扩展功能
文章信息
- 王明鹏, 王学江, 陈蕾
- WANG Ming-Peng, WANG Xue-Jiang, CHEN Lei
- 褐藻表面可培养褐藻酸降解菌的原位筛选
- In situ screening of cultivable alginate-degrading microorganism on surface of brown seaweed
- 微生物学通报, 2018, 45(9): 1853-1860
- Microbiology China, 2018, 45(9): 1853-1860
- DOI: 10.13344/j.microbiol.china.180152
-
文章历史
- 收稿日期: 2018-02-27
- 接受日期: 2018-06-12
- 网络首发日期(www.cnki.net): 2018-06-20
2. 中国科学院烟台海岸带研究所 山东 烟台 264003
2. Yantai Institute of Costal Zone Research, Chinese Academy of Sciences, Yantai, Shandong 264003, China
褐藻酸是褐藻细胞壁的主要组成成分,含量占藻体干重的20%-40%[1]。作为一种天然大分子多糖,褐藻酸可被加工成稳定剂、增稠剂和螯合剂广泛应用于食品、化妆品及制药工业领域。褐藻酸是由2种单体成分——甘露糖醛酸(M)和古罗糖醛酸(G)以1, 4-糖苷键连接而成的线性聚合物[2-3]。同纤维素、淀粉等天然多糖可以被酶降解一样,褐藻酸也可被褐藻酸裂解酶降解成为具有特殊生物学功能的小分子寡糖。据报道,褐藻寡糖具有抗菌、抗氧化、免疫调节、食品保鲜以及促进植物生长和抗逆等多种生物学功能,是一种应用价值极高、应用前景广阔的功能寡糖[4-11]。目前,我国的褐藻酸产业相对成熟,已为诸多领域提供了重要的化工原料,而作为褐藻酸的高值化产品——褐藻寡糖始终没能工业化生产,这与现有的褐藻酸裂解酶种类少、活力低、产量小有着密不可分的关系。因此,探寻多元化、高活性、高效率的褐藻酸裂解酶是非常有意义的工作。
对于生产多功能褐藻寡糖来说,褐藻酸裂解酶是非常重要且必不可少的工具酶[12]。尽管酶的底物特异性和酶切方式不同,所有的褐藻酸裂解酶均是通过糖苷键的β消除反应来降解褐藻酸。根据底物特异性,褐藻酸裂解酶分为聚甘露糖醛酸裂解酶和聚古罗糖醛酸裂解酶;而根据酶切方式的不同,褐藻酸裂解酶则分为外切酶和内切酶,尤其内切酶是用来生产不同聚合度褐藻寡糖的常用酶[13-16]。目前,研究者们已经筛选及鉴定出上百种褐藻酸裂解酶,分别来自海藻、无脊椎动物及各种微生物。其中,各种产酶微生物是褐藻酸裂解酶最丰富的来源,根据自然界物种互相作用影响、共同进化的规律,海藻原位生长的微生物理论上可为褐藻酸裂解酶提供最为可靠和高效的筛选来源。
为了尽可能多地筛选产酶菌株,提高筛选效率,良好的筛选方法必不可少。目前,产褐藻酸裂解酶菌株的筛选标准主要是观察其在褐藻酸为唯一碳源培养基上的生长情况以及产透明圈的大小。常用来产透明圈的试剂有氯化十六烷吡啶(Cetylpyridinium chloride,CPC)和氯化钙(CaCl2),但培养基中的琼脂成分往往会干扰试剂发挥作用,影响透明圈的辨识。此外,CPC和CaCl2的反应时间较长,最少需要30 min,并且形成的透明圈边界不够清晰明确,有时较难辨别。最近,Sawant等[17]报道了一种利用革兰氏碘液在褐藻酸平板上形成透明圈来筛选产酶菌株的方法,该方法的原理是碘液与大分子褐藻酸结合形成黑褐色复合物,但不与降解后的褐藻酸反应,因此在产酶菌株周围形成透明圈。用革兰氏碘液代替CPC和CaCl2的优势明显,首先反应时间短,倾倒革兰氏碘液2-3 min后即可形成透明圈,并且透明圈边缘清晰,与培养基背景对比鲜明,辨识度高。
本文采用了革兰氏碘液显色的方法筛选褐藻表面的产酶菌株,结合牛津杯法对各菌株的产酶性质进行初步的量化及比较,可初步判断潜在的优势产酶菌株,为下一步研究缩小范围、提高筛选效率。
1 材料与方法 1.1 主要试剂和仪器及培养基基因组DNA提取试剂盒、2×EasyPfu PCR SuperMix、蛋白定量试剂盒,北京全式金生物技术有限公司;革兰氏染液,南京建成科技有限公司;革兰氏碘液,Sigma公司。PCR仪,Bioneer公司;台式高速冷冻离心机,青岛科易仪器有限公司;pH值测定仪,上海梅特勒-托利多仪器有限公司。
