扩展功能
文章信息
- 张增虎, 唐丽丽, 张永雨
- ZHANG Zeng-Hu, TANG Li-Li, ZHANG Yong-Yu
- 海洋中藻菌相互关系及其生态功能
- Algae-bacteria interactions and their ecological functions in the ocean
- 微生物学通报, 2018, 45(9): 2043-2053
- Microbiology China, 2018, 45(9): 2043-2053
- DOI: 10.13344/j.microbiol.china.180178
-
文章历史
- 收稿日期: 2018-03-12
- 接受日期: 2018-05-04
- 网络首发日期(www.cnki.net): 2018-06-14
藻类(包括原核藻类和真核藻类)是海洋中最主要的初级生产者,虽然生物量仅占全球植物的1%-2%,但每年贡献了全球40%的固碳量[1]。细菌利用和转化藻类10%-50%的光合作用产物[2],是生物地球化学循环重要的引擎,与藻类共同构成了海洋生态系统的重要调控者。
海洋中藻类和细菌共存了2亿多年[3],相互已经密不可分。藻类为细菌提供营养物质,促进细菌生长;细菌矿化藻源物质,释放无机盐,为藻类生长提供无机营养,这种互利共生的方式维持了藻菌间长期密切的关系。此外,长久共存促进了藻菌间的协同进化,如藻菌间的水平基因转移,藻菌在代谢上发生补充和简化,催化了彼此形成更为密切的关系。
自然界细菌是以群落的形式存在的,藻际环境中存在与周围环境截然不同的细菌群落。而且由于不同的藻类可产生不同性质的有机物而形成不同的藻际微环境,同种藻类不同生长时期也会分泌不同的物质,使得藻际细菌群落处在不断演替的动态过程中。目前,随着分子生态学的发展,大量研究发现藻际微生物群落的构建存在一定的规律和特点,这也使得藻菌关系的研究放大到藻与菌群的关系上,有利于更好地认识藻菌关系的生态功能。
本文综述了藻菌间复杂的相互作用、藻际环境中细菌群落构建规律以及藻菌的生态功能,为进一步研究和开发利用藻菌关系提供基础。
1 藻际环境——藻菌相互作用的主战场类似于土壤生态系统中的根际环境,藻周围存在着藻菌互作的微小区域——藻际环境[4]:藻细胞向周围释放营养物质,形成以藻为核心的独特的富营养区域。藻际环境区域的尺寸与藻细胞大小有关,同时也依赖于藻类的生长速率、藻源物质的分泌速率及扩散率等[5]。
藻细胞向胞外释放物质存在两种方式[5]:一种是被动方式,如气体、溶剂分子、疏水化合物等能够透过细胞膜,或在胞外合成大分子物质;另一种是主动方式,如小的极性或带电分子(如单糖和氨基酸)需要藻细胞主动运输到细胞膜外。物质的主动释放会消耗能量,但藻类可通过这些物质吸引特定的细菌来构建附生或共生有益菌群。藻物质的释放与藻生长阶段有关,在生长早期,藻细胞易分泌溶解性高、可利用性强、分子量低的分子,如氨基酸、碳水化合物和有机酸,而其中某些物质具有潜在趋化特性[6],可以吸引细菌;生长末期,藻细胞常常会释放高分子量的分子,如多糖、核酸、蛋白质和脂质等。
藻际细菌来源方式有所不同。无运动能力的细菌进入藻际环境是随机性的,藻菌接触的概率也是随机的,如在106 cells/mL细菌浓度(细菌直径1 μm)和103 cells/mL的藻细胞浓度(藻细胞直径15 μm)的环境里,细菌每天可遇到0.003 5个藻细胞或者一个藻细胞每天遇到3.5个细菌[5]。有运动能力的细菌,则可以主动到达藻际环境中,某些浮游细菌甚至可以追踪运动的藻类[7]。此外,研究发现细菌存在运动性质的转变,化合物可诱导细菌群体发生运动性的转变,这也可能与藻有关[8]。
藻际细菌有不同的存在形式。有些细菌可以内寄生在藻体内,甚至随着藻的繁殖而传递到子代,从而形成长久关系。有些细菌可营附着生活,形成生物膜,而这种关系依赖于藻的生长状态,随时间也会发生变化。还有一些细菌与藻保持动态的作用关系,没有与藻体稳定接触,存在于藻际环境中。
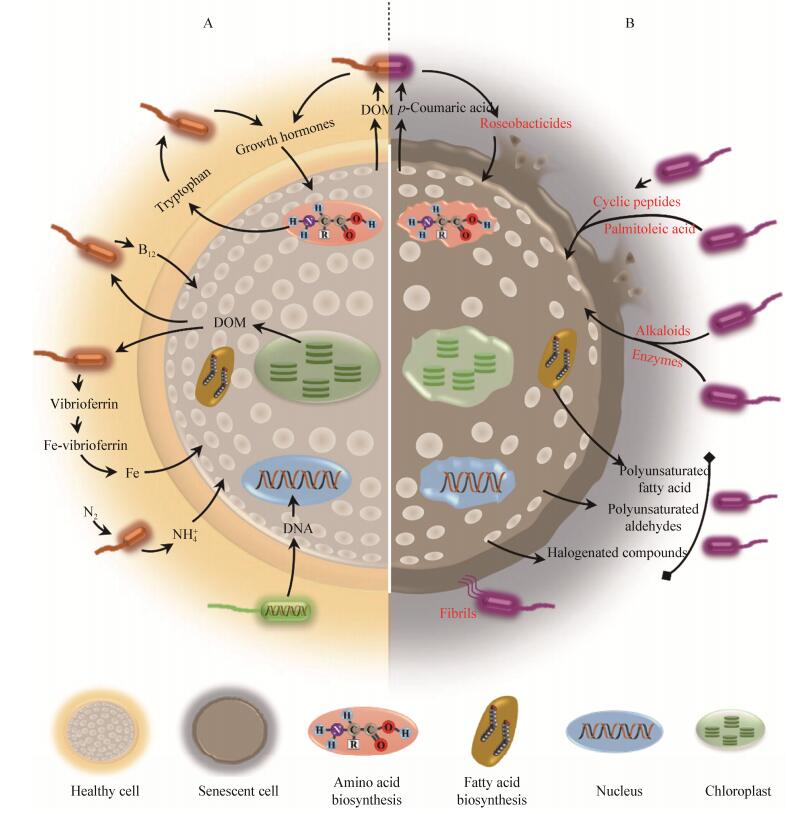
2 藻菌错综复杂的相互关系藻菌间最根本的关系是资源供求关系,藻菌之间的营养物质循环可维持藻菌长久稳定的生存[9],例如笔者通过长期共培养实验发现,在不补充添加任何营养物质的前提下,无菌聚球藻存活时间仅为1-2个月,而有些外加了菌群的聚球藻能存活长达1年之久。在复杂的自然环境下,健康的藻细胞需要产生化合物来吸引有益菌促进自身生长或分泌抗菌物质排斥有害菌的侵入,当藻释放营养物质孕育大量细菌后,这些藻际细菌又反作用于藻类,对藻生长产生促进或抑制的作用,随着藻细胞生长状态的变化,促进和抑制作用也发生转变。长此以往,藻菌之间形成了密切而又复杂的互作关系(图 1)。
藻类为细菌提供营养物质,无论是藻释放的溶解有机物(Dissolved organic matter,DOM)还是藻源颗粒有机物(Particulate organic matter,POM)皆可被细菌所用。藻源物质种类多样,大部分都可作为细菌的营养物质,如碳水化合物(多糖和半活性多糖等)、含硫有机物(Dimethylsulphoniopropionate,DMSP等)和含氮物质(氨基酸等)。藻类通过释放这些化合物吸引细菌,建立密切的相互关系。此外,藻类还可以分泌物质来刺激细菌DNA合成,增加其生物膜形成能力[11],进而维持建立起的互作关系。
藻类可以产生抗菌物质,杀死或排斥部分细菌。这类物质主要包括脂肪酸、多不饱和醛以及卤代化合物等。藻类产生的有抑菌活性的脂肪酸种类多样,例如硅藻Navicula delognei产生的十六碳三烯酸、十六碳四烯酸、十八碳四烯酸、二十碳五烯酸[12]等;多不饱和醛(Polyunsaturated aldehydes,PUAs)是一种氧化脂类,多发现于硅藻的次级代谢产物中,对浮游动物、植物和细菌都有毒性效应[13];海洋藻类可以产生多种卤代代谢物(包括卤代烷、短链碳氢化合物、萜烯和酚类化合物),这些物质也有抑菌活性[14]。此外,藻类还可以通过抑制细菌的密度感应系统,间接地抑制细菌的代谢活动[15]。
2.2 细菌对藻类生长的促进及抑制细菌为藻类生长营造了良好的环境,例如藻际细菌消耗掉氧,缓解了藻生长过程中的氧气压力;细菌对有机氮等物质的重新矿化为藻提供大量的营养盐;细菌为维生素依赖型的藻类提供维生素[16];为铁依赖型的藻类提供铁载体[17];有时细菌作为临时磷库为藻类提供所需要的磷[18];细菌也可以产生植物生长素(吲哚-3-乙酸)促进藻的生长[19]。除了藻类生长,细菌还有助于形成或维持藻类的形态,例如细菌的存在促进了绿藻Ulva mutabilis[20]形态的形成;对于微藻,细菌也可以维持藻细胞(硅藻)长时间培养下的形态[21]。
有些细菌能够直接杀藻,噬纤维菌Cytophaga sp.可直接杀死硅藻和部分甲藻[22],但是通过这种直接接触杀藻的细菌为数不多。细菌代谢活动多样,一些分泌物质具有抑藻或溶藻的活性,例如笔者实验室从一株近海沉积物细菌Microbulbifer sp.的代谢产物中分离鉴定出一种卤代联苯类物质,显示出了很强的杀藻活性(暂未发表数据)。但由于溶藻物质的分离纯化比较复杂,目前主要确定的物质包括氨基酸、多肽、环肽、水解酶和生物碱等物质(表 1)。抑藻或溶藻的效应一般包括降低藻类光合作用和丧失藻细胞壁完整性[25]。除此之外,细菌可以通过抑制藻细胞分裂[52]、抑制藻细胞的运动[53]和抑制藻类游动孢子的发芽和生长[54]等方式抑藻或溶藻。细菌溶藻效果会受到不同条件的限制,比如细菌溶藻活性与细菌的生长时期有一定关系[55],有的细菌的毒性效果还与温度相关,例如Ruegeria sp. R11在25 ℃杀藻,在18 ℃不杀藻[56],有些溶藻物质的释放还会受到密度感应的调节[57]。
| 物质分类 Substance classification |
溶藻物质 Algicides |
溶藻细菌 Algicidal bacteria |
相关藻类 Related algaes |
文献 References |
| 氨基酸Amino acid | β-氰基-L-丙氨酸β-Cyano-L-alanine | Vibrio sp. C-979 | Oscillatoria amphibian NIES-361 | [23] |
| L-赖氨酸L-lysine | Streptomyces sp. S-9 | Microcystis aeruginosa NIES-112 | [24] | |
| L-缬氨酸L-valine | Streptomyces jiujiangensis | Microcystis aeruginosa FACHB-905 | [25] | |
| 脂肪酸Fatty acid环肽/多肽衍生物 Cyclic peptides/peptide derivates |
十六碳烯酸Palmitoleic acid | Vibrio sp. BS02 | Alexandrium tamarense ATGD98-006 | [26] |
| 五肽Pentapeptide | Sphingomonas sp. M-174 | Microcystis viridis NIES-102 | [27] | |
| Sphingomonas sp. M-174 | Microcystis aeruginosa NIES-298 | [27] | ||
