扩展功能
文章信息
- 蒋承州, 黄勇, 段燕文, 朱湘成
- JIANG Cheng-Zhou, HUANG Yong, DUAN Yan-Wen, ZHU Xiang-Cheng
- 后基因组时代基因组重排在微生物菌种选育中的应用及展望
- Progress in genome shuffling in the post genomic era for microbial strains improvement
- 微生物学通报, 2018, 45(11): 2494-2502
- Microbiology China, 2018, 45(11): 2494-2502
- DOI: 10.13344/j.microbiol.china.171064
-
文章历史
- 收稿日期: 2017-12-20
- 接受日期: 2018-02-09
- 网络首发日期(www.cnki.net): 2018-03-16
2. 新药组合生物合成国家地方联合工程研究中心 湖南 长沙 410205;
3. 组合生物合成与天然产物药物湖南省工程研究中心 湖南 长沙 410205
2. National Engineering Research Center of Combinatorial Biosynthesis for Drug Discovery, Changsha, Hunan 410205, China;
3. Hunan Engineering Research Center of Combinatorial Biosynthesis and Natural Product Drug Discovery, Changsha, Hunan 410205, China
微生物代谢产物与社会发展息息相关,比如乳酸、丙酸、乙醇和丙二醇等初级代谢产物是替代传统石油化工实现绿色制造和生物质能源开发的重要基础[1];而结构和活性多样化的次级代谢产物则是新药开发的主要源泉[2]。作为发酵工业核心的微生物菌种,因在人工培育条件下极易出现耐受性差、生物利用度低和产能不足等缺陷,是微生物代谢产物开发及其产业化所必须面对的重大挑战。微生物菌种选育就是为了获得产量提高、遗传稳定、适应人工条件的发酵友好型高产菌株。传统的随机诱变育种无需了解微生物及其代谢产物的遗传背景,但选育过程费时费力且正突变效率较低[3]。而基于合成生物学技术的代谢工程育种虽然在产量提升和衍生物开发等方面卓有成效[4],但其成功与否严格取决于微生物的遗传可操作性和对目标代谢产物生物合成及调控机制的认知,并且受限于特定代谢途径的局部调控而无法全面提升微生物的性能;此外,在微生物复杂的基因转录和代谢网络调控背景下如何确定有效的基因靶标也是该技术的关键瓶颈[5]。随着基因组学和生物信息技术的快速发展,针对微生物的探索已正式步入了后基因组时代,即不再局限于对单一基因或蛋白质的研究,而是在基因组和系统水平上全面分析多个基因或蛋白质的功能。基因组重排(Genome shuffling)[6]利用多轮递推原生质体融合对微生物进行全基因组范围的基因片段重组和交换,以累积有益突变来实现目标微生物的人工定向进化。这种实用高效的技术不仅是传统育种方法的有效补充和延伸,还为我们通过组学研究(基因组、转录组和蛋白组等)系统地解析微生物基因转录表达和代谢网络提供了平台,将有助于特定基因靶标的甄选,为通过合成生物学技术实现更为高效的微生物育种和精准人工调控创造条件。本文将对基因组重排在微生物菌种选育中的应用研究进行介绍,尤其针对近年来围绕其开展的组学研究进展进行详细阐述。
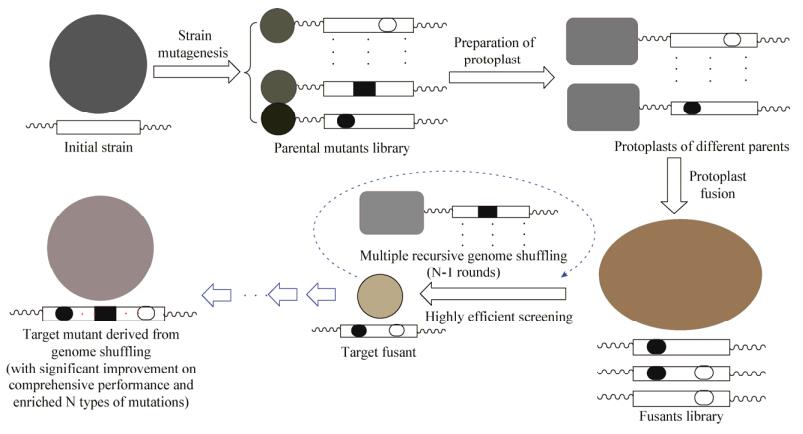
1 基因组重排技术在微生物菌种选育中的应用简介基于基因组重排的微生物菌种选育主要包括4大步骤[7](图 1):(1)构建具有基因和表型多样性的亲本文库。主要通过物理(辐射、紫外线等)、化学(化学诱变剂、生物制剂等)和核糖体工程等多元化手段来诱导随机基因突变并筛选获得性状或/和性能改良的正向突变菌文库[8],以及其它具有特定表型的不同种属的微生物菌种等。一般具备特征筛选标记(如抗生素抗性等)或/和表型(如耐酸、耐热)的亲本更利于后续的原生质体融合及筛选,并可在基因组重排过程中得到有效地累积;(2)原生质体的高效制备。由于原生质体制备尚无普适性方法,主要采用酶解法去除细胞壁,因此酶的种类和用量、酶解时间、温度和pH等都会直接影响原生质体的质量[7],需要对上述诸多条件进行优化才能获得高质量的原生质体;(3)原生质体的理性融合。一般需要盐类(硝酸盐、氯化物、葡聚糖硫酸盐等)、电场、多聚化合物(聚乙二醇、聚乙烯醇等)[7]等外界诱导或刺激来有效地促进融合,而融合模型的理性设计与后续的突变菌筛选密不可分,对目标菌种的定向进化有直接影响。以原生质体灭活融合为例,在利用物理或化学方法损伤原生质体生理结构使其失去再生能力后,只有采取不同机制(高温灭活、紫外线灭活、化学试剂灭活等)灭活的亲本之间才能发生互补融合并恢复再生,因此该融合模型十分适用于缺少遗传标记和显著表型的亲本[8]。