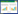扩展功能
文章信息
- 王艺雅, 张其中
- WANG Yi-Ya, ZHANG Qi-Zhong
- 一株光合细菌的分离鉴定及该菌对氨氮和亚硝态氮的去除作用
- Isolation and identification of a photosynthetic bacterium strain and removal of ammonia nitrogen and nitrite by the bacterium species in water
- 微生物学通报, 2019, 46(10): 2512-2528
- Microbiology China, 2019, 46(10): 2512-2528
- DOI: 10.13344/j.microbiol.china.180847
-
文章历史
- 收稿日期: 2018-10-30
- 接受日期: 2019-03-14
- 网络首发日期: 2019-04-23
中国是水产养殖大国,自1991年起,我国食用鱼类养殖产量高于全球其余国家和地区的合计产量;自2002年起,我国一直是世界上鱼和鱼产品最大的出口国[1]。2016年,我国水产养殖产量高达5 142.39万t,占全国水产品总产量的74.51%,水产养殖产值为8 953.57亿元[2]。我国多采用集约化高密度养殖方式饲养,致使大量残饵和水产动物排泄物进入水体,成为养殖池塘水体中氮的主要来源。养殖水体的氮污染问题已经成为制约我国水产养殖业发展的重要因素之一。
氨氮(NH4+-N)是水体氮污染中最常见的污染物,主要以离子态(NH4+)和分子态(NH3)两种形式存在,水体中有毒性作用的主要是分子氨[3]。我国渔业用水水质标准规定分子氨浓度不得超过0.02 mg/L[4]。氨氮可使动物肝肾受损、体表及内脏充血[5],也能致使水生动物鳃组织受损、鳃小片弯曲、粘连或融合[6]。因此去除养殖水体氨氮是保障水产动物健康养殖的关键。目前,去除水体氨氮的方法主要分为物化法和生物脱氮[7]两大类,其中物化法由于投入大量化学药剂易造成养殖水体的二次污染[8],而生物脱氮是目前应用最为广泛的环境友好无污染技术,其原理是利用微生物的硝化作用将水体的含氮污染物转化成为分子态氮气离开水体,从而达到水体脱氮目的。
亚硝态氮是硝化作用和反硝化作用的中间产物,也是养殖水体常见污染物之一。亚硝态氮进入鱼体内与血红蛋白结合,将二价铁(Fe2+)氧化为高铁(Fe3+),让血红蛋白失去载氧能力[9],使鱼体缺氧,免疫力下降,从而引发疾病,甚至窒息死亡[10]。去除亚硝态氮主要有两种途径,一种是通过硝化作用将亚硝态氮转化为无毒害作用的硝态氮,另一种是通过反硝化作用转化为氮气离开养殖水体进入大气[11]。一般条件下,水产养殖水体中亚硝态氮(以氮计)浓度宜控制在0.1 mg/L以下[12]。
亚硝酸盐还原酶(Nitrite reductase)是氮循环反硝化过程中的关键限速酶[13]。目前发现亚硝酸盐还原酶常见的有5种类型,nirK基因编码的含铜型亚硝酸盐还原酶(Cu-Type nitrite reductase)[14]、nirS基因编码的细胞色素cd1亚硝酸盐还原酶(Cytochrome cd1 nitrite reductase)[15]、nasB基因编码的NADH依赖性亚硝酸盐还原酶(NADH dependent nitrite reductase)[16]、nrfA基因编码的细胞色素c亚硝酸盐还原酶(Cytochrome c nitrite reductase)[17]和nii1基因编码的铁氧化还原蛋白依赖性亚硝酸盐还原酶(Ferredoxin-dependent nitrite reductase)[18]。含铜型亚硝酸盐还原酶和细胞色素cd1亚硝酸盐还原酶在结构上存在差异,但功能相同[19],催化亚硝酸盐还原成一氧化氮。编码亚硝酸盐还原酶的nirS基因和nirK基因是研究反硝化细菌的分子标记[20]。
光合细菌是地球上最早出现的具有原始光能合成体系的原核生物[21],能充分利用光能以及各种有机物进行不放氧的光合作用[22],广泛存在于海洋、湖泊、水田、污泥以及某些极端生境中[23]。20世纪60年代,日本学者小林正泰发现光合细菌能有效净化有机废水,开创了用光合细菌法处理污水的先河[24]。随后一系列研究显示,光合细菌不仅可用于各种污水脱氮[25-29],还能去除养殖水体中氨氮和亚硝态氮[30-34]。
目前发现多种光合细菌具有降解氨氮的能力,如粪红假单胞菌(Rhodopesudomonas faecalis)可使自然水体中初始氨氮浓度从1.753 mg/L下降到0.711 mg/L[35];浑球红假单胞菌(R. sphaeroides)对鱼池水的氨氮去除率高达99%[36];海洋紫色硫细菌(Marichromatium gracile) YL28对水体氨氮最大去除量为9.64 mmol/L,对氨氮含量低于3.21 mmol/L的水体氨氮去除率可达97.61%以上[37];纤细红螺菌(Rhodospirillum tneue) GH1、球形红假单胞菌(R. sphaeroides) GH2和GH3、沼泽红假单胞菌(R. palustris) GH4四株混合菌在微氧具光照条件下对养殖水体的氨氮去除率为89.2%[38];研究还发现部分光合细菌能降解亚硝态氮,如球形红细菌(Rhodobacter sphaeroides) P4以苹果酸作为碳源时,对起始亚硝酸盐浓度为1 mmol/L的亚硝态氮去除率为99.8%[39];胶质红假单胞菌(R. gelatinosa) H1对亚硝态氮浓度为2.9 mg/L的彭泽鲫养殖水体亚硝态氮去除率为99.79%,球形红假单胞菌(R. sphaeroides) PSB-3对该水质亚硝氮去除率为86.21%,沼泽红假单胞菌(R. palustris) H10对该水质亚硝氮去除率为93.79%[40]。可见自然界确实存在不少能有效去除水体中氨氮和亚硝态氮的光合细菌,但是它们的降解效率各不相同,因此,有必要从自然水体中筛选出高效降解氨氮或亚硝态氮的光合细菌,以满足改善养殖水质、保障水产养殖业健康发展的需要。
本研究旨在鉴定分离自养殖水体中一株能去除水体氨氮和亚硝态氮的光合细菌的菌种,明确其去除氨氮和亚硝态氮能力,为养殖水体生物脱氮提供备选菌株。
1 材料与方法 1.1 材料 1.1.1 供试菌株标准菌株沙氏外硫红螺菌(Ectothiorhodospira shaposhnikovii)购于美国典型培养物保藏中心(ATCC),产品编号:43036;SP3菌株分离自广东省养殖水体。
1.1.2 培养基RCVBN培养基的配制参照文献[41],van Niel培养基的配制参照文献[42]。
1.1.3 主要试剂和仪器亚硝酸钠、氯化铵,天津大茂公司;苹果酸,上海试剂三厂;生物素,Sigma公司。水平核酸电泳仪、荧光定量PCR仪、凝胶成像分析系统,Bio-Rad公司;低温高速离心机,Sigma公司;全波长酶标仪,BioTek公司;自动蒸汽灭菌锅,北京发恩科贸有限公司。
1.2 方法 1.2.1 光合细菌的富集、分离和保存将广东养殖水体采集水样带回实验室后立即进行富集试验。取50 mL养殖水体水样接种到装有200 mL van Niel培养基的锥形瓶中,再加入一定量的液体石蜡以隔绝空气,在30 ℃和光照2 000 lx条件下富集静置培养10-15 d,直至培养液呈红色。取10 mL红色富集培养液加到含新灭菌的90 mL van Niel培养基的锥形瓶中,再加入一定量的液体石蜡(高于培养液1 cm左右)在30 ℃和光照2 000 lx条件下静置培养至培养液变红,以上步骤重复富集3次。
取富集培养液1 mL,用0.9%的生理盐水进行10倍稀释,终浓度分别为原始菌浓度的10-5、10-6、10-7、10-8和10-9倍,用双层培养基(双层均为含2%琼脂的RCVBN固体培养基)平板法分离单菌落,用接种环蘸取稀释后的富集培养菌液在固体培养基上划线接种,然后再倒一层已灭菌的2%琼脂的RCVBN培养基,冷却凝固后在30 ℃和光照2 000 lx条件下倒置培养7 d。待红色菌落长出后,挑单菌落于相同固体培养基平板上划线接种再分离纯化培养7 d,获得单一菌种的单菌落,并在RCVBN液体培养基中于30 ℃和光照2 000 lx条件下进行扩大培养,得到菌液SP3。
保种方法:(1)取500 μL SP3纯培养菌液加入500 μL浓度为40%的甘油中,-80 ℃保存。(2)取SP3菌液100 mL,在5 000 r/min、4 ℃下离心10 min,倒掉上清液并用已灭菌的0.9%生理盐水离心洗涤,重复3次。去上清,将离心后的SP3菌体接种到500 mL已灭菌的RCVBN液体培养基中,并加入一定量的液体石蜡,在30 ℃和光照2 000 lx条件下培养7 d后室温保存,每20-30 d按上述步骤转接培养,以保持菌种生长活力。
1.2.2 光合细菌SP3的菌种鉴定(1) 分子生物学鉴定:提取SP3菌株DNA,用通用引物[43] (27F:5′-AGAGTTTGATCCTGGCT CAG-3′;1492R:5′-TA CGGTTACCTTGTTACGAC TT-3′)进行PCR扩增。PCR反应体系(25 μL):10×KOD buffer 2.5 μL,2.5 mmol/L dNTP mix 2.5 μL,25 mmol/L MgSO4 1.5 μL,F和R引物(20 μmol/L)各1 μL,模板DNA 1 μL,KOD-plus-neo酶0.5 μL,以及ddH2O 15 μL。PCR反应条件:94 ℃ 4 min;98 ℃ 30 s,56 ℃ 30 s,68 ℃ 1 min,30个循环;68 ℃ 7 min。PCR产物经纯化后通过加尾、T/A克隆到pMD18-T载体,再转化到E. coli DH5α感受态细胞中,经蓝白斑筛选鉴定为阳性克隆。菌液送华大基因科技有限公司测序。将16S rRNA基因序列用软件MEGA 6.0通过邻接法(Neighbor-Joining,N-J法)构建系统发育进化树,自举分析(1 000次)进行置信度检测,初步确定菌株SP3可能的属和种。
(2) 形态学观察:观察菌落的典型特征,挑取分离纯化所得的SP3菌体,通过革兰氏染色在光学显微下观察该菌株的形态。
(3) 生理生化实验:参照《常见细菌系统鉴定手册》[44]。
1.2.3 菌株SP3生长条件优化(1) 最适pH试验:将RCVBN液体培养基的pH分别调至6.0、6.5、7.0、7.5、8.0、8.5、9.0、9.5、10.0、10.5、11.0,分别设3个平行。接种培养方式同1.2.1节保种方法(2)。每间隔24 h测定OD600[45]。
