donova MV. Steroid Bioconversions[A]//Barredo JL, Herráiz I. Microbial Steroids: Methods and Protocols[M]. New York, NY: Humana Press, 2017: 1-13 https://www.ncbi.nlm.nih.gov/pubmed/28710617
|
| [17] |
Antunes LCM, Davies JE, Finlay BB. Chemical signaling in the gastrointestinal tract[J]. F1000 Biology Reports, 2011, 3: 4. |
| [18] |
Fragkaki AG, Angelis YS, Koupparis M, et al. Structural characteristics of anabolic androgenic steroids contributing to binding to the androgen receptor and to their anabolic and androgenic activities applied modifications in the steroidal structure[J]. Steroids, 2009, 74(2): 172-197. DOI:10.1016/j.steroids.2008.10.016 |
| [19] |
Wilbrink MH. Microbial sterol side chain degradation in Actinobacteria[D]. Groningen: Doctoral Dissertation of University of Groningen, 2011
|
| [20] |
Moreau RA, Nyström L, Whitaker BD, et al. Phytosterols and their derivatives: Structural diversity, distribution, metabolism, analysis, and health-promoting uses[J]. Progress in Lipid Research, 2018, 70: 35-61. DOI:10.1016/j.plipres.2018.04.001 |
| [21] |
Piironen V, Lindsay DG, Miettinen TA, et al. Plant sterols: biosynthesis, biological function and their importance to human nutrition[J]. Journal of the Science of Food and Agriculture, 2000, 80(7): 939-966. DOI:10.1002/(SICI)1097-0010(20000515)80:7<939::AID-JSFA644>3.0.CO;2-C |
| [22] |
Hofmann AF, Hagey LR. Bile acids: chemistry, pathochemistry, biology, pathobiology, and therapeutics[J]. Cellular and Molecular Life Sciences, 2008, 65(16): 2461-2483. DOI:10.1007/s00018-008-7568-6 |
| [23] |
Schüle C, Eser D, Baghai TC, et al. Neuroactive steroids in affective disorders: target for novel antidepressant or anxiolytic drugs?[J]. Neuroscience, 2011, 191: 55-77. DOI:10.1016/j.neuroscience.2011.03.025 |
| [24] |
Eser D, Schüle C, Baghai TC, et al. Neuroactive steroids and affective disorders[J]. Pharmacology Biochemistry and Behavior, 2006, 84(4): 656-666. DOI:10.1016/j.pbb.2006.05.020 |
| [25] |
Melcangi RC, Panzica G, Garcia-Segura LM. Neuroactive steroids: focus on human brain[J]. Neuroscience, 2011, 191: 1-5. DOI:10.1016/j.neuroscience.2011.06.024 |
| [26] |
Yang Y. Research on study on bioconversion of phytosteril into steroid medicine intermediates by mycobacter[D]. Hefei: Doctoral Dissertation of Hefei University of Technology, 2009 (in Chinese) 杨英.微生物转化植物甾醇制备甾体药物关键中间体研究[D].合肥: 合肥工业大学博士学位论文, 2009 http://cdmd.cnki.com.cn/Article/CDMD-10359-2010037032.htm |
| [27] |
Wang XJ, Feng JH, Zhang DL, et al. Characterization of new recombinant 3-ketosteroid-Δ1-dehydrogenases for the biotransformation of steroids[J]. Applied Microbiology and Biotechnology, 2017, 101(15): 6049-6060. DOI:10.1007/s00253-017-8378-2 |
| [28] |
Parshikov IA, Sutherland JB. Biotransformation of steroids and flavonoids by cultures of Aspergillus niger[J]. Applied Biochemistry and Biotechnology, 2015, 176(3): 903-923. |
| [29] |
Donova MV. Transformation of steroids by actinobacteria: a review[J]. Applied Biochemistry and Microbiology, 2007, 43(1): 1-14. |
| [30] |
Gao Q, Qiao YQ, Shen YB, et al. Screening for strains with 11α-hydroxylase activity for 17α-hydroxy progesterone biotransformation[J]. Steroids, 2017, 124: 67-71. DOI:10.1016/j.steroids.2017.05.009 |
| [31] |
Savinova OS, Solyev PN, Vasina DV, et al. Biotransformation of progesterone by the ascomycete Aspergillus niger N402[J]. Biochemistry (Moscow), 2018, 83(1): 26-31. DOI:10.1134/S0006297918010030 |
| [32] |
Wei Q. Screeing and transformation conditions of 11α-hydroxylation of androst-4-ene-3, 17-dione by Beauveria bassiana[D]. Wuhan: Master's Thesis of Huazhong Agricultural University, 2007 (in Chinese) 魏琦.雄烯二酮11α羟化的球孢白僵菌菌株筛选及其转化工艺研究[D].武汉: 华中农业大学硕士学位论文, 2007 http://cdmd.cnki.com.cn/Article/CDMD-10504-2007209435.htm |
| [33] |
Yeh CH, Kuo YS, Chang CM, et al. Deletion of the gene encoding the reductase component of 3-ketosteroid 9α-hydroxylase in Rhodococcus equi USA-18 disrupts sterol catabolism, leading to the accumulation of 3-oxo-23, 24-bisnorchola-1, 4-dien-22-oic acid and 1, 4-androstadiene-3, 17-dione[J]. Microbial Cell Factories, 2014, 13: 130. |
| [34] |
Zhang CG. The clone and expression of 3-ketosteroid-1(2)-dehydrogenase from Arthrobacter simplex and Mycobacterium sp.[D]. Shanghai: Master's Thesis of East China University of Science and Technology, 2011 (in Chinese) 张成刚.简单节杆菌与分枝杆菌3-酮基甾体-1(2)位脱氢酶的克隆表达[D].上海: 华东理工大学硕士学位论文, 2011 http://cdmd.cnki.com.cn/Article/CDMD-10251-1011174942.htm |
| [35] |
Li Y, Lu F, Sun T, et al. Expression of ksdD gene encoding 3-ketosteroid-Δ1-dehydrogenase from Arthrobacter simplex in Bacillus subtilis[J]. Letters in Applied Microbiology, 2007, 44(5): 563-568. DOI:10.1111/j.1472-765X.2007.02134.x |
| [36] |
Naghibi F, Tabatabai-Yazdi M, Noori-Daloii M, et al. Microbial chain degradation of cholesterol by Arthrobacter simplex[J]. Journal of Sciences Islamic Republic of Iran, 1995, 6(4): 207-210. |
| [37] |
Owen RW, Mason AN, Bilton RF. The degradation of β-sitosterol by Pseudomonas sp. NCIB 10590 under aerobic conditions[J]. Journal of Steroid Biochemistry, 1985, 23(3): 327-332. DOI:10.1016/0022-4731(85)90412-1 |
| [38] |
Göbel M, Kassel-Cati K, Schmidt E, et al. Degradation of aromatics and chloroaromatics by Pseudomonas sp. strain B13: cloning, characterization, and analysis of sequences encoding 3-oxoadipate:succinyl-coenzyme A (CoA) transferase and 3-oxoadipyl-CoA thiolase[J]. Journal of Bacteriology, 2002, 184(1): 216-223. DOI:10.1128/JB.184.1.216-223.2002 |
| [39] |
Kaschabek SR, Kuhn B, Muller D, et al. Degradation of aromatics and chloroaromatics by Pseudomonas sp. strain B13: purification and characterization of 3-oxoadipate: succinyl-coenzyme A (CoA) transferase and 3-oxoadipyl-CoA thiolase[J]. Journal of Bacteriology, 2002, 184(1): 207-215. DOI:10.1128/JB.184.1.207-215.2002 |
| [40] |
Holert J, Yücel O, Jagmann N, et al. Identification of bypass reactions leading to the formation of one central steroid degradation intermediate in metabolism of different bile salts in Pseudomonas sp. strain Chol1[J]. Environmental Microbiology, 2016, 18(10): 3373-3389. DOI:10.1111/1462-2920.13192 |