| 十二肽Dodecapeptide | Vibrio shiloi AK1 | Zooxanthellae | [28] | |
| 环肽Cyclo-(Pro-Gly) | Stenotrophomonas sp. F6 | Microcystis aeruginosa 9110 | [29] | |
| Bacillus sp. Ts-12 | Phaeocystis globosa | [30] | ||
| 酶类Enzymes | 丝氨酸蛋白酶Serine protease | Pseudoalteromonas sp. A28 | Skeletonema costatum NIES-324 | [31] |
| 几丁质酶Chitinase | Chitinimonas prasina LY03 | Thalassiosira pseudonana | [32] | |
| 胺类Amines | 羟胺Hydroxylamine | Arthrobacter sp. Q1 | Chlorella vulgaris | [33] |
| 2-异丁氧基苯基胺(2-Isobutoxyphenyl)amine | Brevibacterium sp. BS01 | Alexandrium tamarense ATGD98-006 | [34] | |
| 生物碱类 Alkaloids |
灵菌红素Prodigiosin | Hahella sp. KA22 | Microcystis aeruginosa FACHB-1752 | [35] |
| 1-甲基-β-咔啉1-Methyl-carboline | Pseudomonas sp. K44-1 | Anabaena cylindrica NIES-19 | [36] | |
| 噻唑生物碱Bacillamide | Bacillus sp. SY-1 | Cochlodinium polykrikoides | [37] | |
| 2-戊基-4-喹啉醇2-Pentyl-4-quinolinol | Alteromonas sp. | Amphora coffeaeformis | [38] | |
| Alteromonas sp. | Navicula sp. | [38] | ||
| 3-甲基吲哚3-Methylindole | Aeromonas sp. GLY-2107 | Microcystis aeruginosa 9110 | [39] | |
| l-乙酰基-β-咔啉1-Acetyl-β-carboline | Brachybacterium sp. YS-3 | Alexandrium catenella | [40] | |
| 2, 3-吲哚醌2, 3-Indolinedione | Shewanella sp. Lzh-2 | Microcystis aeruginosa 9110 | [41] | |
| Shewanella sp. Lzh-2 | Synechococcus sp. BN60 | [41] | ||
| Pseudomonas C55a-2 | Chaetoceros ceratosporum C-16 | [42] | ||
| 反式-3-吲哚丙烯酸Trans-3-indoleacrylic acid | Rhodococcus sp. p52 | Microcystis aeruginosa FACHB 927 | [43] | |
| 喹诺酮Quinolone | Alteromonos KNS-16 | Heterosigma akashiwo | [44] | |
| 密度感应相关 Quorum sensing |
密度感应前体Quorum-sensing precursor | Pseudoalteromonas piscicida | Emiliania huxleyi DHB624 | [45] |
| 密度感应分子N-acylhomoserine lactones | Roseobacter clade bacteria | Skeletonema costatum CCMP 1332 | [46] | |
| 其他Others | 1-羟基吩嗪1-Hydroxyphenazine | Pseudomonas aeruginosa | Chlorella sp., Klebsormidium sp. | [47] |
| 类胡萝卜素Deinoxanthin | Deinococcus sp. Y35 | Alexandrium tamarense ATGD98-006 | [48] | |
| 三萜皂苷Triterpenoid saponin | Streptomyces sp. L74 | Microcystis aeruginosa FACHB 905 | [49] | |
| 苯甲酸Benzoic acid | Thalassospira sp. ZR-2 | Karenia mikimotoi OUC151001 | [50] | |
| 蛋白质Protein | Vibrio sp. DHQ25 | Alexandrium tamarense | [51] | |
| Pseudoalteromonas sp. DHY3 | Alexandrium tamarense | [51] |
藻的聚集沉降是海洋中形成“海雪”驱动深海碳输出作用的主要因素之一,是海洋中的一个重要生态现象与过程。一些细菌可以引起藻类的聚集和沉降,例如微氏海链藻Thalassiosira weissflogii的附着细菌Marinobacter adhaerens HP15可以导致微氏海链藻形成透明胞外聚合颗粒物(Transparent exopolymer particles,TEP),而TEP是硅藻聚集的主要粘合剂,增加了藻的粘性,引起了藻的聚集[58]。除了通过影响藻细胞的粘性导致藻的聚集外,有些细菌具有特殊的细胞结构特点,如具有菌毛结构[59]、细菌丝状排列[60]、细菌表面带电[60]以及自身细胞表面含凝胶物质等,都可能通过与藻发生作用从而形成聚集。例如Malfatti和Azam利用原子力显微镜观察发现大洋水体中大多数的聚球藻在纳米尺度上与其他细菌紧密联系,彼此通过菌毛或者细胞表面凝胶类物质形成细胞聚集体[59];笔者实验室也发现聚球藻Synechococcus sp. PCC7002可与近海微生物群落形成较大的聚集体并向下沉降和贴壁(图 2)。近年来,科学家揭示在寡营养的大洋环境中,聚球藻与某些细菌种群的相互作用可能是驱动大洋碳输出作用的关键因素[61],藻菌聚集体的形成与沉降可能是这种相互作用的重要方式。除此之外,这种用细菌促使藻聚集的方法还受到能源微藻养殖领域的青睐[60],并展示出潜在的应用前景。

|
| 图 2 有/无菌培养条件下聚球藻Synechococcus sp. PCC7002的聚集贴壁现象 Figure 2 Cell aggregation and adherence of Synechococcus sp. PCC7002 under axenic/non-axenic culture conditions 注:图片所示为将培养液倾倒后粘附于培养瓶底部的藻体. A:无菌培养条件;B:有菌培养条件. Note: The pictures show the algal mat adhered to the bottom of the flasks after the interior algal liquid was poured out. A: Under axenic culture condition; B: Under non-axenic culture condition. |
|
|
藻菌间存在“双重”关系(Jekyll-and-Hyde),即共生和敌对关系共存。细菌Dinoroseobacter shibae和微小原甲藻Prorocentrum minimum共培养时,存在共生和敌对2个阶段,微小原甲藻生长初期,D. shibae为其提供维生素B12,促进藻的生长,而大约21 d后,D. shibae与微小原甲藻之间转化为敌对关系,细菌开始溶藻[62]。这种现象也存在于其他藻类和细菌(玫瑰杆菌类群)中,例如赫氏颗石藻Emiliania huxleyi和海洋细菌Phaeobacter inhibens,赫氏颗石藻生长初期为细菌提供营养物质,细菌为藻提供生长激素,是互利共生关系,当赫氏颗石藻衰亡时,藻细胞释放p-香豆酸,引起细菌合成溶藻物质(Roseobacticides),两者即转化为敌对关系[63],细菌开始溶藻。其他研究指出细菌Phaeobacter inhibens产生的植物激素(吲哚-3-乙酸)既可以益藻又可以杀藻,可能是控制藻菌关系转变的重要因子[64]。藻菌间的这种“双重”关系可能对海洋藻类的生理、藻华动力过程和生物地球化学循环有重要的影响。
3 藻际环境中的细菌群落构建自然环境下,细菌是以群落的形式存在的[65],单菌和单藻之间的关系几乎不存在。藻际环境虽然微小,但因为其内部的营养物质特殊,形成了与周围环境十分不同的藻际细菌群落。随着分子生态学的发展,人们对藻际微生物群落的研究不断深入,发现了藻际微生物群落构建具有一定的规律和特点。