(2) 最适盐度试验:将RCVBN液体培养基里加入NaCl,浓度分别为0%、0.5%、1%、1.5%、2%、3%、4%、5%、6%、7%、8%、9%、10% (质量体积比),分别设3个平行。接种方式同上。每间隔24 h测定OD600。
1.2.4 不同氮源对SP3生长的影响(1) 不同浓度氯化铵对SP3生长的影响:将RCVBN液体培养基中的氮源去除,以氯化铵作为氮源,分别配制含0.25、0.5、1.0、1.5 g/L氯化铵的培养基,以不含氮源的RCVBN液体培养基作为对照,分别设3个平行。接种方式同上,在30 ℃和光照2 000 lx条件下培养7 d,每间隔24 h测定OD600。
(2) 不同浓度亚硝酸钠对SP3生长的影响:将RCVBN液体培养基中的氮源去除,以亚硝酸钠作为氮源,分别配制含0.5、1.0、2.0、4.0 mg/L的低浓度亚硝酸钠培养基和0.25、0.5、1.0、1.5 g/L的高浓度亚硝酸钠培养基,以不含氮源的RCVBN液体培养基作为对照,分别设3个平行。接种培养方式同上,每间隔24 h测定OD600。
(3) SP3去除氨氮能力:将RCVBN液体培养基中的氮源去除,以0.25 g/L氯化铵作为氮源配制培养基,接种培养方式同上,以不加SP3菌液的培养基对照组,分别设3个平行。每间隔24 h取100 mL菌液,5 000 r/min离心10 min,离心后菌体立即冻存于液氮罐中,采用纳氏比色法[46]测定上清中氨氮的含量。氨氮累计去除量c=(A0-At)- (K0-Kt),氨氮累计去除率C=c/A0×100%,式中A0表示实验组起始(第0天)氨氮含量(3次重复的平均值),At表示实验组当天氨氮含量,K0表示对照组起始(第0天)氨氮含量(3次重复的平均值),Kt表示对照组当天氨氮含量(3次重复的平均值)。
取实验组菌液1 mL,用0.9%生理盐水将其稀释至10-7。取0.1 mL稀释的光合细菌菌液均匀涂布于底层琼脂浓度为2%的RCVBN培养基,分别设3个平行组,烘干水分后,加入琼脂浓度为2%的RCVBN培养基,将平板置于30 ℃和光照2 000 lx倒置培养7 d后,测定菌落数。
1.2.5 SP3去除亚硝态氮能力将RCVBN液体培养基中的氮源去除,以2 mg/L亚硝酸钠作为氮源配制培养基,接种培养方式同上,3个平行组。对照组不加SP3菌液,3个平行组。每间隔24 h于超净工作台中取100 mL培养中的菌液,5 000 r/min离心10 min,离心后菌体立即冻存于液氮罐中,采用分光光度法[47]{New York State Department of Environmental Conservation, 2009 #82}测定上清中亚硝态氮的含量。
亚硝态氮累计去除量r=(N0-Nt)-(M0-Mt)和亚硝态氮累计去除率R=r/N0×100%,式中N0表示实验组起始(第0天)亚硝态氮含量(3次重复的平均值),Nt表示实验组当天亚硝态氮含量,M0表示对照组起始(第0天)亚硝态氮含量(3次重复的平均值),Mt表示对照组当天亚硝态氮含量(3次重复的平均值)。
1.2.6 SP3菌株的亚硝酸盐还原酶基因及其在去除氨氮和亚硝态氮中的表达水平动态(1) SP3菌株亚硝酸盐还原酶基因的确定:以菌株SP3的DNA为模板,用文献资料中已报道的编码亚硝酸盐还原酶基因保守序列的引物(NirS-F,NirS-R,NirK-F,NirK-R)进行PCR扩增。PCR反应体系参照本文1.2.2,扩增产物经1.2%琼脂糖凝胶电泳有单一明显条带后,切胶回收产物进行TA克隆,再转化到E. coli DH5α感受态细胞中,蓝白斑筛选阳性克隆,菌液PCR验证成功后将该菌液送至华大基因有限公司测序。测序结果在NCBI数据库中比对。
(2) Genome walking获得SP3菌株nirS基因全序列及生物信息学分析:根据测序获得亚硝酸盐还原酶基因序列,分别设计3条退火温度较高的特异性引物(SP1,SP2,SP3),依据TaKaRa Genome Walking Kit的说明书对菌株SP3的DNA进行3次巢式PCR获得已知亚硝酸盐还原酶的侧翼序列,PCR产物回收纯化后经克隆连接转化,筛选阳性克隆进行菌落PCR后送华大基因有限公司测序。将新获得的序列和已经得到的nirS基因序列拼接得到nirS基因全序列。利用http://emboss.bioinformatics.nl/网站得到序列展示图,通过Protparam服务器(http://us.expasy.org/tools/protparam.html)预测nirS蛋白质的基本理化性质。PROST服务器(https://psort.hgc.jp/form.html)预测nirS蛋白质亚细胞定位。在http://swissmodel.expasy.org/里模拟并预测nirS的三维结构,在https://blast.ncbi.nlm.nih.gov/Blast.cgi网站预测SP3菌株nirS蛋白功能域。
1.2.7 检测nirS表达水平的荧光定量PCR方法的建立引物设计及合成:根据SP3菌株nirS基因全序列,设计一对近3′端特异性引物RT-NirS-F/ RT-NirS-R(表 1)用于荧光定量PCR,扩增片段长度为157 bp。以已确立的hrdB作为内参基因[50]。引物由华大基因科技有限公司合成。
| 引物名称 Primer name | 核酸序列 Nucleotide sequence (5′→3′) | 目的基因 Target gene | 来源或参考文献 Sources or reference |
| NirS-F | CCTAYTGGCCGCCRCART | nirS | [48] |
| NirS-R | CGTTGAACTTRCCGGT | nirS | [48] |
| NirK-F | GGMATGGTKCCSTGGCA | nirK | [49] |
| NirK-R | GCCTCGATCAGRT | nirK | [49] |
| SP1 | GATGAGCGCCACCGAATCGT | 1st Genome walking of nirS | This study |
| SP2 | GGTGATCGCCGCCAGCTTGC | 2nd Genome walking of nirS | This study |
| SP3 | GGTATAGTCGACCAGCTGGATCT | 3rd Genome walking of nirS | This study |
| RT-NirS-F | CCTTGTTGTATTCGCCCTGC | RT-PCR of nirS | This study |
| RT-NirS-R | AACCCCGAAACCGACATCTC | RT-PCR of nirS | This study |
| RT-hrdB-F | AGGACGGCGACAGCGAGTTC | RT-PCR of hrdB | [50] |
| RT-hrdB-R | CCGAAGCGCATGGAGACG | RT-PCR of hrdB | [50] |
目的基因引物扩增效率和特异性的确定:取2 mg/L亚硝酸钠培养3 d的SP3菌体,提取总RNA逆转录获得cDNA,并10倍梯度稀释作为模板,3个重复。用RT-NirS-F/RT-NirS-R引物,荧光定量PCR反应体系:SYBR Green Realtime PCR Master Mix 10 μL,RT-NirS-R (10 μmol/L) 0.4 μL,RT-NirS-F (10 μmol/L) 0.4 μL,cDNA 1 μL以及ddH2O 8.2 μL。荧光定量PCR反应条件:95 ℃ 1 min;95 ℃ 15 s,59 ℃ 20 s,75 ℃ 1 min,40个循环。熔解曲线分析:起始温度65 ℃,每10 s升温0.5 ℃,80个循环。根据标准曲线确定引物扩增效率,熔解曲线确定引物特异性。
1.2.8 SP3菌株nirS在去除氨氮和亚硝态氮过程中的表达水平动态模板制备:Trizol法提取冻存于液氮罐中0.25 g/L氯化铵培养的菌株SP3菌体样品、2 mg/L亚硝酸钠培养的SP3菌体实验组样品和不加氮源培养的菌株SP3菌体对照组样本的总RNA,琼脂糖凝胶核酸电泳检验RNA提取质量。选取高质量的RNA,用酶标仪检测RNA的纯度(OD260/OD280)。参照Promega公司M-MLV Reverse Transcriptase的说明书将高质量的RNA逆转录成cDNA。
荧光定量PCR:在mRNA水平检测菌株SP3的nirS在去除氨氮和亚硝态过程中的表达动态,以hrdB作为内参基因,同一批样品同时对2个基因进行荧光定量PCR,3个重复,收集荧光信号获得Ct值。依据相对荧光定量计算公式2-∆∆Ct进行数据分析处理。
1.3 数据的统计分析用IBM SPSS Statistics 22.0软件进行数据统计分析,采用OriginPro 8.0软件进行数据作图。
2 结果与分析 2.1 菌株鉴定 2.1.1 16S rRNA基因序列测序得到1 438 bp的l6S rRNA基因序列(GenBank登录号为MH325212.1)。将菌株SP3的16S rRNA基因序列在NCBI数据库进行BLAST比对。比对结果表明菌株SP3的16S rRNA基因序列与外硫红螺菌属(Ectothiorhodospira)相似度高达99%。采用邻接法构建系统发育进化树,菌株SP3与外硫红螺菌属的沙氏外硫红螺菌(Ectothiorhodospira shaposhnikovii)聚为一支,置信度为99% (图 1)。
|
| 图 1 基于菌株SP3的16S rRNA基因序列构建的系统发育树 Figure 1 The phylogenetic tree based on 16S rRNA gene sequence 注:分支点上的数字表示构建系统树时1 000次计算时形成该节点的百分比;括号内为菌株的16S rRNA基因序列在GenBank中的登录号. Note: Numbers at the branch nodes are bootstrap values, expressed as percentages of 1 000 replicates; Numbers in parentheses represent the accession numbers in GenBank for the 16S rRNA gene sequences of those type strains. |
|
|
SP3菌株的菌落形态为深红色小圆球状,边缘整齐光滑;菌液呈红色。该菌为革兰氏阴性菌,短杆状。
2.1.3 碳源利用结果如表 2所示,SP3菌株和标准菌株沙氏外硫红螺菌对不同碳源的利用情况一致,可以利用醋酸盐、丙酮酸盐、丙酸盐、丁酸盐、乳酸盐、富马酸盐、琥珀酸盐、苹果酸、果糖、葡萄糖作为碳源,不能利用乙醇和丙醇。
| 碳源 Carbon source | Ectothiorhodospira shaposhnikovii | SP3 |