| [41] |
Holert J, Kulić Ž, Yücel O, et al. Degradation of the acyl side chain of the steroid compound cholate in Pseudomonas sp. strain Chol1 proceeds via an aldehyde intermediate[J]. Journal of Bacteriology, 2013, 195(3): 585-595. DOI:10.1128/JB.01961-12 |
| [42] |
Holert J, Jagmann N, Philipp B. The essential function of genes for a hydratase and an aldehyde dehydrogenase for growth of Pseudomonas sp. strain Chol1 with the steroid compound cholate indicates an aldolytic reaction step for deacetylation of the side chain[J]. Journal of Bacteriology, 2013, 195(15): 3371-3380. DOI:10.1128/JB.00410-13 |
| [43] |
Hawley RJ, Imaeda T, Mann N. Isolation and characterization of nocardia-like variants of Mycobacterium smegmatis[J]. Canadian Journal of Microbiology, 1976, 22(10): 1480-1491. DOI:10.1139/m76-219 |
| [44] |
Atrat P, Hüller E, Hörhold C, et al. Steroid transformation using immobilized microorganisms. Ⅱ. Degradation of the sidechain of cholesterol by immobilized cells of Nocardia erythropolis[J]. Zeitschrift fur Allgemeine Mikrobiologie, 1980, 20(3): 159-166. DOI:10.1002/jobm.3630200302 |
| [45] |
Horinouchi M, Hayashi T, Kudo T. Steroid degradation in Comamonas testosteroni[J]. The Journal of Steroid Biochemistry and Molecular Biology, 2012, 129(1/2): 4-14. |
| [46] |
Shtratnikova VY, Schelkunov MI, Fokina VV, et al. Genome-wide bioinformatics analysis of steroid metabolism-associated genes in Nocardioides simplex VKM Ac-2033D[J]. Current Genetics, 2016, 62(3): 643-656. DOI:10.1007/s00294-016-0568-4 |
| [47] |
Horinouchi M, Hayashi T, Yamamoto T, et al. A new bacterial steroid degradation gene cluster in Comamonas testosteroni TA441 which consists of aromatic-compound degradation genes for seco-steroids and 3-ketosteroid dehydrogenase genes[J]. Applied and Environmental Microbiology, 2003, 69(8): 4421-4430. DOI:10.1128/AEM.69.8.4421-4430.2003 |
| [48] |
Birkenmaier A, Möller HM, Philipp B. Identification of a thiolase gene essential for β-oxidation of the acyl side chain of the steroid compound cholate in Pseudomonas sp. strain Chol1[J]. FEMS Microbiology Letters, 2011, 318(2): 123-130. DOI:10.1111/j.1574-6968.2011.02250.x |
| [49] |
de las Heras LF, Fernández EG, Llorens JMN, et al. Morphological, physiological, and molecular characterization of a newly isolated steroid-degrading actinomycete, identified as Rhodococcus ruber strain Chol-4[J]. Current Microbiology, 2009, 59(5): 548-553. DOI:10.1007/s00284-009-9474-z |
| [50] |
Sih CJ, Tai HH, Tsong YY, et al. Mechanisms of steroid oxidation by microorgansism. XIV. Pathway of cholesterol side-chain degradation[J]. Biochemistry, 1968, 7(2): 808-818. DOI:10.1021/bi00842a039 |
| [51] |
Yam KC, van der Geize R, Eltis LD. Catabolism of aromatic compounds and steroids by Rhodococcus[A]//Alvarez HM. Biology of Rhodococcus[M]. Berlin, Heidelberg: Springer, 2010: 133-169
|
| [52] |
Mohn WW, Wilbrink MH, Casabon I, et al. Gene cluster encoding cholate catabolism in Rhodococcus spp.[J]. Journal of Bacteriology, 2012, 194(24): 6712-6719. DOI:10.1128/JB.01169-12 |
| [53] |
Egorova OV, Gulevskaya SA, Puntus IF, et al. Production of androstenedione using mutants of Mycobacterium sp.[J]. Journal of Chemical Technology and Biotechnology, 2002, 77(2): 141-147. DOI:10.1002/jctb.536 |
| [54] |
Bragin EY, Shtratnikova VY, Dovbnya DV, et al. Comparative analysis of genes encoding key steroid core oxidation enzymes in fast-growing Mycobacterium spp. strains[J]. The Journal of Steroid Biochemistry and Molecular Biology, 2013, 138: 41-53. DOI:10.1016/j.jsbmb.2013.02.016 |
| [55] |
Donova MV, Dovbnya DV, Sukhodolskaya GV, et al. Microbial conversion of sterol-containing soybean oil production waste[J]. Journal of Chemical Technology and Biotechnology, 2005, 80(1): 55-60. DOI:10.1002/jctb.1156 |
| [56] |
Xu LX, Yang HL, Kuang MA, et al. Comparative genomic analysis of Mycobacterium neoaurum MN2 and MN4 substrate and product tolerance[J]. 3 Biotech, 2017, 7(3): 181. DOI:10.1007/s13205-017-0818-2 |
| [57] |
Molchanova MA, Andriushina VA, Savinova TS, et al. Preparation of androsta-1, 4-diene-3, 17-dione from sterols using Mycobacterium neoaurum VKPM As-1656 strain[J]. Bioorganicheskaia Khimiia, 2007, 33(3): 379-384. |
| [58] |
Wovcha MG, Antosz FJ, Knight JC, et al. Bioconversion of sitosterol to useful steroidal intermediates by mutants of Mycobacterium fortuitum[J]. Biochimica et Biophysica Acta (BBA) - Lipids and Lipid Metabolism, 1978, 531(3): 308-321. DOI:10.1016/0005-2760(78)90213-8 |
| [59] |
Josefsen KD, Nordborg A, Sletta H. Bioconversion of phytosterols into androstenedione by Mycobacterium[A]//Barredo JL, Herráiz I. Microbial Steroids: Methods and Protocols[M]. New York, NY: Humana Press, 2017: 177-197
|
| [60] |
Malaviya A, Gomes J. Rapid screening and isolation of a fungus for sitosterol to androstenedione biotransformation[J]. Applied Biochemistry and Biotechnology, 2009, 158(2): 374-386. DOI:10.1007/s12010-008-8416-8 |
| [61] |
Lin YL, Song X, Fu J, et al. Microbial transformation of phytosterol in corn flour and soybean flour to 4-androstene-3, 17-dione by Fusarium moniliforme Sheld[J]. Bioresource Technology, 2009, 100(5): 1864-1867. DOI:10.1016/j.biortech.2008.09.040 |
| [62] |
Kollerov VV, Shutov AA, Fokina VV, et al. Bioconversion of C19- and C21-steroids with parent and mutant strains of Curvularia lunata[J]. Prikladnaia Biokhimiia I Mikrobiologiia, 2010, 46(2): 212-220. |
| [63] |
Wang WY, Ge FL, Ma CH, et al. Heterologous expression and characterization of a 3-ketosteroid-∆1-dehydrogenase from Gordonia neofelifaecis and its utilization in the bioconversion of androst-4, 9(11)-dien-3, 17-dione[J]. 3 Biotech, 2017, 7(1): 19. |
| [64] |
Yuan JD, Chen GY, Cheng SJ, et al. Accumulation of 9α-hydroxy-4-androstene-3, 17-dione by co-expressing kshA and kshB encoding component of 3-ketosteroid-9α-hydroxylase in Mycobacterium sp. NRRL B-3805[J]. Chinese Journal of Biotechnology, 2015, 31(4): 523-533. (in Chinese) 袁家代, 陈贵英, 程世君, 等. 3-甾酮-9α-羟基化酶基因在分枝杆菌中的异源表达与9α-羟基雄烯二酮的制备[J]. 生物工程学报, 2015, 31(4): 523-533. |
| [65] |