3.1 细菌分类水平上的群落构建特征借用分子生态学研究方法,藻际微生物群落的构建存在种属特异性,其中变形菌门和拟杆菌门是藻际环境中最丰富的细菌门类[66],例如,微星鼓藻藻际环境中主要的细菌类群是拟杆菌门[67];硅藻富含属于玫瑰杆菌类群、γ-变形菌纲和黄杆菌纲的细菌[68]。但由于微生物群落构建主要是由藻源物质诱导的[69],所以分泌独特藻源物质的藻种也会吸引其他优势细菌种类来构建群落,例如软骨藻酸产生藻Pseudo-nitzschia australis富集的菌群主要属于厚壁菌门[70]。藻际微生物群落与藻的生长时期有关,不同生长时期,营养物质的浓度和可利用性影响了藻际环境中附着和游离细菌的群落演替,例如,黄杆菌纲的细菌偏好附着生长,易降解藻源高分子量的有机物[71],经常出现在藻类释放高分子量DOC (Dissolved organic carbon,DOC)和POC (Particulate organic carbon,POC)的生长阶段,而玫瑰杆菌类群的细菌却不同,可在游离和附着状态下转化,既能利用生物高分子量的物质又能利用藻类和黄杆菌纲细菌产生的活性低分子量的DOC,它们可以存在于藻类生长的各个时期。
3.2 细菌功能水平上的群落构建特征虽然在高的分类水平(门或纲)藻际微生物群落总是富集类似的细菌类群(如变形菌门和拟杆菌门等),但往往在种水平上,即使是相似的藻环境,藻际细菌却存在很大差异[72]。随着宏基因组学技术的应用,人们开始把微生物功能与分类区别开来,对藻际微生物群落的构建也有了新的认识。Burke等[73]研究发现在相似的藻际环境中,虽然藻际微生物在种水平上相似度很低,但在功能基因组成上却存在高的相似度,为此,他们引入了竞争性机会模型(Competitive lottery model)来揭示藻际微生物群落构建规律:在同一个生态环境中,类似功能的细菌能够占据相同的生态位,且占据过程是随机的。它们结合竞争性机会模型并把探讨藻际微生物群落构建的关注点由细菌的分类转向了细菌的功能基因,合理解释了相似的藻环境中细菌种类差异很大而功能基因相似度高的问题,为探讨藻际微生物群落构建规律提供了新的思路。
4 藻菌关系的生态作用藻类和细菌是微生物环[74]的重要组成部分,也是海洋生物泵固碳和微型生物碳泵储碳[75]的重要驱动者。虽然藻菌关系发生在微尺度环境,但对海洋生态过程(如藻华过程、水质修复等)及海洋生物地球化学循环(如碳循环等)有着重要的作用。
4.1 藻菌关系与藻华过程藻菌复杂互作关系渗透在藻华的整个过程中:一方面,藻华期间为细菌提供了丰富的营养物质,抚育了大量的细菌,藻华暴发初期和中前期,藻释放活性的低分子量DOC,刺激了细菌的生长代谢,细菌的丰度随着藻丰度的升高而升高;藻华衰退阶段,藻数量开始下降,但细菌可以利用衰亡藻细胞释放的有机物质,维持了高的丰度;藻际细菌群落组成受到藻源物质变化的影响而发生变化。另一方面,藻际细菌是藻华过程中重要的生物因子,藻际细菌有助于藻类形态的形成,矿化了藻源有机物,为藻提供了丰富的营养盐,共生固氮菌可为藻类提供生物可利用的氮元素[76],维生素产生菌和铁载体产生菌可为藻类提供关键的微量元素,而藻华末期,溶藻菌分泌溶藻物质加速了藻华的衰退。复杂的藻菌互作关系在藻华生消过程中发挥了重要作用。1998年,Lovejoy等[77]发现了Pseudoalteromonas sp. Y能引起链状裸甲藻(Gymnodinium catenatum)、海洋卡盾藻(Chattonella marina)和赤潮异弯藻(Heterosigma akashiwo)的裂解和死亡,指出细菌对有害藻华有重要的调控作用。近年来随着溶藻细菌研究的不断深入,科学家们已经发现多种溶藻细菌(如Streptomyces、Bacillus、Ruegeria、Pseudoalteromonas、Aeromonas、Shewanella、Pseudomonas、Vibrio和Hahella属等)通过分泌溶藻物质(如蛋白酶、生物碱、氨基酸、羟胺、苯甲酸、环肽与氢醌等)能对藻华肇事种进行裂解,例如Heterosigma akashiwo、Microcystis aeruginosa、Phaeocystis globosa、Skeletonema costatum、Lingulodinium polyedrum、Prorocentrum minimum、Chattonella marina、Emiliania huxleyi和Alexandrium tamarense等藻种。基于溶藻菌的杀藻效能,“以菌治藻”被认为是一种具有潜力的赤潮治理方式。
4.2 藻菌共生与水质修复早在1957年,Oswald等就描述了藻菌共生体系在富营养废水修复中的应用[78],藻菌共生体系不仅具有改善水质、降低污染、提供饵料微藻、抑制有害菌或病原虫和节约成本的特点,与单独使用藻或菌处理废水相比,它还具有修复效果好、稳定性强及循环性高的优势[79]。藻从环境中获取无机氮磷等营养盐,降低污水中过高的硝酸盐和磷酸盐浓度,同时为藻菌共生环境提供氧气(异养细菌降解有机物时最重要的电子受体);藻共生细菌分解代谢环境中难降解的有机质,部分细菌可通过分泌化合物促进藻的生长,例如,产吲哚-3-乙酸(植物激素)的细菌——固氮螺菌(Azospirillum brasilens),可以促进小球藻(Chlorella vulgaris,废水治理时常用的藻种)的生长,也有细菌如硝化细菌和反硝化细菌等有去除氨氮效果,可加强氨氮的去除;藻菌间的互利关系维持了代谢活动的稳定和不断的循环。除了对常见的富营养废水的净化和修复外,藻菌互利关系还有利于降解其他有机污染物(如黑油、乙腈、苯酚、萘、苯并芘、二苯并呋喃及偶氮化合物等)[80]。近年来,海水养殖业的迅猛发展,饵料的排放、残饵的分解以及抗生素的使用,使水体中氮磷营养盐及有机污染物等严重超标,而且养殖废水已成为一类重要的环境污染源,藻菌共生体系对近海养殖水体的修复往往有良好的效果,有着重要的应用潜力[80]。
4.3 藻菌关系与海洋碳循环藻类是海洋重要的初级生产者[81],藻际环境中有机物极其丰富,是微观尺度上的重要有机碳库。藻类释放的DOC和POC绝大多数进入了微生物环,还有部分沉降后埋藏在沉积物中或输送到深海而长期存储[82]。细菌是微生物环的主要驱动者,可通过转化藻源物质成为细菌自身生物量,再进而传递到较高营养水平的生物体中。此外,藻际细菌可通过呼吸作用将有机碳转化为CO2释放到环境中去,或者通过微型生物碳泵机制将POC转化为DOC分泌到海洋水体当中,其中相当一部分DOC具有生物惰性,以惰性溶解有机碳形式可在海洋中储存数千年之久[83]。有些藻菌关系还会影响碳的输出:细菌可以引起藻类聚集,提高藻的沉降速率,从而增加海洋生物泵碳输出效率;可以提高藻类多糖的分泌能力和有毒物质的产量,影响藻类对溶解有机物、TEP和含蛋白颗粒物的分泌[84];反之,藻类输出的有机碳发生改变,细菌利用和转化生成的有机碳也会发生改变[85],进而影响微生物驱动的海洋碳循环过程。
5 展望自“藻际环境”概念提出以来,藻菌关系的研究已近50年。人们已经发现了许多相关的功能基因,鉴定了一些关键的物质,藻菌复杂的相互关系也越来越清晰。高通量测序技术和生物信息学方法的发展目前也已成为深入解析藻菌复杂关系的重要工具。然而,目前针对藻菌关系研究方面还有一些薄弱环节值得进一步深入开展:(1)藻际环境中,不仅存在藻菌之间的互作关系,还包括细菌-细菌、细菌-病毒[86]和细菌与其他宿主之间的关系,这些混合因素也是影响藻际微生物群落构建的重要方面;(2)藻际环境中细菌群落与周边环境差异极大,包含有很多潜在的新物种[87],发掘藻际环境新型微生物类群有助于获得新型天然产物或发现新颖的藻菌关系;(3)目前解析藻类对全球气候变化或海洋酸化的响应是一个研究热点[88],然而往往忽视了细菌在这一过程中对藻类生理生态功能的调控作用;(4)藻类光合作用与细菌代谢转化分别是驱动生物泵固碳和微型生物碳泵储碳的主体[89],藻类通过光合作用形成的有机碳经微生物代谢转化,有多大比例能以惰性有机碳的形式在海洋中长期储存目前较少了解。海洋中藻菌关系的深入研究,不仅是开发“以菌治藻”赤潮治理模式、应用于富营养水体修复、微生物新资源和新型天然产物挖掘等的重要理论基础,而且有助于深入了解藻菌驱动的海洋碳循环过程与机制。
| [1] |
Falkowski PG. The role of phytoplankton photosynthesis in global biogeochemical cycles[J]. Photosynthesis Research, 1994, 39(3): 235-258. DOI:10.1007/BF00014586 |
| [2] |
Fuhrman JA, Azam F. Thymidine incorporation as a measure of heterotrophic bacterioplankton production in marine surface waters: evaluation and field results[J]. Marine Biology, 1982, 66(2): 109-120. DOI:10.1007/BF00397184 |
| [3] |