作为基因组重排的核心过程,原生质体融合可以突破种属间界限,极大地扩充了基因组重排的范畴和多样性;(4)基因组重排突变菌的高效筛选和验证。主要是基于融合模型中不同亲本的差异化特性(如抗生素抗性、发酵环境耐受性、产量差异等)进行针对性的筛选,从而在融合子中富集多种有益的突变。目前常用方法主要包括互补筛选(营养缺陷型互补、抗性互补、代谢互补抑制)、差异筛选(物理特性、生理特性、不同抗性)、荧光标记筛选[9]等,其中抗生素抗性和包括酸碱、温度、化合物(底物、产物和副产物等)等在内的条件耐受性,因其鲜明的筛选标记近来在基因组重排的筛选中广为应用[10-12]。在筛选到目标融合子后,还需对其进行性能和遗传稳定性等多方面的综合评价及验证,最终才能获得预期的满足应用开发需要的工业改良菌种。
|
| 图 1 基因组重排的技术流程概括 Figure 1 The general process of genome shuffling |
|
|
基因组重排在微生物育种中最主要的应用就是提升代谢产物产量。表 1对近5年来在涉及化工、食品、医药、生物能源等工业领域中应用基因组重排提升各类微生物代谢产物产量的主要实例进行了汇总,充分展示了该技术与传统诱变育种方法的相辅相成及多元化应用。以核糖体工程诱变为例,其特有的抗生素筛选标记与基因组重排联用后相得益彰,在生物质能源丁醇和乙醇[12],抗生素药物多拉菌素[13]、阿维拉霉素[14]和诺西肽[15],以及天然食品防腐剂ε-多聚赖氨酸(ε-PL)[16]等重要代谢产物的产量提升中得到了高效应用。更为重要的是,基因组重排有效地规避了微生物基因工程改造所有的必需条件,对许多特殊种类或无法进行遗传操作的微生物而言,仍是最有效的菌种选育方法。比如在具有生物降解和环保功效的耐冷微生物约氏不动杆菌(Acinetobacter johnsonii)中增强低温碱性脂肪酶的合成[17],在红树林内生真菌琉球曲霉(Aspergillus luchuensis)中提高洛伐他汀的产量[18],在丙酮丁醇梭菌(Clostridium acetobutylicum)和蜡样芽孢杆菌(Bacillus cereus)共生体系中提高生物能源丁醇的产量[19]等。此外,由于基因组重排的多轮递推及突破种属限制等特性,其对于目标菌种的累积进化所导致的产量提升叠加效应是单一应用代谢工程或其它菌种选育方法所无法比拟的。例如对ε-PL的产量提升中,首先在禾粟链霉菌(Streptomyces graminearus)中通过UV和亚硝基胍诱变及随后的基因组重排将ε-PL的产量提高了88%[11];随后采用同样策略分别处理稠李链霉菌(Streptomyces padanus)、灰褐链霉菌(Streptomyces griseofuscus)、吸水链霉菌(Streptomyces hygroscopicus)和白色链霉菌(Streptomyces albulus),使ε-PL产量均有不同程度的提高;最终对所有5类突变菌再次进行基因组重排,将ε-PL产量进一步提升了近80%,获得与原始菌种相比底物耐受性明显增强及ε-PL产量提升超过3倍的高产菌株[20]。
| 代谢产物 Metabolite |
亲本诱变 Parental mutagenesis |
微生物种属 Species |
最终产量(提升程度) Final production (increased level) |
| ε-多聚赖氨酸ε-Poly-L-lysine | UV+NTG | 禾粟链霉菌Streptomyces graminearus | 13.5 g/L (88%)[11] |
| 丙酮-丁醇-乙醇Acetone-butanol-ethanol | UV+NTG+MW | 丙酮丁醇梭菌Clostridium acetobutylicum | 22.21 g/L (34.53%)[12] |
| 多拉菌素Doramectin | UV+NTG | 阿维链霉菌Streptomyces avermitilis | 992 mg/L (11.2 folds)[13] |
| 阿维拉霉素Avilamycin | 60Co-γ ray | 绿色产色链霉菌Streptomyces viridochromogenes | 1.4 g/L (36.8 folds)[14] |
| 诺西肽Nosiheptide | 60Co-γ ray+LiCl | 活跃链霉菌Streptomyces actuosus | 1.54 g/L (9.2 folds)[15] |
| ε-多聚赖氨酸ε-Poly-L-lysine | ARTP+RE | 白色链霉菌Streptomyces albulus | 56.5 g/L (1.5 folds)[16] |
| 低温碱性脂肪酶Low-temperature alkalophilic lipase | UV+DES | 约氏不动杆菌Acinetobacter johnsonii | 7 U/mL (~3.0 folds)[17] |