Li WJ, Ge FL, Zhang QY, et al. Identification of gene expression profiles in the actinomycete Gordonia neofelifaecis grown with different steroids[J]. Genome, 2014, 57(6): 345-353. DOI:10.1139/gen-2014-0030 |
| [66] |
Liu YC, Ge FL, Chen GY, et al. Gordonia neofelifaecis sp. nov., a cholesterol side-chain-cleaving actinomycete isolated from the faeces of Neofelis nebulosa[J]. International Journal of Systematic and Evolutionary Microbiology, 2011, 61(1): 165-169. DOI:10.1099/ijs.0.020321-0 |
| [67] |
Chaudhari PN, Chaudhari BL, Chincholkar SB. Cholesterol biotransformation to androsta-1, 4-diene-3, 17-dione by growing cells of Chryseobacterium gleum[J]. Biotechnology Letters, 2010, 32(5): 695-699. DOI:10.1007/s10529-010-0206-z |
| [68] |
Zhang QY, Ren Y, He JZ, et al. Multiplicity of 3-ketosteroid Δ1-dehydrogenase enzymes in Gordonia neofelifaecis NRRL B-59395 with preferences for different steroids[J]. Annals of Microbiology, 2015, 65(4): 1961-1971. DOI:10.1007/s13213-015-1034-0 |
| [69] |
Gordon RE, Mihm JM. A comparative study of some strains received as nocardiae[J]. Journal of Bacteriology, 1957, 73(1): 15-27. |
| [70] |
Larkin MJ, Kulakov LA, Allen CC. Biodegradation and Rhodococcus–masters of catabolic versatility[J]. Current Opinion in Biotechnology, 2005, 16(3): 282-290. DOI:10.1016/j.copbio.2005.04.007 |
| [71] |
Bonds AC, Sampson NS. More than cholesterol catabolism: regulatory vulnerabilities in Mycobacterium tuberculosis[J]. Current Opinion in Chemical Biology, 2018, 44: 39-46. DOI:10.1016/j.cbpa.2018.05.012 |
| [72] |
Ouellet H, Johnston JB, de Montellano PRO. Cholesterol catabolism as a therapeutic target in Mycobacterium tuberculosis[J]. Trends in Microbiology, 2011, 19(11): 530-539. DOI:10.1016/j.tim.2011.07.009 |
| [73] |
García JL, Uhía I, Galán B. Catabolism and biotechnological applications of cholesterol degrading bacteria[J]. Microbial Biotechnology, 2012, 5(6): 679-699. DOI:10.1111/j.1751-7915.2012.00331.x |
| [74] |
Kendall SL, Withers M, Soffair CN, et al. A highly conserved transcriptional repressor controls a large regulon involved in lipid degradation in Mycobacterium smegmatis and Mycobacterium tuberculosis[J]. Molecular Microbiology, 2007, 65(3): 684-699. DOI:10.1111/j.1365-2958.2007.05827.x |
| [75] |
Neu TR. Significance of bacterial surface-active compounds in interaction of bacteria with interfaces[J]. Microbiological Reviews, 1996, 60(1): 151-166. |
| [76] |
Bicca FC, Fleck LC, Ayub MAZ. Production of biosurfactant by hydrocarbon degrading Rhodococcus ruber and Rhodococcus erythropolis[J]. Revista de Microbiologia, 1999, 30(3): 231-236. DOI:10.1590/S0001-37141999000300008 |
| [77] |
Donova MV, Nikolaeva VM, Egorova OV. Enzymes involved in modification of the steroid nucleus of industrial mycobacterial strains: isolation, functions, and properties[J]. Prikladnaia Biokhimiia I Mikrobiologiia, 2005, 41(5): 514-520. |
| [78] |
Huang CL, Chen YR, Liu WH. Production of androstenones from phytosterol by mutants of Mycobacterium sp.[J]. Enzyme and Microbial Technology, 2006, 39(2): 296-300. DOI:10.1016/j.enzmictec.2005.10.017 |
| [79] |
Gulla V, Banerjee T, Patil S. Bioconversion of soysterols to androstenedione by Mycobacterium fortuitum subsp. fortuitum NCIM 5239, a mutant derived from total sterol degrader strain[J]. Journal of Chemical Technology and Biotechnology, 2010, 85(8): 1135-1141. DOI:10.1002/jctb.2410 |
| [80] |
Seidel L, Hörhold C. Selection and characterization of new microorganisms for the manufacture of 9-OH-AD from sterols[J]. Journal of Basic Microbiology, 1992, 32(1): 49-55. DOI:10.1002/jobm.3620320115 |
| [81] |
Stahl DA, Urbance JW. The division between fast- and slow-growing species corresponds to natural relationships among the mycobacteria[J]. Journal of Bacteriology, 1990, 172(1): 116-124. DOI:10.1128/jb.172.1.116-124.1990 |
| [82] |
Zhang RJ, Liu XC, Wang YS, et al. Identification, function, and application of 3-ketosteroid Δ1-dehydrogenase isozymes in Mycobacterium neoaurum DSM 1381 for the production of steroidic synthons[J]. Microbial Cell Factories, 2018, 17(1): 77. DOI:10.1186/s12934-018-0916-9 |
| [83] |
Yang F, Wang H, Song S, et al. Complete genome sequence of Mycobacterium sp. Neoaurum HGMS2[DB/OL]. (2018-08-01). https://www.ncbi.nlm.nih.gov/nuccore/NZ_CP031414.1
|
| [84] |
Su LQ, Shen YB, Xia ML, et al. Overexpression of cytochrome P450 125 in Mycobacterium: a rational strategy in the promotion of phytosterol biotransformation[J]. Journal of Industrial Microbiology & Biotechnology, 2018, 45(10): 857-867. |
| [85] |
Liu HH, Xu LQ, Yao K, et al. Engineered 3-ketosteroid 9α-hydroxylases in Mycobacterium neoaurum: an efficient platform for production of steroid drugs[J]. Applied and Environmental Microbiology, 2018, 84(14): e02777-17. |
| [86] |
Liu HH, Xu LQ, Yao K, et al. Characterization and engineering of 3-ketosteroid 9α-hydroxylases in Mycobacterium neoaurum ATCC 25795 for the development of androst-1, 4-diene-3, 17-dione and 9α-hydroxy-androst-4-ene-3, 17-dione--producing strains[J]. Applied and Environmental Microbiology, 2018, 84(14): e02777-17. |
| [87] |
Li XM, Chen X, Wang Y, et al. New product identification in the sterol metabolism by an industrial strain Mycobacterium neoaurum NRRL B-3805[J]. Steroids, 2018, 132: 40-45. DOI:10.1016/j.steroids.2018.02.001 |
| [88] |
Rodríguez-García A, Fernández-Alegre E, Morales A, et al. Complete genome sequence of 'Mycobacterium neoaurum' NRRL B-3805, an androstenedione (AD) producer for industrial biotransformation of sterols[J]. Journal of Biotechnology, 2016, 224: 64-65. DOI:10.1016/j.jbiotec.2016.03.021 |
| [89] |
Shtratnikova VY, Schelkunov MI, Dovbnya DV, et al. Complete genome sequence of Mycobacterium sp. strain VKM Ac-1817D, capable of producing 9α-Hydroxy-androst-4-ene-3, 17-dione from phytosterol[J]. Genome Announcements, 2015, 3(1): e01447-14. |
| [90] |
Shtratnikova VY, Bragin EY, Dovbnya DV, et al. Complete genome sequence of sterol-transforming Mycobacterium neoaurum strain VKM Ac-1815D[J]. Genome Announcements, 2014, 2(1): e01177-13. |
| [91] |