| 醋酸盐 Acetate | + | + |
| 丙酮酸盐 Pyruvate | + | + |
| 丙酸盐 Propionate | + | + |
| 丁酸盐 Butyrate | + | + |
| 乳酸盐 Lactate | + | + |
| 富马酸盐 Fumarate | + | + |
| 琥珀酸盐 Succinate | + | + |
| 苹果酸 Malate | + | + |
| 果糖 Fructose | + | + |
| 葡萄糖 Glucose | + | + |
| 乙醇 Ethanol | - | - |
| 丙醇 Propanol | - | - |
| 注:有机酸以钠盐的形式提供. +:阳性反应;-:阴性反应. Note: Organic acids are provided in the form of sodium salt. +: Positive reaction; -: Negative reaction. | ||
菌株SP3和标准菌株均能以Na2S2O3·5H2O、Na2S·9H2O、Na2SO3作为无机电子供体(表 3)。这一结果同样符合《伯杰氏细菌鉴定手册》(8版)中E. shaposhnikovii对硫化合物的利用。
| 电子供体 Electron donor | Ectothiorhodospira shaposhnikovii | SP3 |
| Na2S2O3·5H2O | + | + |
| Na2S·9H2O | + | + |
| Na2SO3 | + | + |
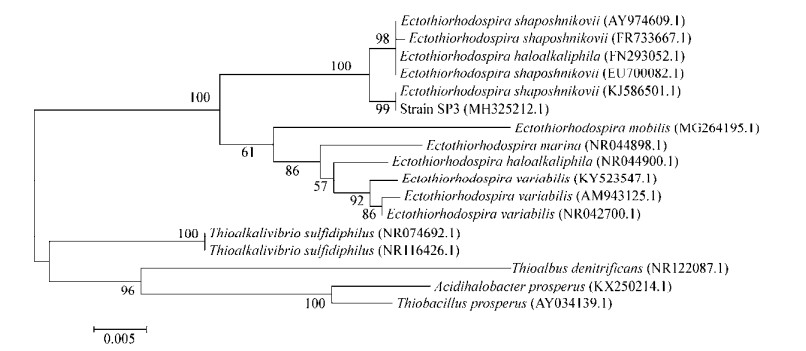
在不同pH的培养条件下,SP3菌株生长状态不同。当pH 7.5时,OD600为2.100±0.015,OD600达到最大值。当pH > 9.5时,OD600低于0.680±0.001。由图 2可知,SP3菌株适宜生长的pH范围为6.0-8.5。
|
| 图 2 不同pH对SP3生长影响 Figure 2 Effects of different pH values on growth of SP3 |
|
|
在不同盐度下,SP3菌株生长状态不同。当NaCl (质量体积比)浓度为0.5%时,OD600为2.970±0.013,OD600达到最大值。当NaCl (质量体积比)浓度为4%时,OD600为0.232±0.012。由图 3可知SP3菌株适宜生长的盐度为0-3%。

|
| 图 3 不同浓度NaCl对SP3生长的影响 Figure 3 Effects of different concentrations of NaCl on growth of SP3 |
|
|
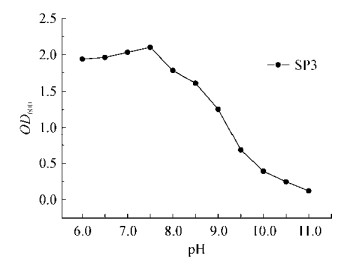
如图 4所示,以不加氮源(0 g/L)的培养基为对照组,菌株SP3以氯化铵为唯一氮源时,OD600由高到低依次是氯化铵浓度为0.25、0.125、0.5、1.0、1.5 g/L。菌株SP3在氯化铵浓度为0.25 g/L的培养基中生长状态最佳,OD600在6 d达到最高,为2.812±0.001。菌株SP3在氯化铵浓度为1.5 g/L的培养基中生长状态相对最差,OD600在6 d达到最高,为2.435±0.004。

|
| 图 4 不同浓度NH4Cl对菌株SP3生长的影响 Figure 4 Effects of different concentrations of NH4Cl on growth of strain SP3 |
|
|
以不加氮源的培养基作为对照组,当亚硝酸钠作为唯一氮源时,培养基中亚硝酸钠浓度大于0.25 g/L会抑制SP3菌株生长(图 5),而以低浓度的亚硝酸钠作为氮源,菌液呈粉红色,OD600由高到低依次是培养基中亚硝酸钠浓度为2.0、1.0、0.5、4.0 mg/L (图 6)。SP3菌株在亚硝酸钠浓度为2.0 mg/L的培养基中生长状态最佳,OD600在6 d达到最高,为2.266±0.004。SP3菌株在亚硝酸钠浓度为4.0 mg/L的培养基中生长状态相对最差,OD600在6 d达到最高,为0.837±0.003。
|
| 图 5 高浓度NaNO2对菌株SP3生长的影响 Figure 5 Effect of high concentration of NaNO2 on growth of strain SP3 |
|
|
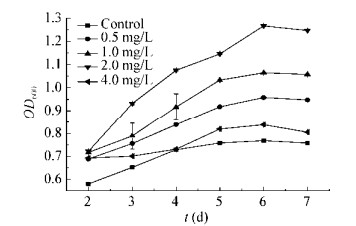
|
| 图 6 低浓度NaNO2对菌株SP3生长的影响 Figure 6 Effects of low concentration of NaNO2 on the growth of strain SP3 |
|
|
光合细菌初始浓度为8.6×109 CFU/mL (双层平板计数的菌落平均值)。接种前(0天),培养液中的氨氮含量为84.15 mg/L±0.58 mg/L。单因素方差分析结果表明,接种后第1天到第4天氨氮去除效果最显著,4 d相互间氨氮累计去除量和累计去除率差异显著(P < 0.05),接种第1天氨氮去除量最大,为50.13±1.89 mg/L,氨氮去除率为65.52%±0.02%。接种后第5天到第7天氨氮累计去除量和累计去除率相互间差异不显著;接种后第7天,氨氮累计去除量为79.45±0.29 mg/L,氨氮累计去除率为94.42%±0.00% (表 4)。
| 时间 Time (d) | 氨氮含量 Ammonia nitrogen content (mg/L) | 累计去除量c Cumulative removal (mg/L) | 累计去除率C Cumulative removal percentage (%) | |
| 对照组 Control group | 实验组 Test group | |||
| 0 | 84.15±0.58 | 84.15±0.58 | ||
| 1 | 83.65±0.29 | 28.52±1.89 | 50.13±1.89a | 65.52±0.02a |
| 2 | 82.65±1.04 | 18.41±2.04 | 64.24±2.04b | 76.35±0.03b |
| 3 | 81.91±1.75 | 10.42±1.44 | 71.48±1.44c | 84.95±0.02c |
| 4 | 81.57±1.32 | 6.27±1.32 | 75.30±1.32d | 89.49±0.02d |
| 5 | 81.32±1.32 | 2.95±0.76 | 78.37±0.76e | 93.13±0.01e |
| 6 | 80.66±1.25 | 1.29±0.50 | 79.37±0.50e | 94.32±0.01e |
| 7 | 80.08±1.32 | 0.63±0.29 | 79.45±0.29e | 94.42±0.00e |
| 注:数据后不同小写字母分别表示存在显著性差异(P < 0.05). Note: Different lowercase letters are significantly at 0.05 probability level. | ||||
在同种光合细菌初始浓度条件下,接种前(0天)培养液中的亚硝态氮含量为1.31±0.00 mg/L。单因素方差分析结果表明,接种后第1天到第3天亚硝态氮去除效果最显著,这3天相互间亚硝态氮累计去除量和累计去除率差异显著(P < 0.05),接种第1天,亚硝态氮去除量最大,为1.00±0.75 mg/L,亚硝态氮去除率最高,为76.44%±0.06%。接种后第3天和第4天相互间的亚硝态氮累计去除量和累计去除率差异不显著;接种后第4天,氨氮累计去除量为1.25±0.00 mg/L,累计去除率为95.20%±0.00%。接种后第5天,培养液亚硝氮浓度低于最低检测限0.003 mg/L (表 5)。
| 时间 Time (d) | 亚硝态氮含量 Nitrogen content (mg/L) | 累计去除量r Cumulative removal (mg/L) | 累计去除率R Cumulative removal percentage (%) | |
| 对照组 Control group | 实验组 Test group | |||
| 0 | 1.31±0.00 | 1.31±0.00 | ||
| 1 | 1.30±0.01 | 0.30±0.08 | 1.00±0.75a | 76.44±0.06a |
| 2 | 1.30±0.01 | 0.17±0.03 | 1.14±0.03b | 86.57±0.02b |
| 3 | 1.29±0.00 | 0.05±0.02 | 1.24±0.02c | 94.75±0.01c |
| 4 | 1.29±0.00 | 0.04±0.01 | 1.25±0.00c | 95.20±0.00c |
| 5 | 1.29±0.00 | 低于检测限 | ||
| 注:数据后不同小写字母分别表示存在水平显著性差异(P < 0.05). Note: Different lowercase letters are significantly at 0.05 probability level. | ||||
以菌株SP3基因组DNA为模板,先用针对两种亚硝酸盐还原酶基因nirS和nirK序列的引物扩增获得PCR产物,克隆和测序获得889 bp的片段。以此为基础,采用Genome Walking三重巢式PCR扩增获得的目标片段克隆测序,通过与已知的889 bp中的重叠部分比对,将新克隆测序片段与已知片段拼接在一起,获得1 404 bp的序列(图 7A),通过BLAST比对发现该序列与nirS基因的相似度最高,属于亚硝酸盐还原酶基因nirS。针对基因nirK的引物没有扩增出目标条带。nirS基因开放阅读框为1 299 bp,编码432个氨基酸(图 7A),其功能结构域的三维结构如图 7 B,预测nirS蛋白质相对分子质量为46.99 kD,理论等电点(PI)为9.56,该蛋白是一种胞浆蛋白。

|
| 图 7 菌株SP3 nirS基因及其编码的氨基酸序列(A)和功能结构域三维结构(B) Figure 7 The strain SP3 nirS gene sequence and deduced amino acid sequence (A) as well as the deduced three dimensional structure (B) of the functional domain encoded by nirS gene 注:A中黄色atg编码启始密码子,黄色taa编码终止密码子;翻译终止密码子用星号(*)表示;下划线编码B中功能结构域. Note: In A, the yellow atg is the initiation codon, the yellow taa encodes the terminator; The translation stop codon is represented by an asterisk (*); The underline encodes the functional domain of B. |