ALG培养基(g/L):褐藻酸钠5.0,硫酸铵5.0,磷酸氢二钾2.0,硫酸镁1.0,琼脂20.0,pH 7.2-7.4。
1.2 样品采集及菌株筛选新鲜铜藻样品采自山东半岛荣成市,运回实验室后4 ℃低温保存。用灭过菌的剪刀将铜藻样品剪成碎片,取10 g铜藻碎片置于无菌培养皿中,并加入10 mL无菌水,30 ℃培养至铜藻表面有菌落长出。将铜藻表面菌落刮下并用无菌水进行10倍梯度稀释。取10-4、10-5和10-6三个浓度涂布褐藻酸为唯一碳源的平板(ALG培养基),置于30 ℃培养3-4 d后,挑选形态不同的菌落进行编号并转移至新的ALG平板上。新平板继续在30 ℃培养形成可见菌落,而原始平板用革兰氏碘液进行显色反应,周围能够形成透明圈的菌落被认为是产褐藻酸裂解酶的菌落,挑选这些菌落并记录编号,用于后续研究。
1.3 菌种鉴定及系统发育学分析用基因组DNA提取试剂盒提取上述挑选出的产酶菌株基因组DNA。以基因组DNA为模板,用27F (5′-AGAGTTTGATCCTGGCTCAG-3′)和1492R (5′-TACGGTTACCTTGTTACGACTT-3′)为引物PCR扩增各菌株的16S rRNA基因序列。PCR扩增体系(50 μL):2×EasyPfu PCR SuperMix 25 μL,上、下游引物(10 μmol/L)各0.4 μL,样品基因组DNA模板10 ng,超纯水补足至50 μL。PCR反应条件:95 ℃ 5 min;95 ℃ 45 s,55 ℃ 45 s,72 ℃ 90 s,共30个循环;72 ℃ 10 min。扩增的基因片段送至生工生物工程(上海)股份有限公司测序。测序数据经BLAST在线工具进行序列比对,并用邻接法(Neighbor-Joining,NJ)构建系统发育树,分析各菌株的进化关系,确定菌株种类。产酶菌株的16S rRNA基因序列提交至GenBank数据库,获得的登录号为MH000672-MH000678。
菌种形态学鉴定包括革兰氏染色、细胞形态、运动性及芽孢观察等,均按照《伯杰氏细菌鉴定手册》[18]的操作规程进行。菌种的碳源利用和酶反应特性则根据表型芯片系统进行鉴定分析。
1.4 酶活比较产酶菌株经活化后接种至100 mL液体ALG培养基中,30 ℃、200 r/min培养24 h。发酵液经12 000 r/min离心5 min后保留上清液备用。上清液蛋白含量采用蛋白定量试剂盒进行定量检测。取200 μL上清液置于ALG平板表面的无菌牛津杯中,将平板置于30 ℃培养24 h,革兰氏碘液染色。测量透明圈的内、外直径并计算透明圈面积来表征各菌株酶活大小。酶活力单位定义为:1 min透明圈面积增加0.1 mm2为1个酶活力单位(Enzyme unit,EU),每毫克蛋白质的酶活力单位称为酶的比活(EU/mg)。
2 结果与分析 2.1 褐藻酸降解菌株的筛选及鉴定 2.1.1 褐藻酸降解菌株的筛选从铜藻样品中共筛选得到81株菌,其中28个菌落能够产生如图 1中所示的透明圈。随后对28株菌进行分子鉴定,经16S rRNA基因测序,去除序列相同的重复菌株后,共得到7株产褐藻酸裂合酶的菌。如图 1B所示,7株产酶菌分属5个不同的属:芽孢杆菌属(Bacillus)的产酶菌株最多,有3株;节杆菌属(Arthrobacter)、德库菌属(Desemzia)、短杆菌属(Brachybacterium)和链孢子囊菌属(Streptosporangium)各1株。
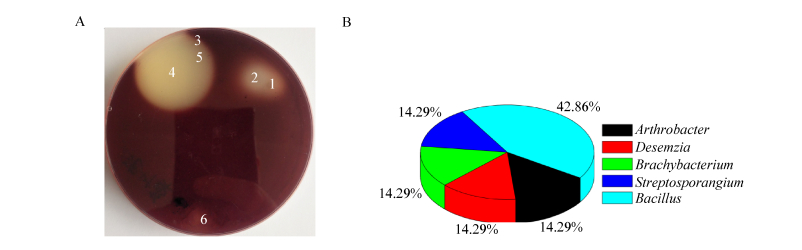
|
| 图 1 产酶菌株筛选结果 Figure 1 The screening results of the alginate lyase-excreting bacteria strains 注:A:筛菌平板实物照片;B:产酶菌株种属占比图. Note: A: One photo of screening plate; B: Proportion of the alginate lyase-excreting bacteria strains. |
|
|