Falkowski PG, Katz ME, Knoll AH, et al. The evolution of modern eukaryotic phytoplankton[J]. Science, 2004, 305(5682): 354-360. DOI:10.1126/science.1095964 |
| [4] |
Bell W, Mitchell R. Chemotactic and growth responses of marine bacteria to algal extracellular products[J]. The Biological Bulletin, 1972, 143(2): 265-277. DOI:10.2307/1540052 |
| [5] |
Seymour JR, Amin SA, Raina JB, et al. Zooming in on the phycosphere: the ecological interface for phytoplankton-bacteria relationships[J]. Nature Microbiology, 2017, 2: 17065. DOI:10.1038/nmicrobiol.2017.65 |
| [6] |
Labeeuw L, Khey J, Bramucci AR, et al. Indole-3-acetic acid is produced by Emiliania huxleyi coccolith-bearing cells and triggers a physiological response in bald cells[J]. Frontiers in Microbiology, 2016, 7: 828. |
| [7] |
Locsei JT, Pedley TJ. Bacterial tracking of motile algae assisted by algal cell's vorticity field[J]. Microbial Ecology, 2009, 58(1): 63-74. DOI:10.1007/s00248-008-9468-6 |
| [8] |
Sule P, Belas R. A novel inducer of Roseobacter motility is also a disruptor of algal symbiosis[J]. Journal of Bacteriology, 2013, 195(4): 637-646. DOI:10.1128/JB.01777-12 |
| [9] |
Christie-Oleza JA, Sousoni D, Lloyd M, et al. Nutrient recycling facilitates long-term stability of marine microbial phototroph–heterotroph interactions[J]. Nature Microbiology, 2017, 2: 17100. DOI:10.1038/nmicrobiol.2017.100 |
| [10] |
Amin SA, Parker MS, Armbrust EV. Interactions between diatoms and bacteria[J]. Microbiology and Molecular Biology Reviews, 2012, 76(3): 667-684. DOI:10.1128/MMBR.00007-12 |
| [11] |
Murray RE, Cooksey KE, Priscu JC. Stimulation of bacterial DNA synthesis by algal exudates in attached algal-bacterial consortia[J]. Applied and Environmental Microbiology, 1986, 52(5): 1177-1182. |
| [12] |
Findlay JA, Patil AD. Antibacterial constituents of the diatom Navicula delognei[J]. Journal of Natural Products, 1984, 47(5): 815-818. DOI:10.1021/np50035a010 |
| [13] |
Vidoudez C, Nejstgaard JC, Jakobsen HH, et al. Dynamics of dissolved and particulate polyunsaturated aldehydes in mesocosms inoculated with different densities of the diatom Skeletonema marinoi[J]. Marine Drugs, 2011, 9(3): 345-358. DOI:10.3390/md9030345 |
| [14] |
Paul C, Pohnert G. Production and role of volatile halogenated compounds from marine algae[J]. Natural Product Reports, 2011, 28(2): 186-195. DOI:10.1039/C0NP00043D |
| [15] |
Natrah FMI, Kenmegne MM, Wiyoto W, et al. Effects of micro-algae commonly used in aquaculture on acyl-homoserine lactone quorum sensing[J]. Aquaculture, 2011, 317(1/4): 53-57. |
| [16] |
Cruz-López R, Maske H. The vitamin B1 and B12 required by the marine dinoflagellate Lingulodinium polyedrum can be provided by its associated bacterial community in culture[J]. Frontiers in Microbiology, 2016, 7: 560. |
| [17] |
Amin SA, Green DH, Hart MC, et al. Photolysis of iron-siderophore chelates promotes bacterial-algal mutualism[J]. Proceedings of the National Academy of Sciences of the United States of America, 2009, 106(40): 17071-17076. DOI:10.1073/pnas.0905512106 |
| [18] |
Jiang L, Yang LY, Xiao L, et al. Quantitative studies on phosphorus transference occuring between Microcystis aeruginosa and its attached bacterium (Pseudomonas sp.)[J]. Hydrobiologia, 2007, 281(1): 161-165. |
| [19] |
De-Bashan LE, Antoun H, Bashan Y. Involvement of indole-3-acetic acid produced by the growth-promoting bacterium Azospirillum spp. in promoting growth of Chlorella vulgaris[J]. Journal of Phycology, 2008, 44(4): 938-947. DOI:10.1111/jpy.2008.44.issue-4 |
| [20] |
Wichard T. Exploring bacteria-induced growth and morphogenesis in the green macroalga order Ulvales (Chlorophyta)[J]. Frontiers in Plant Science, 2015, 6: 86. |
| [21] |
Windler M, Bova D, Kryvenda A, et al. Influence of bacteria on cell size development and morphology of cultivated diatoms[J]. Phycological Research, 2014, 62(4): 269-281. DOI:10.1111/pre.2014.62.issue-4 |
| [22] |