Majumdar G, Mbau R, Singh V, et al. Genome-wide transposon mutagenesis in Mycobacterium tuberculosis and Mycobacterium smegmatis[A]//Reeves A. In Vitro Mutagenesis[M]. New York, NY: Humana Press, 2017: 321-335 https://link.springer.com/protocol/10.1007/978-1-4939-6472-7_21
|
| [92] |
Li XF, Mei H, Chen F, et al. Transcriptome landscape of Mycobacterium smegmatis[J]. Frontiers in Microbiology, 2017, 8: 2505. DOI:10.3389/fmicb.2017.02505 |
| [93] |
Li Q, Ge FL, Tan YY, et al. Genome-wide transcriptome profiling of Mycobacterium smegmatis MC2 155 cultivated in minimal media supplemented with cholesterol, androstenedione or glycerol[J]. International Journal of Molecular Sciences, 2016, 17(5): 689. DOI:10.3390/ijms17050689 |
| [94] |
Galán B, Uhía I, García-Fernández E, et al. Mycobacterium smegmatis is a suitable cell factory for the production of steroidic synthons[J]. Microbial Biotechnology, 2017, 10(1): 138-150. |
| [95] |
Uhía I, Galán B, Kendall SL, et al. Cholesterol metabolism in Mycobacterium smegmatis[J]. Environmental Microbiology Reports, 2012, 4(2): 168-182. DOI:10.1111/j.1758-2229.2011.00314.x |
| [96] |
Mohan A, Padiadpu J, Baloni P, et al. Complete genome sequences of a Mycobacterium smegmatis laboratory strain (MC2 155) and isoniazid-resistant (4XR1/R2) mutant strains[J]. Genome Announcements, 2015, 3(1): e01520-14. |
| [97] |
Shtratnikova VY, Schelkunov MI, Pekov YA, et al. Complete genome sequence of steroid-transforming Nocardioides simplex VKM Ac-2033D[J]. Genome Announcements, 2015, 3(1): e01406-14. |
| [98] |
Holert J, Alam I, Larsen M, et al. Genome sequence of Pseudomonas sp. strain Chol1, a model organism for the degradation of bile salts and other steroid compounds[J]. Genome Announcements, 2013, 1(1): e00014-12. |
| [99] |
Xu LX, Wang XL, Yang HL, et al. Whole-genome sequencing and analysis of a adrostenedione yielding strains Mycobacterium neoaurum MN4[J]. Acta Microbiologica Sinica, 2016, 56(8): 1358-1367. (in Chinese) 徐玲霞, 王筱兰, 杨慧林, 等. 雄烯二酮高产菌新金分枝杆菌MN4的全基因组测序及序列分析[J]. 微生物学报, 2016, 56(8): 1358-1367. |
| [100] |
Camus JC, Pryor MJ, Médigue C, et al. Re-annotation of the genome sequence of Mycobacterium tuberculosis H37Rv[J]. Microbiology, 2002, 148(10): 2967-2973. DOI:10.1099/00221287-148-10-2967 |
| [101] |
Cole S, Brosch R, Parkhill J, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence[J]. Nature, 1998, 393(6685): 537-544. DOI:10.1038/31159 |
| [102] |
Liu M, Xiong LB, Tao XY, et al. Integrated transcriptome and proteome studies reveal the underlying mechanisms for sterol catabolism and steroid production in Mycobacterium neoaurum[J]. Journal of Agricultural and Food Chemistry, 2018, 66(34): 9147-9157. DOI:10.1021/acs.jafc.8b02714 |
| [103] |
Schnappinger D, Ehrt S, Voskuil MI, et al. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages[J]. Journal of Experimental Medicine, 2003, 198(5): 693-704. DOI:10.1084/jem.20030846 |
| [104] |
van der Geize R, Yam K, Heuser T, et al. A gene cluster encoding cholesterol catabolism in a soil actinomycete provides insight into Mycobacterium tuberculosis survival in macrophages[J]. Proceedings of the National Academy of Sciences of the United States of America, 2007, 104(6): 1947-1952. DOI:10.1073/pnas.0605728104 |
| [105] |
Petrusma M, Dijkhuizen L, van der Geize R. Rhodococcus rhodochrous DSM 43269 3-ketosteroid 9α-hydroxylase, a two-component iron-sulfur-containing monooxygenase with subtle steroid substrate specificity[J]. Applied and Environmental Microbiology, 2009, 75(16): 5300-5307. DOI:10.1128/AEM.00066-09 |
| [106] |
Rosłoniec KZ, Wilbrink MH, Capyk JK, et al. Cytochrome P450 125 (CYP125) catalyses C26-hydroxylation to initiate sterol side-chain degradation in Rhodococcus jostii RHA1[J]. Molecular Microbiology, 2009, 74(5): 1031-1043. DOI:10.1111/j.1365-2958.2009.06915.x |
| [107] |
García-Fernández E, Frank DJ, Galán B, et al. A highly conserved mycobacterial cholesterol catabolic pathway[J]. Environmental Microbiology, 2013, 15(8): 2342-2359. DOI:10.1111/1462-2920.12108 |
| [108] |
Luengo JM, Olivera ER. Identification and characterization of the genes and enzymes belonging to the bile acid catabolic pathway in Pseudomonas[A]//Barredo JL, Herráiz I. Microbial Steroids: Methods and Protocols[M]. New York, NY: Humana Press, 2017: 109-142
|
| [109] |
Bergstrand LH, Cardenas E, Holert J, et al. Delineation of steroid-degrading microorganisms through comparative genomic analysis[J]. mBio, 2016, 7(2): e00166-16. |
| [110] |
Wipperman MF, Sampson NS, Thomas ST. Pathogen roid rage: cholesterol utilization by Mycobacterium tuberculosis[J]. Critical Reviews in Biochemistry and Molecular Biology, 2014, 49(4): 269-293. DOI:10.3109/10409238.2014.895700 |
| [111] |
Griffin JE, Gawronski JD, DeJesus MA, et al. High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism[J]. PLoS Pathogens, 2011, 7(9): e1002251. DOI:10.1371/journal.ppat.1002251 |
| [112] |
Yao K, Wang FQ, Zhang HC, et al. Identification and engineering of cholesterol oxidases involved in the initial step of sterols catabolism in Mycobacterium neoaurum[J]. Metabolic Engineering, 2013, 15: 75-87. DOI:10.1016/j.ymben.2012.10.005 |
| [113] |
Uhía I, Galán B, Morales V, et al. Initial step in the catabolism of cholesterol by Mycobacterium smegmatis mc2155[J]. Environmental Microbiology, 2011, 13(4): 943-959. DOI:10.1111/j.1462-2920.2010.02398.x |
| [114] |
Guevara G, de las Heras LF, Perera J, et al. Functional characterization of 3-Ketosteroid 9α-hydroxylases in Rhodococcus ruber strain Chol-4[J]. The Journal of Steroid Biochemistry and Molecular Biology, 2017, 172: 176-187. DOI:10.1016/j.jsbmb.2017.06.011 |
| [115] |
Petrusma M, Dijkhuizen L, van der Geize R. Structural features in the KshA terminal oxygenase protein that determine substrate preference of 3-ketosteroid 9α-hydroxylase enzymes[J]. Journal of Bacteriology, 2012, 194(1): 115-121. DOI:10.1128/JB.05838-11 |
| [116] |
Petrusma M, Hessels G, Dijkhuizen L, et al. Multiplicity of 3-ketosteroid-9α-hydroxylase enzymes in Rhodococcus rhodochrous DSM43269 for specific degradation of different classes of steroids[J]. Journal of Bacteriology, 2011, 193(15): 3931-3940. DOI:10.1128/JB.00274-11 |