对各菌株16S rRNA基因序列进行系统发育学分析,结果如图 2所示,与3株芽孢杆菌亲缘关系最近的菌株分别为特基拉芽孢杆菌(Bacillus tequilensis)、甲基营养型芽孢杆菌(Bacillus methylotrophicus)和地衣芽孢杆菌(Bacillus licheniformis),其余亲缘关系最近的菌株分别是未定德库菌(Desemzia incerta)、烟草节杆菌(Arthrobacter nicotianae)、人参短杆菌(Brachybacterium ginsengisoli)以及胜利链霉菌(Streptosporangium shengliensis)。

|
| 图 2 基于16S rRNA基因序列的产酶菌株系统发育树 Figure 2 Neighbor-joining phylogenetic tree of the alginate lyase-excreting bacteria based on 16S rRNA genes sequence 注:括号内为GenBank登录号;分支点数字为1 000次重复后获得的置信值;标尺表示100个核苷酸中有2个被替换. Note: Numbers in parentheses are GenBank accession numbers; Numbers at the branches are bootstrap values obtained after 1 000 replicates; The scale bar represent 2 substitutions per 100 nucleotide positions. |
|
|
对7株菌进行了革兰氏染色形态观察和生理生化特性验证,作为辅助鉴定结果。结果如图 3所示,菌体多为杆状或短杆状,均为革兰氏阳性细菌,各菌体形态及生化特性(表 1)与文献报道的其亲缘关系最近的菌株基本一致[19-25]。
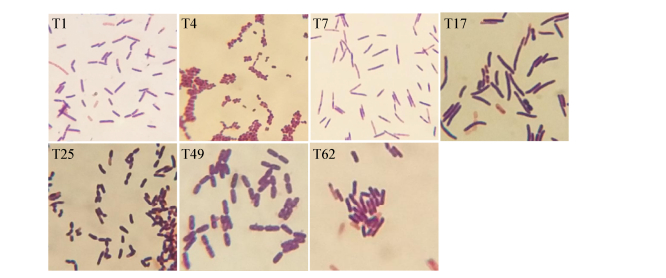
|
| 图 3 产酶菌株形态观察 Figure 3 The morphology of the alginate lyase-excreting bacteria strains |
|
|
| 生理生化指标 Characteristics |
T1 | T4 | T7 | T17 | T25 | T49 | T62 |
| 菌落颜色 Colony colour |
黄色 Yellow |
白色 White |
白色 White |
黄色 Yellow |
透明 Opaque |
白色 White |
透明 Opaque |
| 需氧性 Aerobic |
+ | + | +/- | + | + | + | + |
| 运动性 Motile |
- | - | + | + | + | + | - |
| 芽孢 Spore-forming |
- | - | + | + | - | + | - |
| 脲酶 Urease |
- | - | + | - | - | - | - |
| 硝酸盐还原性 Nitrate reduction |
- | - | + | + | - | + | + |
| 葡萄糖 D-Glucose |
+ | + | + | + | + | + | + |
| 果糖 D-Fructose |
- | + | + | + | + | + | + |
| 阿拉伯糖 L-Arabinose |
+ | - | + | + | - | + | + |
| 半乳糖 Galactose |
+ | + | - | + | - | - | + |
| 甘露糖 D-Mannose |
- | + | + | + | + | + | + |
| 麦芽糖 D-Maltose |
+ | + | + | + | + | + | + |
| 鼠李糖 Rhamnose |
- | + | + | + | - | - | - |
| 淀粉 Starch |
- | + | + | + | - | + | + |
| 山梨醇 Sorbitol |
+ | - | + | + | - | + | - |
| 甘露醇 D-Mannitol |
- | - | + | + | - | + | - |
| 注:+:阳性;-:阴性. Note: +: Positive; -: Negative. | |||||||
结合革兰氏碘液显色反应及牛津杯法,对7株产酶菌株的酶活做了初步的检测及比较。如图 4所示,各菌株的透明圈直径大小不一,除了T4、T7菌株透明圈较小,直径分别为1.2 cm和1.5 cm之外,其他菌株的透明圈直径均在2 cm左右(表 2)。与蛋白含量结果整合计算后,如图 4B所示,T1菌株的酶比活力最大,其次为T17、T49、T25和T62菌株,相对酶活为T1菌株的54%-81%,而T4和T7菌株酶比活力较小,相对酶活为T1菌株的21%-37%。