Imai I, Ishida Y, Hata Y. Killing of marine-phytoplankton by a gliding bacterium Cytophaga sp, isolated from the coastal sea of Japan[J]. Marine Biology, 1993, 116(4): 527-532. DOI:10.1007/BF00355470 |
| [23] |
van Tol HM, Amin SA, Armbrust EV. Ubiquitous marine bacterium inhibits diatom cell division[J]. The ISME Journal, 2017, 11: 31-42. DOI:10.1038/ismej.2016.112 |
| [24] |
Mayali X, Franks PJS, Tanaka Y, et al. Bacteria-induced motility reduction in Lingulodinium polyedrum (Dinophyceae)[J]. Journal of Phycology, 2008, 44(4): 923-928. DOI:10.1111/jpy.2008.44.issue-4 |
| [25] |
Twigg MS, Tait K, Williams P, et al. Interference with the germination and growth of Ulva zoospores by quorum-sensing molecules from Ulva-associated epiphytic bacteria[J]. Environmental Microbiology, 2014, 16(2): 445-453. DOI:10.1111/1462-2920.12203 |
| [26] |
Mitsutani A, Yamasaki I, Kitaguchi H, et al. Analysis of algicidal proteins of a diatom-lytic marine bacterium Pseudoalteromonas sp. strain A25 by two-dimensional electrophoresis[J]. Phycologia, 2001, 40(3): 286-291. DOI:10.2216/i0031-8884-40-3-286.1 |
| [27] |
Mayers TJ, Bramucci AR, Yakimovich KM, et al. A bacterial pathogen displaying temperature-enhanced virulence of the microalga Emiliania huxleyi[J]. Frontiers in Microbiology, 2016, 7: 892. |
| [28] |
Paul C, Pohnert G. Interactions of the algicidal bacterium Kordia algicida with diatoms: regulated protease excretion for specific algal lysis[J]. PLoS One, 2011, 6(6): e21032. DOI:10.1371/journal.pone.0021032 |
| [29] |
Yoshikawa K, Adachi K, Nishijima M, et al. β-cyanoalanine production by marine bacteria on cyanide-free medium and its specific inhibitory activity toward cyanobacteria[J]. Applied and Environmental Microbiology, 2000, 66(2): 718-722. DOI:10.1128/AEM.66.2.718-722.2000 |
| [30] |
Yamamoto Y, Kouchiwa T, Hodoki Y, et al. Distribution and identification of actinomycetes lysing cyanobacteria in a eutrophic lake[J]. Journal of Applied Phycology, 1998, 10(4): 391-397. DOI:10.1023/A:1008077414808 |
| [31] |
Zhang BH, Chen W, Li HQ, et al. L-valine, an antialgal amino acid from Streptomyces jiujiangensis JXJ 0074T[J]. Applied Microbiology and Biotechnology, 2016, 100(10): 4627-4636. DOI:10.1007/s00253-015-7150-8 |
| [32] |
Li D, Zhang HJ, Fu LJ, et al. A novel algicide: evidence of the effect of a fatty acid compound from the marine bacterium, Vibrio sp. BS02 on the harmful dinoflagellate, Alexandrium tamarense[J]. PLoS One, 2014, 9(3): e91201. DOI:10.1371/journal.pone.0091201 |
| [33] |
Hibayashi R, Imamura N. Action mechanism of a selective anti-cyanobacterial compound, argimicin A[J]. The Journal of Antibiotics, 2003, 56(2): 154-159. DOI:10.7164/antibiotics.56.154 |
| [34] |
Banin E, Khare SK, Naider F, et al. Proline-rich peptide from the coral pathogen Vibrio shiloi that inhibits photosynthesis of zooxanthellae[J]. Applied and Environmental Microbiology, 2001, 67(4): 1536-1541. DOI:10.1128/AEM.67.4.1536-1541.2001 |
| [35] |
Lin SQ, Geng MX, Liu XL, et al. On the control of Microcystis aeruginosa and Synechococccus species using an algicidal bacterium, Stenotrophomonas F6, and its algicidal compounds cyclo-(Gly-Pro) and hydroquinone[J]. Journal of Applied Phycology, 2016, 28(1): 345-355. DOI:10.1007/s10811-015-0549-x |
| [36] |
Tan S, Hu XL, Yin PH, et al. Photosynthetic inhibition and oxidative stress to the toxic Phaeocystis globosa caused by a diketopiperazine isolated from products of algicidal bacterium metabolism[J]. Journal of Microbiology, 2016, 54(5): 364-375. DOI:10.1007/s12275-016-6012-0 |
| [37] |
Lee SO, Kato J, Takiguchi N, et al. Involvement of an extracellular protease in algicidal activity of the marine bacterium Pseudoalteromonas sp. strain A28[J]. Applied and Environmental Microbiology, 2000, 66(10): 4334-4339. DOI:10.1128/AEM.66.10.4334-4339.2000 |
| [38] |
Li Y, Lei XQ, Zhu H, et al. Chitinase producing bacteria with direct algicidal activity on marine diatoms[J]. Scientific Reports, 2016, 6: 21984. DOI:10.1038/srep21984 |