| [117] |
van der Geize R, Hessels GI, Nienhuis-Kuiper M, et al. Characterization of a second Rhodococcus erythropolis SQ1 3-ketosteroid 9α-hydroxylase activity comprising a terminal oxygenase homologue, KshA2, active with oxygenase-reductase component KshB[J]. Applied and Environmental Microbiology, 2008, 74(23): 7197-7203. DOI:10.1128/AEM.00888-08 |
| [118] |
Brzostek A, Śliwiński T, Rumijowska-Galewicz A, et al. Identification and targeted disruption of the gene encoding the main 3-ketosteroid dehydrogenase in Mycobacterium smegmatis[J]. Microbiology, 2005, 151(7): 2393-2402. DOI:10.1099/mic.0.27953-0 |
| [119] |
Guevara G, de las Heras LF, Perera J, et al. Functional differentiation of 3-ketosteroid Δ1-dehydrogenase isozymes in Rhodococcus ruber strain Chol-4[J]. Microbial Cell Factories, 2017, 16: 42. DOI:10.1186/s12934-017-0657-1 |
| [120] |
Liu Y, Shen YB, Qiao YQ, et al. The effect of 3-ketosteroid-Δ1-dehydrogenase isoenzymes on the transformation of AD to 9α-OH-AD by Rhodococcus rhodochrous DSM43269[J]. Journal of Industrial Microbiology & Biotechnology, 2016, 43(9): 1303-1311. |
| [121] |
de las Heras LF, van der Geize R, Drzyzga O, et al. Molecular characterization of three 3-ketosteroid-Δ1-dehydrogenase isoenzymes of Rhodococcus ruber strain Chol-4[J]. The Journal of Steroid Biochemistry and Molecular Biology, 2012, 132(3/5): 271-281. |
| [122] |
van der Geize R, Hessels GI, Van Gerwen R, et al. Molecular and functional characterization of kshA and kshB, encoding two components of 3-ketosteroid 9α-hydroxylase, a class IA monooxygenase, in Rhodococcus erythropolis strain SQ1[J]. Molecular Microbiology, 2002, 45(4): 1007-1018. DOI:10.1046/j.1365-2958.2002.03069.x |
| [123] |
Knol J, Bodewits K, Hessels GI, et al. 3-Keto-5α-steroid Δ1-dehydrogenase from Rhodococcus erythropolis SQ1 and its orthologue in Mycobacterium tuberculosis H37Rv are highly specific enzymes that function in cholesterol catabolism[J]. Biochemical Journal, 2008, 410(2): 339-346. DOI:10.1042/BJ20071130 |
| [124] |
Florin C, Köhler T, Grandguillot M, et al. Comamonas testosteroni 3-ketosteroid-Δ4(5α)-dehydrogenase: gene and protein characterization[J]. Journal of Bacteriology, 1996, 178(11): 3322-3330. DOI:10.1128/jb.178.11.3322-3330.1996 |
| [125] |
van Oosterwijk N, Knol J, Dijkhuizen L, et al. Structure and catalytic mechanism of 3-ketosteroid-Δ4-(5α)-dehydrogenase from Rhodococcus jostii RHA1 genome[J]. The Journal of Biological Chemistry, 2012, 287(37): 30975-30983. DOI:10.1074/jbc.M112.374306 |
| [126] |
Ouellet H, Guan SH, Johnston JB, et al. Mycobacterium tuberculosis CYP125A1, a steroid C27 monooxygenase that detoxifies intracellularly generated cholest-4-en-3-one[J]. Molecular Microbiology, 2010, 77(3): 730-742. DOI:10.1111/j.1365-2958.2010.07243.x |
| [127] |
Frank DJ, Waddling CA, La M, et al. Cytochrome P450 125A4, the third cholesterol C-26 hydroxylase from Mycobacterium smegmatis[J]. Biochemistry, 2015, 54(46): 6909-6916. DOI:10.1021/acs.biochem.5b01029 |
| [128] |
Lu R, Schmitz W, Sampson NS. α-methyl acyl CoA racemase provides Mycobacterium tuberculosis catabolic access to cholesterol esters[J]. Biochemistry, 2015, 54(37): 5669-5672. DOI:10.1021/acs.biochem.5b00911 |
| [129] |
Wilbrink MH, van der Geize R, Dijkhuizen L. Molecular characterization of ltp3 and ltp4, essential for C24-branched chain sterol-side-chain degradation in Rhodococcus rhodochrous DSM 43269[J]. Microbiology, 2012, 158(12): 3054-3062. |
| [130] |
Barreiro C, Morales A, Vázquez-Iglesias I, et al. Intra- and extra-cellular proteome analyses of steroid-producer mycobacteria[A]//Barredo JL, Herráiz I. Microbial Steroids: Methods and Protocols[M]. New York, NY: Humana Press, 2017: 73-92
|
| [131] |
Liu M, Zhu ZT, Tao XY, et al. RNA-Seq analysis uncovers non-coding small RNA system of Mycobacterium neoaurum in the metabolism of sterols to accumulate steroid intermediates[J]. Microbial Cell Factories, 2016, 15: 64. DOI:10.1186/s12934-016-0462-2 |
| [132] |
Shell SS, Wang J, Lapierre P, et al. Leaderless transcripts and small proteins are common features of the mycobacterial translational landscape[J]. PLoS Genetics, 2015, 11(11): e1005641. DOI:10.1371/journal.pgen.1005641 |
| [133] |
Sih CJ, Rahim AM. Mechanisms of steroid oxidation by microorganisms Ⅲ: enzymatic mechanism of ring a aromatization[J]. Journal of Pharmaceutical Sciences, 1963, 52(11): 1075-1080. DOI:10.1002/jps.2600521112 |
| [134] |
Wang KC, Sih CJ. Mechanisms of steroid oxidation by microorganisms. Ⅳ. Seco intermediates[J]. Biochemistry, 1963, 2(6): 1238-1243. DOI:10.1021/bi00906a011 |
| [135] |
Tsong YY, Wang KC, Sih CJ. Mechanisms of steroid oxidation by microorganisms. Ⅴ. Reduction of the delta-6, 7 double bond[J]. Biochimica et Biophysica Acta, 1964, 93: 398-401. DOI:10.1016/0304-4165(64)90391-5 |
| [136] |
Coombre RG, Tsong YY, Hamilton PB, et al. Mechanisms of steroid oxidation by microorganisms. Ⅹ. Oxidative cleavage of estrone[J]. The Journal of Biological Chemistry, 1966, 241(7): 1587-1595. |
| [137] |
Gibson DT, Wang KC, Sih CJ, et al. Mechanisms of steroid oxidation by microorganisms. Ⅸ. On the mechanism of ring a cleavage in the degradation of 9, 10-seco steroids by microorganisms[J]. The Journal of Biological Chemistry, 1966, 241(3): 551-559. |
| [138] |
Rahm MA, Sih CJ. Mechanisms of steroid oxidation by microorganisms. Ⅺ. Enzymatic cleavage of the pregnane side chain[J]. The Journal of Biological Chemistry, 1966, 241(15): 3615-3623. |
| [139] |
Sih CJ, Lee SS, Tsong YY, et al. Mechanisms of steroid oxidation by microorganisms. 8. 3, 4-dihydroxy-9, 10-secoandrosta- 1, 3, 5(10)-triene-9, 17-dione, an intermediate in the microbiological degradation of ring a of androst-4-ene-3, 17-dione[J]. The Journal of Biological Chemistry, 1966, 241(3): 540-550. |
| [140] |
Sih CJ. Mechanisms of steroid oxidation by microorganisms[J]. Biochimica et Biophysica Acta, 1962, 62: 541-547. DOI:10.1016/0006-3002(62)90236-6 |
| [141] |
Charney W, Herzog HL. Microbial Transformations of Steroids[M]. New York: Academic Press, 1967: 1-732.