|
|
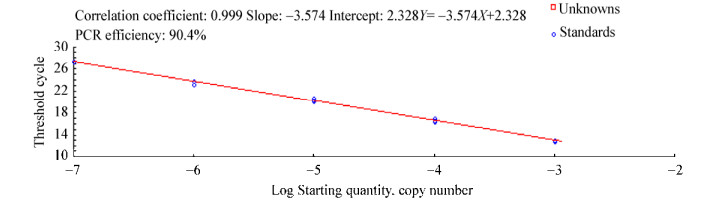
标准曲线方程Y=-3.574X+2.328,相关系数(r)为0.999,且扩增效率为90.4%,满足要求(r > 0.99,110% > Eff > 90%) (图 8)。nirS引物扩增目的片段在温度为94 ℃处出现单一的特异性峰,无杂峰,说明无非特异性扩增及引物二聚体出现(图 9)。
|
| 图 8 nirS引物扩增标准曲线 Figure 8 The standard curve of amplification with nirS primers |
|
|
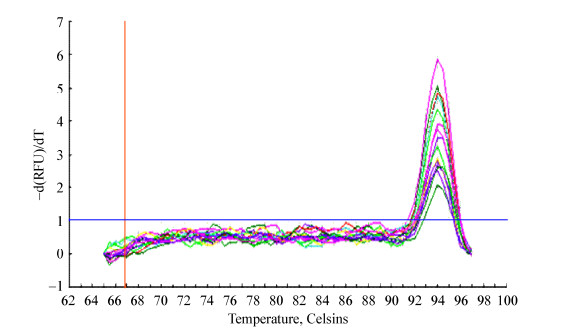
|
| 图 9 nirS引物扩增熔链曲线 Figure 9 The melt curve of products amplified with nirS primers |
|
|
在SP3菌株去除氨氮过程中,nirS表达水平呈先迅速升高达峰值,再降到对照水平这样的模式,接种后第1天到第3天nirS表达量极显著高于对照组(P < 0.01),接种后第4天到第6天,nirS表达量显著高于对照(P < 0.05) (图 10),至第7天nirS表达量与对照间无显著差异。
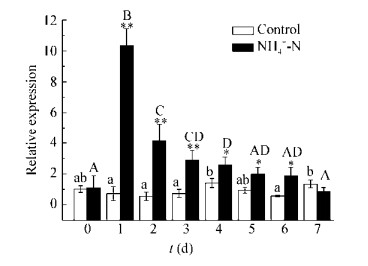
|
| 图 10 受氨氮处理和未处理(对照)的SP3菌株nirS的mRNA表达水平 Figure 10 The mRNA expression levels of nirS in strain SP3 treated with ammonia nitrogen in comparison with those of the SP3 untreated in the control group 注:白色柱:对照组,即不加氮源;黑色柱:氨氮处理组;所有数值均以平均值±标准偏差(n=3)表示;星号:该值与其对照值显著或极显著不同(*:P < 0.05和**:P < 0.01).不同大(小)写字母代表实验组(对照组)值具有显著差异,相同字母表示无显著差异. Note: The white column represents the control group, i.e., no nitrogen source, and the black column represents the ammonia nitrogen treatment group; All values are expressed as mean ± standard deviation (n=3); An asterisk indicates that the value is significantly (*: P < 0.05) or very significantly (**: P < 0.01) different from its control value. Different capital (lowercase) letters represent significant differences among the experimental group (control group) values, and the same letters indicate no significant difference. |
|
|
在SP3菌株去除亚硝态氮的过程中,nirS表达水平也是先迅速升高达峰值,再降到对照水平,接种后第1天到第3天nirS表达量极显著高于对照组(P < 0.01),接种后第4天nirS表达量显著高于对照(P < 0.05),至第5天nirS表达量与对照间无异(图 11)。

|
| 图 11 受亚硝态氮处理和未处理(对照)的SP3菌株nirS的mRNA表达水平 Figure 11 The mRNA expression levels of nirS in strain SP3 treated with nitrite in comparison with those of the SP3 untreated in the control group 注:白色柱:对照组,即不加氮源;黑色柱表示亚硝氮处理组;所有数值均以平均值±标准偏差(n=3)表示;星号:该值与其对照值显著或极显著不同(*:P < 0.05和**:P < 0.01).不同大(小)写字母代表实验组(对照组)值具有显著差异,相同字母表示无显著差异. Note: The white column represents the control group, i.e., no nitrogen source, and the black column represents the nitrite treatment group; All values are expressed as mean ± standard deviation (n=3); An asterisk indicates that the value is significantly (* P < 0.05) or very significantly (** P < 0.01) different from its control value. Different capital (lowercase) letters represent significant differences among the experimental group (control group) values, and the same letters indicate no significant difference. |
|
|
由美国微生物学会发起编写的伯杰氏分类系统是目前国际上普遍接受采用的细菌分类系统。《伯杰氏细菌鉴定手册》最初主要根据细菌的形态学和一系列生理生化指标进行系统分类,在后续再版发行的过程中,逐步增加了16S rRNA基因序列分析、富集和分离方法、DNA中(G+C)mol%等遗传信息、化学分类、生态学、分类学评论以及关键特征鉴定表等更多信息[51]。16S rDNA是细菌遗传物质中编码16S rRNA基因的一段DNA序列,序列保守性较高,已广泛用于细菌属种鉴定[52]。本研究采用伯杰氏分类鉴定方法结合16S rRNA基因序列聚类分析对光合细菌SP3进行种属鉴定。
菌株SP3的16S rRNA基因序列系统发育树分析结果显示其与沙氏外硫红螺菌(E. shaposhnikovii,GenBank登录号KJ566501.1)聚为一支,置信度为99%,由此可推断SP3菌株属于外硫红螺菌属。本次生理生化特性鉴定结果显示,SP3菌株与标准菌株沙氏外硫红螺菌对不同碳源和无机电子受体利用情况一致,也与《伯杰氏细菌鉴定手册》中对沙氏外硫红螺菌记录的生理生化指标、形态学描述和革兰氏染色结果一致。综合形态、生理生化指标和16S rRNA基因序列鉴定结果,确定菌株SP3为沙氏外硫红螺菌(E. shaposhnikovii)。
沙氏外硫红螺菌(E. shaposhnikovii)在分类学上隶属于变形菌门(Proteobacteria) γ-变形菌纲(Gammaproteobacteria)着色菌目(Chromatiales)外硫红螺菌科(Ectothiorhodospiraceae)外硫红螺旋菌属(Ectothiorhodospira)[53]。外硫红螺菌能在缺氧有光照的条件下,以还原的硫化合物作为电子供体,或者以有机化合物为光异养体进行光合自养生长,硫化合物被氧化成为硫酸盐作为一种中间产物和单质硫一起沉积在细胞外[53]。外硫红螺旋菌科分为外硫红螺旋菌属(Ectothiorhodospira)、红硫杆菌属(Thiorhodospira)和盐红螺菌属(Halorhodospira)三个能将硫粒沉积在细胞外部的光合营养细菌属[54]。外硫红螺旋菌属中不同种对盐度的耐受范围不同,H. halophila,H. halochloris和H. abdelmalekii由于极度嗜盐(≥10%)被移出外硫红螺旋菌属,归为盐红螺菌属(Halorhodospira)[55]。活动外硫红螺菌(E. mobilis)的适宜盐度范围为2%-3%[56],菌株SP3最适盐度为0.5%,适宜盐度范围为0%-3%。可见菌株SP3耐盐范围广,能适应绝大多数淡水和海水养殖水体的盐度条件。
pH是水产养殖用水的重要水质指标之一,养殖水体的pH < 5.0或者pH > 9.0都会使养殖鱼类血液pH异常,血液缓冲系统失衡,影响呼吸甚至引起死亡[57]。我国渔业水质标准规定,养殖用淡水的适宜pH值范围是6.5-8.5,海水pH为7.0-8.5[4]。李瑞萍等调查的湖北宜东平原34个分散鱼塘养殖水体pH范围为7.26-8.96,水体酸碱度差异较小且均为弱碱性水体[58]。而养殖水体中的pH不受光合细菌影响[59],并且绝大多数的光合细菌偏好弱碱性环境[60-62]。菌株SP3的适宜生长pH范围是6.0-9.0,与养殖用水的pH范围基本一致,菌株SP3适合在养殖水体中使用。
尽管发现有少数细菌同时具有亚硝酸盐还原酶基因nirS和nirK,如慢生根瘤菌(Bradyrhizobium nitroreducens)[63]和假单胞菌(Pseudomonas stutzeri)[64],但是多数微生物只含有nirS或nirK。本研究表明沙氏外硫红螺菌SP3只含有一种编码亚硝酸盐还原酶基因nirS。在沙氏外硫红螺菌SP3去除氨氮或亚硝态氮过程中,nirS的表达水平的变化模式与氨氮或亚硝态氮的累计去除量的变化模式相一致,即都是第一天迅速上升,然后快速下降,最后与对照无显著区别,这说明nirS在氨氮或亚硝态氮去除过程中发挥了重要作用。该研究结果与Wittorf等的发现一致,即单胞菌(P. stutzeri)去除硝态氮时,其nirS的表达量上调[64]。其原因可能是氨氮可通过氨单加氧酶和羟基氧化还原酶的作用产生亚硝态氮;同时,硝态氮在硝酸还原酶作用下产生亚硝态氮,不同来源的亚硝态氮能被亚硝酸盐还原酶(nirS)降解产生一氧化氮,该分子再经氧化氮还原酶、氧化亚氮还原酶转化为氮气进入空气中,起到脱氮作用,在这里亚硝酸盐还原酶具有限速作用,是整个脱氮过程的关键酶。