| [39] |
Berger PS, Rho J, Gunner H. Bacterial suppression of chlorella by hydroxylamine production[J]. Water Research, 1979, 13(3): 267-273. DOI:10.1016/0043-1354(79)90205-7 |
| [40] |
An XL, Zhang BZ, Zhang HJ, et al. Discovery of an algicidal compound from Brevibacterium sp. BS01 and its effect on a harmful algal bloom-causing species, Alexandrium tamarense[J]. Frontiers in Microbiology, 2015, 6: 1235. |
| [41] |
Yang K, Chen QL, Zhang DY, et al. The algicidal mechanism of prodigiosin from Hahella sp. KA22 against Microcystis aeruginosa[J]. Scientific Reports, 2017, 7: 7750. DOI:10.1038/s41598-017-08132-5 |
| [42] |
Kodani S, Imoto A, Mitsutani A, et al. Isolation and identification of the antialgal compound, harmane (1-methyl-β-carboline), produced by the algicidal bacterium, Pseudomonas sp. K44-1[J]. Journal of Applied Phycology, 2002, 14(2): 109-114. DOI:10.1023/A:1019533414018 |
| [43] |
Jeong SY, Ishida K, Ito Y, et al. Bacillamide, a novel algicide from the marine bacterium, Bacillus sp. SY-1, against the harmful dinoflagellate, Cochlodinium polykrikoides[J]. Tetrahedron Letters, 2003, 44(43): 8005-8007. DOI:10.1016/j.tetlet.2003.08.115 |
| [44] |
Wigglesworlth-Cooksey B, Cooksey KE, Long R. Antibiotic from the marine environment with antimicrobial fouling activity[J]. Environmental Toxicology, 2007, 22(3): 275-280. DOI:10.1002/(ISSN)1522-7278 |
| [45] |
Guo XL, Liu XL, Wu LS, et al. The algicidal activity of Aeromonas sp. strain GLY-2107 against bloom-forming Microcystis aeruginosa is regulated by N-acyl homoserine lactone-mediated quorum sensing[J]. Environmental Microbiolog, 2016, 18(11): 3867-3883. DOI:10.1111/1462-2920.13346 |
| [46] |
Kim YS, Son HJ, Jeong SY. Isolation of an algicide from a marine bacterium and its effects against the toxic dinoflagellate Alexandrium catenella and other harmful algal bloom species[J]. Journal of Microbiology, 2015, 53(8): 511-517. DOI:10.1007/s12275-015-5303-1 |
| [47] |
Li Z, Lin S, Liu X, et al. A freshwater bacterial strain, Shewanella sp Lzh-2, isolated from Lake Taihu and its two algicidal active substances, hexahydropyrrolo[1, 2-a] pyrazine-1, 4-dione and 2, 3-indolinedione[J]. Applied Microbiology and Biotechnology, 2014, 98(10): 4737-4748. DOI:10.1007/s00253-014-5602-1 |
| [48] |
Sakata T, Yoshikawa T, Nishitarumizu S. Algicidal activity and identification of an algicidal substance produced by marine Pseudomonas sp. C55a-2[J]. Fisheries Science, 2011, 77(3): 397-402. DOI:10.1007/s12562-011-0345-8 |
| [49] |
Wang MH, Peng P, Liu YM, et al. Algicidal activity of a dibenzofuran-degrader Rhodococcus sp.[J]. Journal of Microbiology and Biotechnology, 2013, 23(2): 260-266. DOI:10.4014/jmb |
| [50] |
Cho JY. Algicidal activity of marine Alteromonas sp. KNS-16 and isolation of active compounds[J]. Bioscience Biotechnology and Biochemistry, 2012, 76(8): 1452-1458. DOI:10.1271/bbb.120102 |
| [51] |
Harvey EL, Deering RW, Rowley DC, et al. A bacterial quorum-sensing precursor induces mortality in the marine Coccolithophore, Emiliania huxleyi[J]. Frontiers in Microbiology, 2016, 7: 59. |
| [52] |
Ziesche L, Bruns H, Dogs M, et al. Homoserine lactones, methyl oligohydroxybutyrates, and other extracellular metabolites of macroalgae-associated bacteria of the Roseobacter clade: identification and functions[J]. ChemBioChem, 2015, 16(14): 2094-2107. DOI:10.1002/cbic.201500189 |
| [53] |
Dakhama A, De la Noüe J, Lavoie MC. Isolation and idetification of antialgal substances produced by Pseudomonas aeruginosa[J]. Journal of Applied Phycology, 1993, 5(3): 297-306. DOI:10.1007/BF02186232 |
| [54] |
Li Y, Zhu H, Lei XQ, et al. The first evidence of deinoxanthin from Deinococcus sp. Y35 with strong algicidal effect on the toxic dinoflagellate Alexandrium tamarense[J]. Journal of Hazardous Materials, 2015, 290: 87-95. DOI:10.1016/j.jhazmat.2015.02.070 |
| [55] |