|
| [142] |
Conner AH, Nagaoka M, Rowe JW, et al. Microbial conversion of tall oil sterols to C19 steroids[J]. Applied and Environmental Microbiology, 1976, 32(2): 310-311. |
| [143] |
Coulter AW, Talalay P. Studies on the microbiological degradation of steroid ring A[J]. The Journal of Biological Chemistry, 1968, 243(12): 3238-3247. |
| [144] |
Martin CKA. Microbial cleavage of sterol side chains[J]. Advances in Applied Microbiology, 1977, 22: 29-58. DOI:10.1016/S0065-2164(08)70159-X |
| [145] |
Fujimoto Y, Chen CS, Gopalan AS, et al. Microbial degradation of the phytosterol side chain. Ⅱ. Incorporation of
|
| [146] |
Kieslich K. Microbial side-chain degradation of sterols[J]. Journal of Basic Microbiology, 1985, 25(7): 461-474. DOI:10.1002/jobm.3620250713 |
| [147] |
Sedlaczek L, Smith LL. Biotransformations of steroids[J]. Critical Reviews in Biotechnology, 1988, 7(3): 187-236. DOI:10.3109/07388558809146602 |
| [148] |
Szentirmai A. Microbial physiology of sidechain degradation of sterols[J]. Journal of Industrial Microbiology, 1990, 6(2): 101-115. DOI:10.1007/BF01576429 |
| [149] |
Ahmad S, Garg SK, Johri BN. Biotransformation of sterols: selective cleavage of the side chain[J]. Biotechnology Advances, 1992, 10(1): 1-67. DOI:10.1016/0734-9750(92)91351-E |
| [150] |
Björkhem I. Mechanism of degradation of the steroid side chain in the formation of bile acids[J]. Journal of Lipid Research, 1992, 33(4): 455-471. |
| [151] |
Murohisa T, Iida M. Some new intermediates in microbial side chain degradation of β-sitosterol[J]. Journal of Fermentation and Bioengineering, 1993, 76(3): 174-177. DOI:10.1016/0922-338X(93)90003-Q |
| [152] |
Smith KE, Ahmed F, Antoniou T. Microbial transformations of steroids[J]. Biochemical Society Transactions, 1993, 21(4): 1077-1080. DOI:10.1042/bst0211077 |
| [153] |
Mahato SB, Garai S. Advances in microbial steroid biotransformation[J]. Steroids, 1997, 62(4): 332-345. DOI:10.1016/S0039-128X(96)00251-6 |
| [154] |
Fernandes P, Cruz A, Angelova B, et al. Microbial conversion of steroid compounds: recent developments[J]. Enzyme and Microbial Technology, 2003, 32(6): 688-705. DOI:10.1016/S0141-0229(03)00029-2 |
| [155] |
Yang FC, Chen YL, Tang SL, et al. Integrated multi-omics analyses reveal the biochemical mechanisms and phylogenetic relevance of anaerobic androgen biodegradation in the environment[J]. The ISME Journal, 2016, 10(8): 1967-1983. DOI:10.1038/ismej.2015.255 |
| [156] |
Wang PH, Lee TH, Ismail W, et al. An oxygenase-independent cholesterol catabolic pathway operates under oxic conditions[J]. PLoS One, 2013, 8(6): e66675. DOI:10.1371/journal.pone.0066675 |
| [157] |
Crowe AM, Workman SD, Watanabe N, et al. IpdAB, a virulence factor in Mycobacterium tuberculosis, is a cholesterol ring-cleaving hydrolase[J]. Proceedings of the National Academy of Sciences of the United States of America, 2018, 115(15): E3378-E3387. DOI:10.1073/pnas.1717015115 |
| [158] |
Horinouchi M, Hayashi T, Koshino H, et al. Identification of 9α-hydroxy-17-oxo-1, 2, 3, 4, 10, 19-hexanorandrostan-5-oic acid in steroid degradation by Comamonas testosteroni TA441 and its conversion to the corresponding 6-en-5-oyl coenzyme A (CoA) involving open reading frame 28 (ORF28)- and ORF30-encoded acyl-CoA dehydrogenases[J]. Journal of Bacteriology, 2014, 196(20): 3598-3608. DOI:10.1128/JB.01878-14 |
| [159] |
Carere J, McKenna SE, Kimber MS, et al. Characterization of an aldolase-dehydrogenase complex from the cholesterol degradation pathway of Mycobacterium tuberculosis[J]. Biochemistry, 2013, 52(20): 3502-3511. DOI:10.1021/bi400351h |
| [160] |
Lack NA, Yam KC, Lowe ED, et al. Characterization of a carbon-carbon hydrolase from Mycobacterium tuberculosis involved in cholesterol metabolism[J]. The Journal of Biological Chemistry, 2010, 285(1): 434-443. DOI:10.1074/jbc.M109.058081 |
| [161] |
Yam KC, D'angelo I, Kalscheuer R, et al. Studies of a ring-cleaving dioxygenase illuminate the role of cholesterol metabolism in the pathogenesis of Mycobacterium tuberculosis[J]. PLoS Pathogens, 2009, 5(3): e1000344. DOI:10.1371/journal.ppat.1000344 |
| [162] |
Capyk JK, D'angelo I, Strynadka NC, et al. Characterization of 3-ketosteroid 9α-hydroxylase, a rieske oxygenase in the cholesterol degradation pathway of Mycobacterium tuberculosis[J]. The Journal of Biological Chemistry, 2009, 284(15): 9937-9946. DOI:10.1074/jbc.M900719200 |
| [163] |
Horinouchi M, Hayashi T, Koshino H, et al. Identification of 9, 17-dioxo-1, 2, 3, 4, 10, 19-hexanorandrostan-5-oic acid, 4-hydroxy-2-oxohexanoic acid, and 2-hydroxyhexa-2, 4-dienoic acid and related enzymes involved in testosterone degradation in Comamonas testosteroni TA441[J]. Applied and Environmental Microbiology, 2005, 71(9): 5275-5281. DOI:10.1128/AEM.71.9.5275-5281.2005 |
| [164] |
Horinouchi M, Hayashi T, Kudo T. The genes encoding the hydroxylase of 3-hydroxy-9, 10-secoandrosta-1, 3, 5(10)-triene- 9, 17-dione in steroid degradation in Comamonas testosteroni TA441[J]. The Journal of Steroid Biochemistry and Molecular Biology, 2004, 92(3): 143-154. DOI:10.1016/j.jsbmb.2004.09.002 |
| [165] |
Xu LQ, Liu YJ, Yao K, et al. Unraveling and engineering the production of 23, 24-bisnorcholenic steroids in sterol metabolism[J]. Scientific Reports, 2016, 6: 21928. DOI:10.1038/srep21928 |
| [166] |
Yang M, Lu R, Guja KE, et al. Unraveling cholesterol catabolism in Mycobacterium tuberculosis: ChsE4-ChsE5 α2β2 acyl-CoA dehydrogenase initiates β-oxidation of 3-oxo-cholest-4-en-26-oyl CoA[J]. Acs Infect Dis, 2015, 1(2): 110-125. DOI:10.1021/id500033m |
| [167] |
Crowe AM, Casabon I, Brown KL, et al. Catabolism of the last two steroid rings in Mycobacterium tuberculosis and other bacteria[J]. mBio, 2017, 8(2): e00321-17. |
| [168] |
van der Geize R, Grommen AWF, Hessels GI, et al. The steroid catabolic pathway of the intracellular pathogen Rhodococcus equi is important for pathogenesis and a target for vaccine development[J]. PLoS Pathogens, 2011, 7(8): e1002181. DOI:10.1371/journal.ppat.1002181 |
| [169] |
Rohman A, van Oosterwijk N, Thunnissen AMWH, et al. Crystal structure and site-directed mutagenesis of 3-ketosteroid Δ1-dehydrogenase from Rhodococcus erythropolis SQ1 explain its catalytic mechanism[J]. The Journal of Biological Chemistry, 2013, 288(49): 35559-35568. DOI:10.1074/jbc.M113.522771 |
| [170] |
Gilbert S, Hood L, Seah S. Characterization of an aldolase involved in cholesterol side chain degradation in Mycobacterium tuberculosis[J]. Journal of Bacteriology, 2018, 200(2): e00512-17. |
| [171] |
Wrońska N, Brzostek A, Szewczyk R, et al. The role of fadD19 and echA19 in sterol side chain degradation by Mycobacterium smegmatis[J]. Molecules, 2016, 21(5): 598. DOI:10.3390/molecules21050598 |
| [172] |
Nesbitt NM, Yang XX, Fontán P, et al. A thiolase of Mycobacterium tuberculosis is required for virulence and production of androstenedione and androstadienedione from cholesterol[J]. Infection and Immunity, 2010, 78(1): 275-282. DOI:10.1128/IAI.00893-09 |
| [173] |
Schaefer CM, Lu R, Nesbitt NM, et al. FadA5 a thiolase from Mycobacterium tuberculosis: a steroid-binding pocket reveals the potential for drug development against tuberculosis[J]. Structure, 2015, 23(1): 21-33. |