有研究表明,在虾苗的培育过程中加入沙氏外硫红螺菌有利于改善水质[65]。沙氏外硫红螺菌对Na2S有较高的耐受度,在Na2S初始浓度为227.6 mg/L的培养基中培养6 d,Na2S转化率达到66.9%[66]。沙氏外硫红螺菌形成的胞外生物聚合物对水体重金属具有很强吸附力,在铜、锌离子初始浓度分别为2 000 mg/L的两种水溶液中,铜离子的去除率达到96.5%,锌离子的去除率达到95%[67]。林启存等分离获得的沙氏外硫红螺菌株对养殖水体中的COD、总氮、氨氮、亚硝酸氮去除率分别为57.38%、45.34%、48.63%和27.91%[68]。本研究发现菌株SP3能以氯化铵、亚硝酸钠为氮源,并且在铵盐中能够生长良好,同时也可以高效去除水体中的氨氮和亚硝态氮,SP3经过7 d对水体氨氮去除率达到94.42%,经过5 d将培养基中2 mg/L的亚硝酸钠去除到检测限(0.003 mg/L)以下,表明菌株SP3有极好的去除水体氮源的潜能。SP3菌株去除氮源的能力较林启存等[68]分离的外硫红螺菌强,这说明SP3是一株良好的可用于净化养殖水环境的沙氏外硫红螺菌。
菌株SP3具有广泛的盐度和pH适应范围,能用于淡水和海水养殖环境有效去除氨氮和亚硝态氮,在养殖水环境净化中有广阔应用前景。
| [1] |
Food and Agriculture Organization of the United Nations. State of World Fisheries and Aquaculture[M]. Rome: Food and Agriculture Organization of the United Nations, 2018: 26-54. (in Chinese) 联合国粮食及农业组织. 世界渔业和水产养殖状况[M]. 罗马: 联合国粮食及农业组织, 2018: 26-54. |
| [2] |
Preparation by the Ministry of Agriculture, Fisheries and Fisheries. 2017 China Fishery Statistical Yearbook[M]. Beijing: China Agriculture Press, 2017: 1-15. (in Chinese) 农业部渔业渔政管理局. 2017中国渔业统计年鉴[M]. 北京: 中国农业出版社, 2017: 1-15. |
| [3] |
Emerson K, Russo RC, Lund RE, et al. Aqueous ammonia equilibrium calculations: effect of pH and temperature[J]. Journal of the Fisheries Board of Canada, 1975, 32(12): 2379-2383. DOI:10.1139/f75-274 |
| [4] |
Ministry of Agriculture and Rural Affairs of the People's Republic of China. GB 11607-1989 Water quality standard for fisheries[S]. Beijing: Standards Press of China, 1989 (in Chinese) 农业部. GB 11607-1989渔业水质标准[S].北京: 中国标准出版社, 1989 |
| [5] |
Qiao SF, Liu HY, Jin XY, et al. Ammonia accumulation hazards and bioavailability in aquaculture waters[J]. Hebei Fisheries, 2006(1): 20-22. (in Chinese) 乔顺风, 刘恒义, 靳秀云, 等. 养殖水体氨氮积累危害与生物利用[J]. 河北渔业, 2006(1): 20-22. DOI:10.3969/j.issn.1004-6755.2006.01.009 |
| [6] |
Zhang JF, Li RW, Liu JF, et al. Accumulation of ammonia nitrogen: Harm and bilolgical control[J]. Hebei Fisheries, 2009(6): 41-44. (in Chinese) 张进凤, 李瑞伟, 刘杰凤, 等. 淡水养殖水体氨氮积累危害及生物控制的研究现状[J]. 河北渔业, 2009(6): 41-44. DOI:10.3969/j.issn.1004-6755.2009.06.019 |
| [7] |
Peng L, Ngo HH, Guo WS, et al. A novel mechanistic model for nitrogen removal in algal-bacterial photo sequencing batch reactors[J]. Bioresource Technology, 2018, 267: 502-509. DOI:10.1016/j.biortech.2018.07.093 |
| [8] |
Luo LH. Research process on the now existing denitrogenation of highly concentrated ammonium-nitrogen wastewater[J]. Sichuan Chemical Industry, 2011, 14(6): 38-42. (in Chinese) 罗龙海. 高浓度氨氮废水处理技术研究进展[J]. 四川化工, 2011, 14(6): 38-42. DOI:10.3969/j.issn.1672-4887.2011.06.013 |
| [9] |
Dong YB, Dai YY. Research survey of toxic effects of nitrite to aquatic animals[J]. Journal of Aquaculture, 2011, 32(4): 28-32. (in Chinese) 董玉波, 戴媛媛. 亚硝酸盐氮对水产经济动物毒性影响的研究概况[J]. 水产养殖, 2011, 32(4): 28-32. DOI:10.3969/j.issn.1004-2091.2011.04.008 |
| [10] |
Wang SQ, Wang FZ, Wang S. Talking about the toxicity of nitrite and its control measures[J]. Fishery Guide to be Rich, 2007(20): 59. (in Chinese) 王圣强, 王甫珍, 王苏. 浅谈亚硝酸盐的毒性及防治措施[J]. 渔业致富指南, 2007(20): 59. |
| [11] |
Chen H, Zhang DM, Wang LG, et al. Biological characteristics and phylogenetic analysis of a denitrifying photosynthetic bacterium[J]. Acta Microbiologica Sinica, 2011, 51(2): 249-255. (in Chinese) 陈慧, 张德民, 王龙刚, 等. 一株反硝化光合细菌的生物学特性及系统发育分析[J]. 微生物学报, 2011, 51(2): 249-255. |
| [12] |
Gao MH, Ma LB, Ge LA, et al. Nitrite uptake mechanism and the influencing factors of accumulation in aquatic animals[J]. South China Fisheries Science, 2008, 4(4): 73-79. (in Chinese) 高明辉, 马立保, 葛立安, 等. 亚硝酸盐在水生动物体内的吸收机制及蓄积的影响因素[J]. 南方水产, 2008, 4(4): 73-79. DOI:10.3969/j.issn.2095-0780.2008.04.014 |
| [13] |
Jones CM, Stres B, Rosenquist M, et al. Phylogenetic analysis of nitrite, nitric oxide, and nitrous oxide respiratory enzymes reveal a complex evolutionary history for denitrification[J]. Molecular Biology and Evolution, 2008, 25(9): 1955-1966. DOI:10.1093/molbev/msn146 |
| [14] |
Yakimov MM, La Cono V, Smedile F, et al. Contribution of crenarchaeal autotrophic ammonia oxidizers to the dark primary production in Tyrrhenian deep waters (Central Mediterranean Sea)[J]. The ISME Journal, 2011, 5(6): 945-961. DOI:10.1038/ismej.2010.197 |
| [15] |
Theerachat M, Virunanon C, Chulalaksananukul S, et al. NirK and nirS nitrite reductase genes from non-agricultural forest soil bacteria in Thailand[J]. World Journal of Microbiology and Biotechnology, 2011, 27(4): 999-1003. DOI:10.1007/s11274-010-0521-5 |
| [16] |
Olmo-Mira MF, Cabello P, Pino C, et al. Expression and characterization of the assimilatory NADH-nitrite reductase from the phototrophic bacterium Rhodobacter capsulatus E1F1[J]. Archives of Microbiology, 2006, 186(4): 339-344. DOI:10.1007/s00203-006-0149-x |