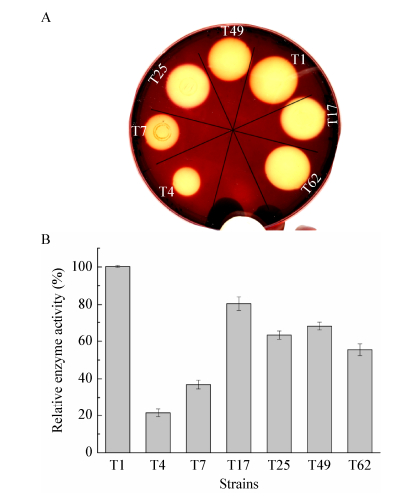
|
| 图 4 产酶菌株的酶活定量分析 Figure 4 Quantification of alginate lyase activity 注:A:革兰氏碘液染色显示产酶透明圈;B:产酶菌株的酶活定量检测及比较分析. Note:A: Gram's iodine method showing the clearance zone; B: Quantification of alginate lyase activity. |
|
|
| 菌株编号 Strain ID |
革兰氏染色 Gram staining |
透明圈直径 Diameter of clearance zone (cm) |
| T1 | G+ | 2.1 |
| T4 | G+ | 1.2 |
| T7 | G+ | 1.5 |
| T17 | G+ | 2.0 |
| T25 | G+ | 1.9 |
| T49 | G+ | 1.9 |
| T62 | G+ | 2.0 |
最近有学者报道,在不同褐藻表面筛选到不同种属的褐藻酸降解菌株。Dong等[26]报道了在采自挪威的海带表面筛选到来自5个不同属的21株产酶菌株,其中有11株菌所产的褐藻酸裂解酶最适温度均在20-30 ℃,是罕见的冷适应性酶,具有特殊的应用潜力。Martin等[27]在泡叶藻表面筛选到14株产褐藻酸裂解酶的菌株,其中Marinomonas属的菌株从未被报道有产褐藻酸裂解酶的能力。本研究从铜藻表面筛选到7株产酶菌,其中有3株产酶能力不同的芽孢杆菌。据报道,芽孢杆菌不像弧菌和假单胞菌等对海参有害[28]。因此,来源于芽孢杆菌属的褐藻酸裂解酶可用于处理海藻来制备海参饵料。除芽孢杆菌属外,其他菌属的菌株从未有产褐藻酸裂解酶的报道。我们首次筛选到的产酶新菌种有巨大的应用价值。
菌种筛选向来是一件费时费力的工作,筛选方法的简单易行对于提高筛选效率至关重要。本文采用革兰氏碘液显色反应来指示产酶菌株,并结合牛津杯法,通过测量透明圈的直径及计算透明圈的面积来表征菌株产酶能力的大小。该方法与传统的二硝基水杨酸(DNS)法测量还原糖增加量以及测量褐藻寡糖在235 nm吸光度相比,可实现同一平板、同一时间内多个菌株的比较,具有操作简单、反应快速、处理量大、无需仪器设备等优点。如图 5所示,要比较6株产酶菌在3种因素、5个梯度条件下的酶活力,传统分光光度法至少需要90支试管来进行反应,随后排队上机检测也需耗费大量时间和精力;而平板显色法只需15个平板即可,反应后倾倒碘液,显色反应瞬间完成,十分高效便捷。本研究表明平板显色法省时省力,非常适合大规模筛选产酶菌株,并可对不同菌株产酶条件、酶活力等进行初步比较,结果与传统分光光度法基本一致[29],从而快速确定出最佳的一个或者几个酶活力高的优势菌株进行深入研究。
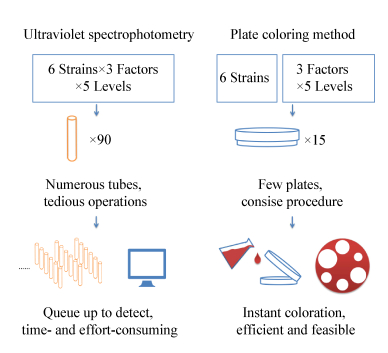
|
| 图 5 褐藻酸裂解酶酶活分析方法的比较 Figure 5 Comparison of different analysis methods of alginate lyase activity |
|
|
本文介绍了一种快速显色筛选褐藻酸降解菌株的方法,为今后筛选产褐藻酸裂解酶菌株工作提供了一个可行的范例。后续通过进一步改进和完善,希望能将该方法推广应用于褐藻酸降解菌的大规模筛选,最终实现产酶菌株酶活力的快速批量比较,提高筛选识别高酶活菌株的效率。
| [1] |
Zhu BW, Yin H. Alginate lyase: review of major sources and classification, properties, structure-function analysis and applications[J]. Bioengineered, 2015, 6(3): 125-131. DOI:10.1080/21655979.2015.1030543 |
| [2] |
Ji MH. Seaweed Chemistry[M]. Beijing: Science Press, 1997. (in Chinese) 纪明侯. 海藻化学[M]. 北京: 科学出版社, 1997. |