| [174] |
Biswas R, Dutta A, Dutta D, et al. Crystal structure of dehydratase component HadAB complex of mycobacterial FAS-Ⅱ pathway[J]. Biochemical and Biophysical Research Communications, 2015, 458(2): 369-374. DOI:10.1016/j.bbrc.2015.01.119 |
| [175] |
Yang M, Guja KE, Thomas ST, et al. A distinct MaoC-like enoyl-CoA hydratase architecture mediates cholesterol catabolism in Mycobacterium tuberculosis[J]. ACS Chemical Biology, 2014, 9(11): 2632-2645. DOI:10.1021/cb500232h |
| [176] |
Capyk JK, Casabon I, Gruninger R, et al. Activity of 3-ketosteroid 9α-hydroxylase (KshAB) indicates cholesterol side chain and ring degradation occur simultaneously in Mycobacterium tuberculosis[J]. The Journal of Biological Chemistry, 2011, 286(47): 40717-40724. DOI:10.1074/jbc.M111.289975 |
| [177] |
Xu XW, Gao XQ, Feng JX, et al. Influence of temperature on nucleus degradation of 4-androstene-3, 17-dione in phytosterol biotransformation by Mycobacterium sp.[J]. Letters in Applied Microbiology, 2015, 61(1): 63-68. DOI:10.1111/lam.12428 |
| [178] |
Marsheck WJ, Kraychy S, Muir RD. Microbial degradation of sterols[J]. Applied Microbiology, 1972, 23(1): 72-77. |
| [179] |
Yao K, Xu LQ, Wang FQ, et al. Characterization and engineering of 3-ketosteroid-Δ1-dehydrogenase and 3-ketosteroid-9α-hydroxylase in Mycobacterium neoaurum ATCC 25795 to produce 9α-hydroxy-4-androstene-3, 17-dione through the catabolism of sterols[J]. Metabolic Engineering, 2014, 24: 181-191. DOI:10.1016/j.ymben.2014.05.005 |
| [180] |
Pandey AK, Sassetti CM. Mycobacterial persistence requires the utilization of host cholesterol[J]. Proceedings of the National Academy of Sciences of the United States of America, 2008, 105(11): 4376-4380. DOI:10.1073/pnas.0711159105 |
| [181] |
Rathor N, Chandolia A, Saini NK, et al. An insight into the regulation of mce4 operon of Mycobacterium tuberculosis[J]. Tuberculosis, 2013, 93(4): 389-397. DOI:10.1016/j.tube.2013.03.007 |
| [182] |
Casali N, Riley LW. A phylogenomic analysis of the Actinomycetales mce operons[J]. BMC Genomics, 2007, 8: 60. DOI:10.1186/1471-2164-8-60 |
| [183] |
Zhang F, Xie JP. Mammalian cell entry gene family of Mycobacterium tuberculosis[J]. Molecular and Cellular Biochemistry, 2011, 352(1/2): 1-10. |
| [184] |
Kumar A, Bose M, Brahmachari V. Analysis of expression profile of mammalian cell entry (mce) operons of Mycobacterium tuberculosis[J]. Infection and Immunity, 2003, 71(10): 6083-6087. DOI:10.1128/IAI.71.10.6083-6087.2003 |
| [185] |
Mohn WW, van der Geize R, Stewart GR, et al. The actinobacterial mce4 locus encodes a steroid transporter[J]. The Journal of Biological Chemistry, 2008, 283(51): 35368-35374. DOI:10.1074/jbc.M805496200 |
| [186] |
Arruda S, Bomfim G, Knights R, et al. Cloning of an M. tuberculosis DNA fragment associated with entry and survival inside cells[J]. Science, 1993, 261(5127): 1454-1458. DOI:10.1126/science.8367727 |
| [187] |
Klepp LI, Forrellad MA, Osella AV, et al. Impact of the deletion of the six mce operons in Mycobacterium smegmatis[J]. Microbes and Infection, 2012, 14(7/8): 590-599. |
| [188] |
García-Fernández J, Papavinasasundaram K, Galán B, et al. Molecular and functional analysis of the mce4 operon in Mycobacterium smegmatis[J]. Environmental Microbiology, 2017, 19(9): 3689-3699. DOI:10.1111/1462-2920.13869 |
| [189] |
Nikolayeva VM, Egorova OV, Dovbnya DV, et al. Extracellular 3β-hydroxysteroid oxidase of Mycobacterium vaccae VKM Ac-1815D[J]. The Journal of Steroid Biochemistry and Molecular Biology, 2004, 91(1/2): 79-85. |
| [190] |
Doukyu N, Shibata K, Ogino H, et al. Purification and characterization of Chromobacterium sp. DS-1 cholesterol oxidase with thermal, organic solvent, and detergent tolerance[J]. Applied Microbiology and Biotechnology, 2008, 80(1): 59-70. |
| [191] |
Doukyu N, Aono R. Two moles of O2 consumption and one mole of H2O2 formation during cholesterol peroxidation with cholesterol oxidase from Pseudomonas sp. strain ST-200[J]. Biochemical Journal, 1999, 341(3): 621-627. |
| [192] |
Uhía I, Galán B, Medrano FJ, et al. Characterization of the KstR-dependent promoter of the gene for the first step of the cholesterol degradative pathway in Mycobacterium smegmatis[J]. Microbiology, 2011, 157(9): 2670-2680. DOI:10.1099/mic.0.049213-0 |
| [193] |
Thomas ST, Vanderven BC, Sherman DR, et al. Pathway profiling in Mycobacterium tuberculosis: elucidation of cholesterol-derived catabolite and enzymes that catalyze its metabolism[J]. The Journal of Biological Chemistry, 2011, 286(51): 43668-43678. DOI:10.1074/jbc.M111.313643 |
| [194] |
Lamba V, Yabukarski F, Pinney M, et al. Evaluation of the catalytic contribution from a positioned general base in ketosteroid isomerase[J]. Journal of the American Chemical Society, 2016, 138(31): 9902-9909. DOI:10.1021/jacs.6b04796 |
| [195] |
Cha HJ, Jang DS, Jeong JH, et al. Role of conserved Met112 residue in the catalytic activity and stability of ketosteroid isomerase[J]. Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics, 2016, 1864(10): 1322-1327. DOI:10.1016/j.bbapap.2016.06.016 |
| [196] |
Cha HJ, Jang DS, Kim YG, et al. Rescue of deleterious mutations by the compensatory Y30F mutation in ketosteroid isomerase[J]. Molecules and Cells, 2013, 36(1): 39-46. DOI:10.1007/s10059-013-0013-1 |
| [197] |
Fafarman AT, Sigala PA, Schwans JP, et al. Quantitative, directional measurement of electric field heterogeneity in the active site of ketosteroid isomerase[J]. Proceedings of the National Academy of Sciences of the United States of America, 2012, 109(6): E299-E308. DOI:10.1073/pnas.1111566109 |
| [198] |
Schwans JP, Sunden F, Gonzalez A, et al. Evaluating the catalytic contribution from the oxyanion hole in ketosteroid isomerase[J]. Journal of the American Chemical Society, 2011, 133(50): 20052-20055. DOI:10.1021/ja208050t |
| [199] |
Cherney MM, Garen CR, James MNG. Crystal structure of Mycobacterium tuberculosis Rv0760c at 1.50 Å resolution, a structural homolog of Δ5-3-ketosteroid isomerase[J]. Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics, 2008, 1784(11): 1625-1632. DOI:10.1016/j.bbapap.2008.05.012 |
| [200] |
Cho HS, Choi G, Choi KY, et al. Crystal structure and enzyme mechanism of Δ5-3-ketosteroid isomerase from Pseudomonas testosteroni[J]. Biochemistry, 1998, 37(23): 8325-8330. DOI:10.1021/bi9801614 |
| [201] |
Xue LA, Talalay P, Mildvan AS. Studies of the mechanism of the Δ5-3-ketosteroid isomerase reaction by substrate, solvent, and combined kinetic deuterium isotope effects on wild-type and mutant enzymes[J]. Biochemistry, 1990, 29(32): 7491-7500. DOI:10.1021/bi00484a019 |
| [202] |
Kim SW, Cha SS, Cho HS, et al. High-resolution crystal structures of Δ5-3-ketosteroid isomerase with and without a reaction intermediate analogue[J]. Biochemistry, 1997, 36(46): 14030-14036. DOI:10.1021/bi971546+ |
| [203] |
Peng F, Yang F, Cheng XY, et al. Crystal structure and characterization of Δ5-3-ketosteroid isomerase from Mycobacterium strain HGMS2GL[J]. Biophysical Journal, 2018, 114(3 Suppl 1): 583a. |
| [204] |