Luo JF, Wang Y, Tang SS, et al. Isolation and identification of algicidal compound from Streptomyces and algicidal mechanism to Microcystis aeruginosa[J]. PLoS One, 2013, 8(10): e76444. DOI:10.1371/journal.pone.0076444 |
| [56] |
Lu XH, Zhou B, Xu LL, et al. A marine algicidal Thalassospira and its active substance against the harmful algal bloom species Karenia mikimotoi[J]. Applied Microbiology and Biotechnology, 2016, 100(11): 5131-5139. DOI:10.1007/s00253-016-7352-8 |
| [57] |
Wang BX, Yang XR, Lu JL, et al. A marine bacterium producing protein with algicidal activity against Alexandrium tamarense[J]. Harmful Algae, 2012, 13: 83-88. DOI:10.1016/j.hal.2011.10.006 |
| [58] |
Sonnenschein EC, Syit DA, Grossart HP, et al. Chemotaxis of Marinobacter adhaerens and its impact on attachment to the diatom Thalassiosira weissflogii[J]. Applied and Environmental Microbiology, 2012, 78(19): 6900-6907. DOI:10.1128/AEM.01790-12 |
| [59] |
Malfatti F, Azam F. Atomic force microscopy reveals microscale networks and possible symbioses among pelagic marine bacteria[J]. Aquatic Microbial Ecology, 2009, 58(1): 1-14. DOI:10.1007/s00248-008-9442-3 |
| [60] |
Powell RJ, Hill RT. Rapid aggregation of biofuel-producing algae by the bacterium Bacillus sp. strain RP1137[J]. Applied and Environmental Microbiology, 2013, 79(19): 6093-6101. DOI:10.1128/AEM.01496-13 |
| [61] |
Guidi L, Chaffron S, Bittner L, et al. Plankton networks driving carbon export in the oligotrophic ocean[J]. Nature, 2016, 532(7600): 465-470. DOI:10.1038/nature16942 |
| [62] |
Wang H, Tomasch J, Michael V, et al. Identification of genetic modules mediating the Jekyll and Hyde interaction of Dinoroseobacter shibae with the dinoflagellate Prorocentrum minimum[J]. Frontiers in Microbiology, 2015, 6: 1262. |
| [63] |
Seyedsayamdost MR, Wang R, Kolter R, et al. Hybrid biosynthesis of roseobacticides from algal and bacterial precursor molecules[J]. Journal of the American Chemical Society, 2014, 136(43): 15150-15153. DOI:10.1021/ja508782y |
| [64] |
Segev E, Wyche TP, Kim KH, et al. Dynamic metabolic exchange governs a marine algal-bacterial interaction[J]. eLife, 2016, 5: e17473. DOI:10.7554/eLife.17473 |
| [65] |
Li Y, Yang CY, Zheng TL. Bacterial survival modes and community characteristics in natural environment[J]. Chinese Journal of Applied and Environmental Biology, 2013, 19(4): 553-560. (in Chinese) 李祎, 杨彩云, 郑天凌. 自然环境中细菌的生存方式及其群落特征[J]. 应用与环境生物学报, 2013, 19(4): 553-560. |
| [66] |
Zhou J, Lin GH, Cai ZH. Roles of microbes in matter cycles in phycosphere niche[J]. Chinese Journal of Applied Ecology, 2016, 27(8): 2708-2716. (in Chinese) 周进, 林光辉, 蔡中华. 微生物在藻际环境中的物质循环作用[J]. 应用生态学报, 2016, 27(8): 2708-2716. |
| [67] |
Krohn-Molt I, Alawi M, Förstner KU, et al. Insights into microalga and bacteria interactions of selected phycosphere biofilms using metagenomic, transcriptomic, and proteomic approaches[J]. Frontiers in Microbiology, 2017, 8: 1941. DOI:10.3389/fmicb.2017.01941 |
| [68] |
Guannel ML, Horner-Devine MC, Rocap G. Bacterial community composition differs with species and toxigenicity of the diatom Pseudo-nitzschia[J]. Aquatic Microbial Ecology, 2011, 64(2): 117-133. DOI:10.3354/ame01513 |
| [69] |
Teeling H, Fuchs BM, Bennke CM, et al. Recurring patterns in bacterioplankton dynamics during coastal spring algae blooms[J]. eLife, 2016, 5: e11888. DOI:10.7554/eLife.11888 |
| [70] |
Sison-Mangus MP, Jiang S, Kudela RM, et al. Phytoplankton-associated bacterial community composition and succession during toxic diatom bloom and non-bloom events[J]. Frontiers in Microbiology, 2016, 7: 1433. |
| [71] |
Zheng Q, Wang Y, Xie R, et al. Dynamics of heterotrophic bacterial assemblages within Synechococcus cultures[J]. Applied and Environmental Microbiology, 2017, 84(3): e01517-17. |
| [72] |
Burke C, Thomas T, Lewis M, et al. Composition, uniqueness and variability of the epiphytic bacterial community of the green alga Ulva australis[J]. The ISME Journal, 2011, 5(4): 590-600. DOI:10.1038/ismej.2010.164 |
| [73] |