| [3] |
Wong TY, Preston LA, Schiller NL. Alginate lyase: review of major sources and enzyme characteristics, structure-function analysis, biological roles, and applications[J]. Annual Review of Microbiology, 2000, 54: 289-340. DOI:10.1146/annurev.micro.54.1.289 |
| [4] |
Khan S, Tøndervik A, Sletta H, et al. Overcoming drug resistance with alginate oligosaccharides able to potentiate the action of selected antibiotics[J]. Antimicrobial Agents and Chemotherapy, 2012, 56(10): 5134-5141. DOI:10.1128/AAC.00525-12 |
| [5] |
Saigusa M, Nishizawa M, Shimizu Y, et al. In vitro and in vivo anti-inflammatory activity of digested peptides derived from salmon myofibrillar protein conjugated with a small quantity of alginate oligosaccharide[J]. Bioscience, Biotechnology, and Biochemistry, 2015, 79(9): 1518-1527. DOI:10.1080/09168451.2015.1031075 |
| [6] |
Yang JH, Bang MA, Jang CH, et al. Alginate oligosaccharide enhances LDL uptake via regulation of LDLR and PCSK9 expression[J]. The Journal of Nutritional Biochemistry, 2015, 26(11): 1393-1400. DOI:10.1016/j.jnutbio.2015.07.009 |
| [7] |
Zhou R, Shi XY, Bi DC, et al. Alginate-derived oligosaccharide inhibits neuroinflammation and promotes microglial phagocytosis of β-Amyloid[J]. Marine Drugs, 2015, 13(9): 5828-5846. DOI:10.3390/md13095828 |
| [8] |
Zhang B, Wu HX, Yang HC, et al. Cryoprotective roles of trehalose and alginate oligosaccharides during frozen storage of peeled shrimp (Litopenaeus vannamei)[J]. Food Chemistry, 2017, 228: 257-264. DOI:10.1016/j.foodchem.2017.01.124 |
| [9] |
Peng Q, Zhang MM, Gao L, et al. Effects of alginate oligosaccharides with different molecular weights and guluronic to mannuronic acid ratios on glyceollin induction and accumulation in soybeans[J]. Journal of Food Science and Technology, 2018, 55(5): 1850-1858. DOI:10.1007/s13197-018-3101-6 |
| [10] |