Cheng XY, Peng F, Yang F, et al. Crystal structure of delta 5-3-ketosteroid isomerase from Mycobacterium sp.[M/OL]. (2018-01-31). http://www.rcsb.org/structure/5Z3R
|
| [205] |
McLean KJ, Lafite P, Levy C, et al. The structure of Mycobacterium tuberculosis CYP125: molecular basis for cholesterol binding in a P450 needed for host infection[J]. The Journal of Biological Chemistry, 2009, 284(51): 35524-35533. DOI:10.1074/jbc.M109.032706 |
| [206] |
Driscoll MD, McLean KJ, Levy C, et al. Structural and biochemical characterization of Mycobacterium tuberculosis CYP142: evidence for multiple cholesterol 27-hydroxylase activities in a human pathogen[J]. The Journal of Biological Chemistry, 2010, 285(49): 38270-38282. DOI:10.1074/jbc.M110.164293 |
| [207] |
Johnston JB, Ouellet H, Ortiz De Montellano PR. Functional redundancy of steroid C26-monooxygenase activity in Mycobacterium tuberculosis revealed by biochemical and genetic analyses[J]. The Journal of Biological Chemistry, 2010, 285(47): 36352-36360. DOI:10.1074/jbc.M110.161117 |
| [208] |
Johnston JB, Singh AA, Clary AA, et al. Substrate analog studies of the ω-regiospecificity of Mycobacterium tuberculosis cholesterol metabolizing cytochrome P450 enzymes CYP124A1, CYP125A1 and CYP142A1[J]. Bioorganic & Medicinal Chemistry, 2012, 20(13): 4064-4081. |
| [209] |
Capyk JK, Kalscheuer R, Stewart GR, et al. Mycobacterial cytochrome P450 125 (Cyp125) catalyzes the terminal hydroxylation of C27 steroids[J]. The Journal of Biological Chemistry, 2009, 284(51): 35534-35542. DOI:10.1074/jbc.M109.072132 |
| [210] |
Wilbrink MH, Petrusma M, Dijkhuizen L, et al. FadD19 of Rhodococcus rhodochrous DSM43269, a steroid-coenzyme A ligase essential for degradation of C-24 branched sterol side chains[J]. Applied and Environmental Microbiology, 2011, 77(13): 4455-4464. DOI:10.1128/AEM.00380-11 |
| [211] |
Wipperman MF, Yang M, Thomas ST, et al. Shrinking the FadE proteome of Mycobacterium tuberculosis: insights into cholesterol metabolism through identification of an α2β2 heterotetrameric acyl coenzyme A dehydrogenase family[J]. Journal of Bacteriology, 2013, 195(19): 4331-4341. DOI:10.1128/JB.00502-13 |
| [212] |
Taylor RC, Brown AK, Singh A, et al. Characterization of a β-hydroxybutyryl-CoA dehydrogenase from Mycobacterium tuberculosis[J]. Microbiology, 2010, 156(7): 1975-1982. DOI:10.1099/mic.0.038802-0 |
| [213] |
Petrusma M, van der Geize R, Dijkhuizen L. 3-Ketosteroid 9α-hydroxylase enzymes: rieske non-heme monooxygenases essential for bacterial steroid degradation[J]. Antonie van Leeuwenhoek, 2014, 106(1): 157-172. DOI:10.1007/s10482-014-0188-2 |
| [214] |
van der Geize R, Hessels GI, Van Gerwen R, et al. Targeted disruption of the kstD gene encoding a 3-ketosteroid Δ1-dehydrogenase isoenzyme of Rhodococcus erythropolis strain SQ1[J]. Applied and Environmental Microbiology, 2000, 66(5): 2029-2036. DOI:10.1128/AEM.66.5.2029-2036.2000 |
| [215] |
van der Geize R, Hessels GI, Dijkhuizen L. Molecular and functional characterization of the kstD2 gene of Rhodococcus erythropolis SQ1 encoding a second 3-ketosteroid Δ1-dehydrogenase isoenzyme[J]. Microbiology, 2002, 148(10): 3285-3292. DOI:10.1099/00221287-148-10-3285 |
| [216] |
Hu YM, van der Geize R, Besra GS, et al. 3-Ketosteroid 9α-hydroxylase is an essential factor in the pathogenesis of Mycobacterium tuberculosis[J]. Molecular Microbiology, 2010, 75(1): 107-121. DOI:10.1111/j.1365-2958.2009.06957.x |
| [217] |
Wei W, Fan SY, Wang FQ, et al. Accumulation of androstadiene-dione by overexpression of heterologous 3-ketosteroid Δ1-dehydrogenase in Mycobacterium neoaurum NwIB-01[J]. World Journal of Microbiology and Biotechnology, 2014, 30(7): 1947-1954. DOI:10.1007/s11274-014-1614-3 |
| [218] |
Wei W, Wang FQ, Fan SY, et al. Inactivation and augmentation of the primary 3-ketosteroid-Δ1-dehydrogenase in Mycobacterium neoaurum NwIB-01: biotransformation of soybean phytosterols to 4-androstene-3, 17-dione or 1, 4-androstadiene-3, 17-dione[J]. Applied and Environmental Microbiology, 2010, 76(13): 4578-4582. DOI:10.1128/AEM.00448-10 |
| [219] |
Wei W, Fan SY, Wang FQ, et al. A new steroid-transforming strain of Mycobacterium neoaurum and cloning of 3-ketosteroid 9α-hydroxylase in NwIB-01[J]. Applied Biochemistry and Biotechnology, 2010, 162(5): 1446-1456. DOI:10.1007/s12010-010-8919-y |
| [220] |
Penfield JS, Worrall LJ, Strynadka NC, et al. Substrate specificities and conformational flexibility of 3-ketosteroid 9α-hydroxylases[J]. The Journal of Biological Chemistry, 2014, 289(37): 25523-25536. DOI:10.1074/jbc.M114.575886 |
| [221] |
Wang HW, Yang F, Cheng XY, et al. Crystal structure and characterization of 3-ketosteroid-Δ1-dehydrogenase from Mycobacterium strain HGMS2GL[J]. Biophysical Journal, 2018, 114(3 Suppl 1): 583a. |
| [222] |
Donova MV, Dovbnya DV, Koshcheyenko KA. Modified CDs-mediated enhancement of microbial sterol sidechain degradation[A]//Proceedings of the Eighth International Symposium on Cyclodextrins[C]. Budapest, Hungary: Springer, 1996: 527-530
|
| [223] |
Sukhodolskaya GV, Nikolayeva VM, Khomutov SM, et al. Steroid-1-dehydrogenase of Mycobacterium sp. VKM Ac-1817D strain producing 9α-hydroxy-androst-4-ene-3, 17-dione from sitosterol[J]. Applied Microbiology and Biotechnology, 2007, 74(4): 867-873. DOI:10.1007/s00253-006-0728-4 |
| [224] |
Donova MV, Gulevskaya SA, Dovbnya DV, et al. Mycobacterium sp. mutant strain producing 9α-hydroxyandrostenedione from sitosterol[J]. Applied Microbiology and Biotechnology, 2005, 67(5): 671-678. DOI:10.1007/s00253-004-1808-y |
| [225] |
Dresen C, Lin LYC, D'Angelo I, et al. A flavin-dependent monooxygenase from Mycobacterium tuberculosis involved in cholesterol catabolism[J]. The Journal of Biological Chemistry, 2010, 285(29): 22264-22275. DOI:10.1074/jbc.M109.099028 |
| [226] |
Horinouchi M, Yamamoto T, Taguchi K, et al. Meta-cleavage enzyme gene tesB is necessary for testosterone degradation in Comamonas testosteroni TA441[J]. Microbiology, 2001, 147(12): 3367-3375. DOI:10.1099/00221287-147-12-3367 |
| [227] |
Lack N, Lowe ED, Liu J, et al. Structure of HsaD, a steroid-degrading hydrolase, from Mycobacterium tuberculosis[J]. Acta Crystallographica Section F, Structural Biology and Crystallization Communications, 2008, 64: 2-7. DOI:10.1107/S1744309107065931 |
| [228] |
Horinouchi M, Hayashi T, Koshino H, et al. Identification of 9α-hydroxy-17-oxo-1, 2, 3, 4, 10, 19-hexanorandrost-6-en-5-oic acid and β-oxidation products of the C-17 side chain in cholic acid degradation by Comamonas testosteroni TA441[J]. The Journal of Steroid Biochemistry and Molecular Biology, 2014, 143: 306-322. DOI:10.1016/j.jsbmb.2014.04.014 |
| [229] |
Costa S, Giovannini PP, Fantin G, et al. New 9, 10-secosteroids from biotransformations of hyodeoxycholic acid with Rhodococcus spp.[J]. Helvetica Chimica Acta, 2013, 96(6): 1062-1071. DOI:10.1002/hlca.201200330 |
| [230] |
Costa S, Giovannini PP, Fantin G, et al. New 9, 10-secosteroids from biotransformations of bile acids with Rhodococcus ruber[J]. Helvetica Chimica Acta, 2013, 96(11): 2124-2133. DOI:10.1002/hlca.201300114 |
 2019, Vol. 46
2019, Vol. 46