| [17] |
Einsle O, Messerschmidt A, Stach P, et al. Structure of cytochrome c nitrite reductase[J]. Nature, 1999, 400(6743): 476-480. DOI:10.1038/22802 |
| [18] |
Hirasawa M, Tripathy JN, Sommer F, et al. Enzymatic properties of the ferredoxin-dependent nitrite reductase from Chlamydomonas reinhardtii. Evidence for hydroxylamine as a Late intermediate in ammonia production[J]. Photosynthesis Research, 2010, 103(2): 67-77. DOI:10.1007/s11120-009-9512-5 |
| [19] |
Sharma S, Aneja MK, Mayer J, et al. Diversity of transcripts of nitrite reductase genes (nirK and nirS) in rhizospheres of grain legumes[J]. Applied and Environmental Microbiology, 2005, 71(4): 2001-2007. DOI:10.1128/AEM.71.4.2001-2007.2005 |
| [20] |
Braker G, Zhou JZ, Wu LY, et al. Nitrite reductase genes (nirK and nirS) as functional markers to investigate diversity of denitrifying bacteria in Pacific Northwest marine sediment communities[J]. Applied and Environmental Microbiology, 2000, 66(5): 2096-2104. DOI:10.1128/AEM.66.5.2096-2104.2000 |
| [21] |
Xi YF, Zhang MJ, Huang XS, et al. Isolation and identification of a photosynthetic bacteria strain and its ability of denitrification[J]. Journal of Anhui Agricultural Sciences, 2010, 38(27): 14847-14849, 14875. (in Chinese) 席寅峰, 张孟婧, 黄小帅, 等. 1株光合细菌的分离鉴定及其脱氮能力研究[J]. 安徽农业科学, 2010, 38(27): 14847-14849, 14875. DOI:10.3969/j.issn.0517-6611.2010.27.010 |
| [22] |
Hu ZY. Isolation and identification of marine photosynthetic bacteria and their application in the ornamental fish[D]. Qingdao: Master's Thesis of Ocean University of China, 2007 (in Chinese) 胡宗云.海洋光合细菌分离鉴定及其在观赏鱼中的应用[D].青岛: 中国海洋大学硕士学位论文, 2007 http://d.wanfangdata.com.cn/Thesis/Y1112048 |
| [23] |
Ji HP, Li J. The application of photosynthetic bacteria[J]. Progress in Biotechnology, 1997, 17(4): 46-50. (in Chinese) 吉海平, 李君. 光合细菌应用动态[J]. 生物工程进展, 1997, 17(4): 46-50. |
| [24] |
He CH. Research on separation, identification, growth conditions of photosynthetic bacteria and application pretest[D]. Harbin: Master's Thesis of Harbin Institute of Technology, 2009 (in Chinese) 何春华.光合细菌的分离鉴定和生长条件优化及应用初探[D].哈尔滨: 哈尔滨工业大学硕士学位论文, 2009 http://cdmd.cnki.com.cn/article/cdmd-10213-2010027466.htm |
| [25] |
Meng F, Zhang GM, Yang AQ, et al. Bioconversion of wastewater by photosynthetic bacteria: nitrogen source range, fundamental kinetics of nitrogen removal, and biomass accumulation[J]. Bioresource Technology Reports, 2018, 4: 9-15. DOI:10.1016/j.biteb.2018.08.012 |
| [26] |
Huang XF, Li CJ, Zhang FJ. Characteristics of photosynthetic bacteria and its application in wastewater treatment[J]. China Biogas, 2005, 23(1): 29-35. (in Chinese) 黄翔峰, 李春鞠, 章非娟. 光合细菌的特性及其在废水处理中的应用[J]. 中国沼气, 2005, 23(1): 29-35. DOI:10.3969/j.issn.1000-1166.2005.01.007 |
| [27] |
Wang S. Degradation of aniline by photosynthetic bacteria[D]. Hangzhou: Master's Thesis of Zhejiang University of Technology, 2011 (in Chinese) 王帅.光合细菌降解苯胺废水的研究[D].杭州: 浙江工业大学硕士学位论文, 2011 http://cdmd.cnki.com.cn/Article/CDMD-10337-1012286600.htm |
| [28] |
Lu YF. Experimental study of food processing wastewater treatment by photosynthetic bacteria[D]. Harbin: Master's Thesis of Harbin Institute of Technology, 2012 (in Chinese) 卢玉凤.光合细菌处理食品加工废水试验研究[D].哈尔滨: 哈尔滨工业大学硕士学位论文, 2012 http://d.wanfangdata.com.cn/Thesis/D239001 |
| [29] |
Sun T, Li LS. Removal of nitrogen and phosphorus in wastewater using photosynthetic bacteria immobilized on activated carbon[J]. Journal of Agro-Environment Science, 2006, 25(S1): 211-213. (in Chinese) 孙涛, 李兰生. 活性炭固定化对光合细菌去除废水中氮磷的研究[J]. 农业环境科学学报, 2006, 25(S1): 211-213. |
| [30] |
Liu HL, Zhang ZF, Li CL, et al. Effect of photosynthetic bacteria on tilapia based on water quality and disease resistence[J]. Fishery Modernization, 2009, 36(2): 47-51. (in Chinese) 刘慧玲, 张战锋, 李长玲, 等. 光合细菌对罗非鱼鱼苗养殖水质及抗病力的影响[J]. 渔业现代化, 2009, 36(2): 47-51. DOI:10.3969/j.issn.1007-9580.2009.02.011 |
| [31] |
Fang WD, Tang X, He JL, et al. The isolation of PSB strain FP04 and its effect on culture water quality and harvest of Trionyx sinensis[J]. Journal of Aquaculture, 2012, 33(4): 12-16. (in Chinese) 方卫东, 唐旭, 何建林, 等. 光合细菌FP04的筛选及对中华鳖养殖水质和生产效益的影响[J]. 水产养殖, 2012, 33(4): 12-16. DOI:10.3969/j.isssn.1004-2091.2012.04.004 |
| [32] |
Ding AZ, Chen FZ, Lei JQ, et al. Water quality for aquiculture improved by photosynthetic bacteria[J]. Agro-Environmental Protection, 2000, 19(6): 339-341, 344. (in Chinese) 丁爱中, 陈繁忠, 雷剑泉, 等. 光合细菌调控水产养殖业水质的研究[J]. 农业环境保护, 2000, 19(6): 339-341, 344. |
| [33] |
Feng YL. The basic research on application of marine photosynthetic bacteria in aquaculture[D]. Harbin: Master's Thesis of Harbin Institute of Technology, 2017 (in Chinese) 冯亚利.海洋光合细菌在水产养殖中的应用基础研究[D].哈尔滨: 哈尔滨工业大学硕士学位论文, 2017 http://cdmd.cnki.com.cn/Article/CDMD-10213-1017739170.htm |