Zhang YH, Liu H, Yin H, et al. Nitric oxide mediates alginate oligosaccharides-induced root development in wheat (Triticum aestivum L.)[J]. Plant Physiology and Biochemistry, 2013, 71: 49-56. DOI:10.1016/j.plaphy.2013.06.023 |
| [11] |
Zhang YH, Yin H, Zhao XM, et al. The promoting effects of alginate oligosaccharides on root development in Oryza sativa L. mediated by auxin signaling[J]. Carbohydrate Polymers, 2014, 113: 446-454. DOI:10.1016/j.carbpol.2014.06.079 |
| [12] |
Li LY, Guan HS, Jiang XL, et al. Advances in algae tool enzymes: alginate lyases[J]. Chinese Journal of Biotechnology, 2011, 27(6): 838-845. (in Chinese) 李丽妍, 管华诗, 江晓路, 等. 海藻工具酶——褐藻胶裂解酶研究进展[J]. 生物工程学报, 2011, 27(6): 838-845. |
| [13] |
Ertesvåg H. Alginate-modifying enzymes: biological roles and biotechnological uses[J]. Frontiers in Microbiology, 2015, 6: 523. |
| [14] |
Falkeborg M, Cheong LZ, Gianfico C, et al. Alginate oligosaccharides: enzymatic preparation and antioxidant property evaluation[J]. Food Chemistry, 2014, 164: 185-194. DOI:10.1016/j.foodchem.2014.05.053 |
| [15] |
Boucelkha A, Petit E, Elboutachfaiti R, et al. Production of guluronate oligosaccharide of alginate from brown algae Stypocaulon scoparium using an alginate lyase[J]. Journal of Applied Phycology, 2017, 29(1): 509-519. DOI:10.1007/s10811-016-0928-y |
| [16] |
Zhu BW, Hu F, Yuan H, et al. Biochemical characterization and degradation pattern of a unique pH-stable polyM-specific alginate lyase from newly isolated Serratia marcescens NJ-07[J]. Marine Drugs, 2018, 16(4): 129. DOI:10.3390/md16040129 |
| [17] |
Sawant SS, Salunke BK, Kim BS. A rapid, sensitive, simple plate assay for detection of microbial alginate lyase activity[J]. Enzyme and Microbial Technology, 2015, 77: 8-13. DOI:10.1016/j.enzmictec.2015.05.003 |
| [18] |
Bergey DH, Holt JG. Bergey's Manual of Determinative Bacteriology[M]. 9th ed. Baltimore: Williams & Wilkins, 1994.