Burke C, Steinberg P, Rusch D, et al. Bacterial community assembly based on functional genes rather than species[J]. Proceedings of the National Academy of Sciences of the United States of America, 2011, 108(34): 14288-14293. DOI:10.1073/pnas.1101591108 |
| [74] |
Sarmento H, Gasol JM. Use of phytoplankton-derived dissolved organic carbon by different types of bacterioplankton[J]. Environmental Microbiology, 2012, 14(9): 2348-2360. DOI:10.1111/emi.2012.14.issue-9 |
| [75] |
Jiao NZ, Herndl GJ, Hansell DA, et al. Microbial production of recalcitrant dissolved organic matter: long-term carbon storage in the global ocean[J]. Nature Reviews Microbiology, 2010, 8(8): 593-599. DOI:10.1038/nrmicro2386 |
| [76] |
Zehr JP. How single cells work together[J]. Science, 2015, 349(6253): 1163-1164. DOI:10.1126/science.aac9752 |
| [77] |
Lovejoy C, Bowman JP, Hallegraeff GM. Algicidal effects of a novel marine Pseudoalteromonas isolate (class Proteobacteria, gamma subdivision) on harmful algal bloom species of the genera Chattonella, Gymnodinium, and Heterosigma[J]. Applied and Environmental Microbiology, 1998, 64(8): 2806-2813. |
| [78] |
Oswald WJ, Gotaas HB. Photosynthesis in sewage treatment[J]. Transactions of American Society of Civil Engineers, 1957, 122(1): 73-105. |
| [79] |
Subashchandrabose SR, Ramakrishnan B, Megharaj M, et al. Consortia of cyanobacteria/microalgae and bacteria: biotechnological potential[J]. Biotechnology Advances, 2011, 29(6): 896-907. DOI:10.1016/j.biotechadv.2011.07.009 |
| [80] |
Ramanan R, Kim BH, Cho DH, et al. Algae-bacteria interactions: evolution, ecology and emerging applications[J]. Biotechnology Advances, 2016, 34(1): 14-29. DOI:10.1016/j.biotechadv.2015.12.003 |
| [81] |
Liang YT, Zhang YY, Wang NN, et al. Estimating primary production of picophytoplankton using the carbon-based ocean productivity model: a preliminary study[J]. Frontiers in Microbiology, 2017, 8: 1926. DOI:10.3389/fmicb.2017.01926 |
| [82] |
Zhao Z, Gonsior M, Luek J, et al. Picocyanobacteria and deep-ocean fluorescent dissolved organic matter share similar optical properties[J]. Nature Communications, 2017, 8: 15284. DOI:10.1038/ncomms15284 |
| [83] |
Zhang YY, Zhang JH, Liang YT, et al. Carbon sequestration processes and mechanisms in coastal mariculture environments in China[J]. Science China Earth Sciences, 2017, 47(12): 2097-2107. (in Chinese) 张永雨, 张继红, 梁彦韬, 等. 中国近海养殖环境碳汇形成过程与机制[J]. 中国科学:地球科学, 2017, 47(12): 1414-1424. |
| [84] |
Grossart HP, Czub G, Simon M. Algae-bacteria interactions and their effects on aggregation and organic matter flux in the sea[J]. Environmental Microbiology, 2006, 8(6): 1074-1084. DOI:10.1111/emi.2006.8.issue-6 |
| [85] |
Koch BP, Kattner G, Witt M, et al. Molecular insights into the microbial formation of marine dissolved organic matter: recalcitrant or labile?[J]. Biogeosciences Discussions, 2014, 11(2): 3065-3111. DOI:10.5194/bgd-11-3065-2014 |
| [86] |
Zhang YY, Huang CX, Yang J, et al. Interactions between marine microorganisms and their phages[J]. Chinese Science Bulletin, 2011, 56(17): 1770-1777. (in Chinese) 张永雨, 黄春晓, 杨军, 等. 海洋微生物与噬菌体间的相互关系[J]. 科学通报, 2011, 56(14): 1071-1079. |
| [87] |
Tang LL, Zhang ZH, Zhou C, et al. Roseicyclus marinus sp. nov., isolated from a Synechococcus culture, and emended description of the genus Roseicyclus[J]. International Journal of Systematic and Evolutionary Microbiology, 2018, 68: 1781-1786. DOI:10.1099/ijsem.0.002752 |
| [88] |
Mou S, Zhang Y, Li G, et al. Effects of elevated CO2 and nitrogen supply on the growth and photosynthetic physiology of a marine cyanobacterium, Synechococcus sp. PCC7002[J]. Journal of Applied Phycology, 2017, 29(4): 1755-1763. DOI:10.1007/s10811-017-1089-3 |
| [89] |
Zhang Y, Zhao MX, Cui Q, et al. Processes of coastal ecosystem carbon sequestration and approaches for increasing carbon sink[J]. Science China Earth Sciences, 2017, 47(5): 809-820. (in Chinese) 张瑶, 赵美训, 崔球, 等. 近海生态系统碳汇过程、调控机制及增汇模式[J]. 中国科学:地球科学, 2017, 47(4): 438-449. |
 2018, Vol. 45
2018, Vol. 45