| [34] |
Wang H, Wang JW, Du G. Preliminary study on microbial autotrophic nitrogen removal in aquaculture water[J]. Journal of Fisheries Science, 2010, 29(5): 295-298. (in Chinese) 王华, 王京伟, 杜刚. 养殖水体中微生物全程自养脱氮初步研究[J]. 水产科学, 2010, 29(5): 295-298. DOI:10.3969/j.issn.1003-1111.2010.05.011 |
| [35] |
Liu M. Nitrogen and phosphate assimilation capacity of a photosynthetic strain[D]. Wuhan: Master's Thesis of Huazhong Agricultural University, 2011 (in Chinese) 刘敏.一株光合细菌氮磷同化能力的研究[D].武汉: 华中农业大学硕士学位论文, 2011 http://cdmd.cnki.com.cn/article/cdmd-10504-1011404932.htm |
| [36] |
Yu PF, Wang LH, Zhan PR. Separation, characterization and immobilization of photobacteria in PVA vectors and application of it in cleaning water of fish pool[J]. Biotechnology, 1995, 5(3): 35-36, 44. (in Chinese) 于沛芬, 王丽华, 战培荣. 光合细菌的分离、鉴定和固定化及其在净化鱼池水质中的应用研究[J]. 生物技术, 1995, 5(3): 35-36, 44. |
| [37] |
Jiang P, Zhao CG, Jia YQ, et al. Inorganic nitrogen removal by a marine purple sulfur bacterium capable of growth on nitrite as sole nitrogen source[J]. Microbiology China, 2014, 41(5): 824-831. (in Chinese) 蒋鹏, 赵春贵, 贾雅琼, 等. 以亚硝氮为唯一氮源生长的海洋紫色硫细菌去除无机三态氮[J]. 微生物学通报, 2014, 41(5): 824-831. |
| [38] |
Wang L, Liao LH. Separation and identification of photosynthetic bacteria (PSB) and purifying effection aquiculture water[J]. Journal of Microbiology, 2004, 24(2): 7-9, 37. (in Chinese) 王兰, 廖丽华. 光合细菌的分离鉴定及对养殖水的净化研究[J]. 微生物学杂志, 2004, 24(2): 7-9, 37. DOI:10.3969/j.issn.1005-7021.2004.02.003 |
| [39] |
Yu JA, Zhang CK, Chen F, et al. Denitration of photosynthetic bacteria P4 strain[J]. Journal of Shanghai Jiaotong University, 2000, 34(11): 1579-1582, 1597. (in Chinese) 俞吉安, 张承康, 陈峰, 等. 光合细菌P4株的反硝化作用[J]. 上海交通大学学报, 2000, 34(11): 1579-1582, 1597. DOI:10.3321/j.issn:1006-2467.2000.11.031 |
| [40] |
Xu SN, Qiu HD, Lin J, et al. On the isolation, identification and function of purple nonsulfer photosynthetic bacteria[J]. Journal of Fuzhou University (Natural Science), 2004, 32(2): 246-251. (in Chinese) 徐姗楠, 邱宏端, 林娟, 等. 紫色非硫光合细菌的分离鉴定及其功能研究[J]. 福州大学学报:自然科学版, 2004, 32(2): 246-251. |
| [41] |
Li X, Wang YH, Zhang SL, et al. Enhancement of phototrophic hydrogen production by Rhodobacter sphaeroides ZX-5 using a novel strategy - shaking and extra-light supplementation approach[J]. International Journal of Hydrogen Energy, 2009, 34(24): 9677-9685. DOI:10.1016/j.ijhydene.2009.10.020 |
| [42] |
Carlozzi P, Pushparaj B, Degl'Innocenti A, et al. Growth characteristics of Rhodopseudomonas palustris, cultured outdoors, in an underwater tubular photobioreactor, and investigation on photosynthetic efficiency[J]. Applied Microbiology and Biotechnology, 2006, 73(4): 789-795. DOI:10.1007/s00253-006-0550-z |
| [43] |
Jiang HC, Dong HL, Zhang GX, et al. Microbial diversity in water and sediment of Lake Chaka, an athalassohaline lake in northwestern China[J]. Applied and Environmental Microbiology, 2006, 72(6): 3832-3845. DOI:10.1128/AEM.02869-05 |
| [44] |
Dong XZ, Cai MY. Handbook of Identification of Common Bacterial Systems[M]. Beijing: Science Press, 2001: 19-398. (in Chinese) 东秀珠, 蔡妙英. 常见细菌系统鉴定手册[M]. 北京: 科学出版社, 2001: 19-398. |
| [45] |
Liu Y, Wang YJ, Cao F, et al. Research on photosynthetic bacteria culture[J]. Journal of Dalian Institute of Light Industry, 1993, 12(2): 37-41. (in Chinese) 刘茵, 王运吉, 曹方, 等. 光合细菌培养研究[J]. 大连轻工业学院学报, 1993, 12(2): 37-41. |
| [46] |
National Environmental Protection Agency. GB/T 7479-1987. Water quality - Determination of ammonium - Nessler's reagent colorimetric method[S]. Beijing: Standards Press of China, 1987 (in Chinese) 国家环境保护局. GB/T 7479-1987水质铵的测定钠氏试剂比色法[S].北京: 中国标准出版社, 1987 |
| [47] |
State Environmental Protection Administration. GB/T 7493-1987, Water quality-Determination of nitrogen (nitrite); Spectrophotometric method[S]. Beijing: Standards Press of China, 1987 (in Chinese) 国家环境保护总局. GB/T 7493-1987, 水质亚硝酸盐氮的测定分光光度法[S].北京: 中国标准出版社, 1987 |
| [48] |
Sarkar J, Kazy SK, Gupta A, et al. Biostimulation of indigenous microbial community for bioremediation of petroleum refinery sludge[J]. Frontiers in Microbiology, 2016, 7: 1407. |
| [49] |
Zhang X, He LM, Zhang FL, et al. The different potential of sponge bacterial symbionts in N2 release indicated by the phylogenetic diversity and abundance analyses of denitrification genes, nirK and nosZ[J]. PLoS One, 2013, 8(6): e65142. DOI:10.1371/journal.pone.0065142 |
| [50] |