|
| [19] |
Irlinger F, Bimet F, Delettre J, et al. Arthrobacter bergerei sp. nov. and Arthrobacter arilaitensis sp. nov., novel coryneform species isolated from the surfaces of cheeses[J]. International Journal of Systematic and Evolutionary Microbiology,, 2005, 55(1): 457-462. |
| [20] |
Zhang XH, Liu CX, Zhang YJ, et al. Streptosporangium shengliensis sp. nov., a novel actinomycete isolated from a lake sediment[J]. Antonie van Leeuwenhoek, 2014, 105(1): 237-243. DOI:10.1007/s10482-013-0070-7 |
| [21] |
Dunlap CA, Kwon SW, Rooney AP, et al. Bacillus paralicheniformis sp. nov., isolated from fermented soybean paste[J]. International Journal of Systematic and Evolutionary Microbiology, 2015, 65(10): 3487-3492. DOI:10.1099/ijsem.0.000441 |
| [22] |
Gatson JW, Benz BF, Chandrasekaran C, et al. Bacillus tequilensis sp. nov., isolated from a 2000-year-old Mexican shaft-tomb, is closely related to Bacillus subtilis[J]. International Journal of Systematic and Evolutionary Microbiology, 2006, 56(7): 1475-1484. DOI:10.1099/ijs.0.63946-0 |
| [23] |
Stackebrandt E, Schumann P, Swiderski J, et al. Reclassification of Brevibacterium incertum (Breed 1953) as Desemzia incerta gen[J]. International Journal of Systematic Bacteriology, 1999, 49(Pt 1): 185-188. |
| [24] |
Ruiz-García C, Béjar V, Martínez-Checa F, et al. Bacillus velezensis sp. nov., a surfactant-producing bacterium isolated from the river Vélez in Málaga, southern Spain[J]. International Journal of Systematic and Evolutionary Microbiology, 2005, 55(1): 191-195. |
| [25] |
Hoang VA, Kim YJ, Nguyen NL, et al. Brachybacterium ginsengisoli sp. nov., isolated from soil of a ginseng field[J]. International Journal of Systematic and Evolutionary Microbiology, 2014, 64(9): 3063-3068. |
| [26] |
Dong S, Yang J, Zhang XY, et al. Cultivable alginate lyase-excreting bacteria associated with the Arctic brown alga Laminaria[J]. Marine Drugs, 2012, 10(11): 2481-2491. |
| [27] |
Martin M, Barbeyron T, Martin R, et al. The cultivable surface microbiota of the brown alga Ascophyllum nodosum is enriched in macroalgal-polysaccharide-degrading bacteria[J]. Frontiers in Microbiology, 2015, 6: 1487. |
| [28] |
Wang XT, Wang LL, Che J, et al. Improving the quality of Laminaria japonica-based diet for Apostichopus japonicus through degradation of its algin content with Bacillus amyloliquefaciens WB1[J]. Applied Microbiology and Biotechnology, 2015, 99(14): 5843-5853. DOI:10.1007/s00253-015-6583-4 |
| [29] |
Wang MP, Chen L, Zhang ZJ, et al. Screening of alginate lyase-excreting microorganisms from the surface of brown algae[J]. AMB Express, 2017, 7: 74. DOI:10.1186/s13568-017-0361-x |
 2018, Vol. 45
2018, Vol. 45