Yan L, Zhang QZ, Virolle MJ, et al. In conditions of over-expression, WblI, a WhiB-like transcriptional regulator, has a positive impact on the weak antibiotic production of Streptomyces lividans TK24[J]. PLoS One, 2017, 12(3): e0174781. DOI:10.1371/journal.pone.0174781 |
| [51] |
Yang SP, Lin ZH, Cui XH, et al. Current taxonomy of anoxygenic phototrophic bacteria - a review[J]. Acta Microbiologica Sinica, 2008, 48(11): 1562-1566. (in Chinese) 杨素萍, 林志华, 崔小华, 等. 不产氧光合细菌的分类学进展[J]. 微生物学报, 2008, 48(11): 1562-1566. DOI:10.3321/j.issn:0001-6209.2008.11.023 |
| [52] |
Muyzer G, Teske A, Wirsen CO, et al. Phylogenetic relationships of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel electrophoresis of 16S rDNA fragments[J]. Archives of Microbiology, 1995, 164(3): 165-172. DOI:10.1007/BF02529967 |
| [53] |
Brenner DJ, Krieg NR, Staley JT, et al. Bergey's Manual® of Systematic Bacteriology. Vol.2. Part B[M]. 2nd ed. New York: Springer, 2005: 41-48.
|
| [54] |
Brenner DJ, Krieg NR, Staley JT, et al. Bergey's Manual® of Systematic Bacteriology. Vol.2. Part A[J]. New York: Springer, 2005, 131. |
| [55] |
Imhoff JF, Süling J. The phylogenetic relationship among Ectothiorhodospiraceae: a reevaluation of their taxonomy on the basis of 16S rDNA analyses[J]. Archives of Microbiology, 1996, 165(2): 106-113. DOI:10.1007/s002030050304 |
| [56] |
Trüper HG. Ectothiorhodospira mobilis Pelsh, a photosynthetic sulfur bacterium depositing sulfur outside the cells[J]. Journal of Bacteriology, 1968, 95(5): 1910-1920. |
| [57] |
Deng XH. The fishery produces the pH value function and adjustment[J]. Hebei Fisheries, 2008(2): 4-6. (in Chinese) 邓希海. 养殖水体中pH值的作用及调节[J]. 河北渔业, 2008(2): 4-6. DOI:10.3969/j.issn.1004-6755.2008.02.002 |
| [58] |
Li RP, Zhang XX, Liu Z, et al. Distribution characteristics and relationships of pH, nutrients, chlorophyll a and three sulfonamides in pond aquaculture water[J]. Chinese Journal of Environmental Engineering, 2015, 9(6): 2582-2588. (in Chinese) 李瑞萍, 张欣欣, 刘卓, 等. 池塘养殖水体pH、营养盐、叶绿素a及3种磺胺类抗生素分布特征及其相关性分析[J]. 环境工程学报, 2015, 9(6): 2582-2588. |
| [59] |
Wei YL. The culture and application of the photosynthetic bacteria (PSB)[J]. Life Science Instruments, 2007, 5(3): 27-30. (in Chinese) 魏玉利. 光合细菌的培养及应用[J]. 生命科学仪器, 2007, 5(3): 27-30. DOI:10.3969/j.issn.1671-7929.2007.03.006 |
| [60] |
Zhang FF, Zhou K, Zhao YJ, et al. Study on culture condition of photosynthetic bacteria[J]. Tianjin Agricultural Sciences, 2009, 15(2): 9-11. (in Chinese) 张峰峰, 周可, 赵玉洁, 等. 光合细菌培养条件的研究[J]. 天津农业科学, 2009, 15(2): 9-11. DOI:10.3969/j.issn.1006-6500.2009.02.003 |
| [61] |
Xie HG, Wang SF, Chu R. Experimental cultivation of photosynthetic bacteria for wastewater treatment[J]. Environmental Science & Technology, 2007, 30(5): 19-20. (in Chinese) 谢红刚, 王三反, 褚润. 光合细菌培养条件的研究[J]. 环境科学与技术, 2007, 30(5): 19-20. DOI:10.3969/j.issn.1003-6504.2007.05.008 |
| [62] |
Wang XJ, Wang QY. Study on culture condition and application of photosynthetic bacteria[J]. Journal of Xinyang Teachers College (Natural Science Edition), 1994, 7(2): 208-211. (in Chinese) 王小君, 王清义. 光合细菌培养条件及其应用的研究[J]. 信阳师范学院学报:自然科学版, 1994, 7(2): 208-211. |
| [63] |
Jang J, Ashida N, Kai A, et al. Presence of Cu-type (NirK) and cd1-type (NirS) nitrite reductase genes in the denitrifying bacterium Bradyrhizobium nitroreducens sp. nov.[J]. Microbes and Environments, 2018, 33(3): 326-331. DOI:10.1264/jsme2.ME18039 |
| [64] |
Wittorf L, Jones CM, Bonilla-Rosso G, et al. Expression of nirK and nirS genes in two strains of Pseudomonas stutzeri harbouring both types of NO-forming nitrite reductases[J]. Research in Microbiology, 2018, 169(6): 343-347. DOI:10.1016/j.resmic.2018.04.010 |
| [65] |
Wen CQ, Xue M, Liang HF, et al. Beneficial effects of Ectothiorhodospira shaposhnikovii WF on larval cultivation of Litopenaeus vannamei[J]. Beneficial Microbes, 2015, 6(4): 529-533. |
| [66] |
Liu JF, Huang Y, Liu ZH, et al. Isolation, identification and characterization of a Ectothiorhodospira sp. from mangrove[J]. Environmental Science & Technology, 2018, 41(2): 8-12. (in Chinese) 刘杰凤, 黄瑶, 刘正辉, 等. 1株红树林外硫红螺菌的分离鉴定及其特性[J]. 环境科学与技术, 2018, 41(2): 8-12. |
| [67] |
Tian JM. Using the biopolymer of microorganism Ectothiorhodospira shaposhnikovii to remove of heavy metals from waste waters[J]. Journal of Taiyuan University of Technology, 1999, 30(2): 175-178. (in Chinese) 田建民. 用微生物外红硫螺菌属形成的生物聚合物去除废水中的重金属[J]. 太原理工大学学报, 1999, 30(2): 175-178. DOI:10.3969/j.issn.1007-9432.1999.02.019 |
| [68] |
Lin QC, Zhu LM, Shen XH, et al. Isolation and identification of a photosynthetic bacterium[J]. Journal of Zhejiang Agricultural Sciences, 2017, 58(8): 1444-1446. (in Chinese) 林启存, 朱丽敏, 沈小红, 等. 一株光合细菌的分离与鉴定[J]. 浙江农业科学, 2017, 58(8): 1444-1446. |
 2019, Vol. 46
2019, Vol. 46