| 洛伐他汀Lovastatin | UV | 琉球曲霉Aspergillus luchuensis | 57.0 mg/gds (6.0 folds)[18] |
| 丁醇Butanol | NTG | 丙酮丁醇梭菌Clostridium acetobutylicum | 20.1 g/L (23.3%)[19] |
| ε-多聚赖氨酸ε-Poly-L-lysine | UV+NTG | 五种链霉菌杂交Interspecific hybridization | 24.5 g/L (> 3.0 folds)[20] |
| 乙醇Ethanol | UV+LiCl | 树干毕赤酵母Pichia stipitis | 41 g/L (~1.5 folds)[21] |
| 乙醇Ethanol | NA | 酿酒酵母Saccharomyces cerevisiae | 120 g/L (11%)[22] |
| 丁醇Butanol | ARTP | 丙酮丁醇梭菌/蜡样芽孢杆菌共生系统C. acetobutylicum/Bacillus cereus symbiotic system | 15.63 g/L (34%)[23] |
| L-谷氨酸L-glutamic acid | UV+DES | 谷氨酸棒状杆菌Corynebacteria glutamicum | 119 g/L (1.8 folds)[24] |
| 琥珀酸Succinic acid | UV+NTG | 琥珀酸放线杆菌Actinobacillus succinogenes | 95.6 g/L (73%)[25] |
| 糖醇Sugar alcohol | UV+ARTP | 异常毕赤酵母Pichia anomala | 47.1 g/L (32.3%)[26] |
| 丙酸Propionic acid | UV+LiCl | 丙酸杆菌Propionibacterium | 4.01 g/g (25%)[27] |
| 核苷虫草素Cordycepin | UV+HNO2 | 九州虫草Cordyceps kyushuensis | 978.25 μg/g (9.6 folds)[28] |
| 木聚糖酶Xylanase | UV+NTG+EB | 曲霉NRCF5 Aspergillus sp. NRCF5 | 427.5 U/mL (6.13 folds)[29] |
| 核酸酶P1 Nuclease P1 | 60Co-γ ray | 桔青霉Penicillium citrinum | 1 980.2 U/mL (4.7 folds)[30] |
| 转糖苷酶Transglycosidase | 60Co-γ ray | 尼日尔曲霉Aspergillus niger | 14.91 U/mL (194%)[31] |
| 胞外β-葡萄糖苷酶Extracellular β-glucosidase | UV+Ultrasonic | 异酒香酵母Brettanomyces anomalus | 4 790 U/L (~8.0 folds)[32] |
| 纤维素酶Cellulase | UV | 灰绿曲霉Aspergillus glaucus | 70 U/mL (~1.8 folds)[33] |
| 果糖基转移酶Fructosyltransferase | UV+LiCl | 米曲霉Aspergillus oryzae | 353 U/g (3.4 folds)[34] |
| 纳他霉素Natamycin | UV+5-BU | 褐黄孢链霉菌Streptomces gilvosporeus | 4.69 g/L (3.8 folds)[35] |
| 表面活性素Surfactin | UV+NTG+Ion beam | 解淀粉芽孢杆菌Bacillus amyloliquefaciens | 350.1 mg/L (10.3 folds)[36] |
| 他克莫司Tacrolimus | UV+NTG | 筑波链霉菌Streptomyces tsukubaensis | 365.6 mg/L (11.0 folds)[37] |
| 达托霉素Daptomycin | UV+NTG | 玫瑰孢链霉菌Streptomyces roseosporus | 582 mg/L (3.8 folds)[38] |
| 乳酸链球菌素Nisin | UV+DES | 乳酸乳球菌Lactococcus lactis | 4 023 IU/mL (2.4 folds)[39] |
| 普那霉素Pristinamycin | UV | 始旋链霉菌Streptomyces pristinaespiralis | 120 mg/L (6.0 folds)[40] |
| 抗菌肽芬荠素Fengycin | NTG +RE | 解淀粉芽孢杆菌Bacillus amyloliquefaciens | 450.51 mg/L (8.3 folds)[41] |
| 南强菌素Deacetylmycoepoxydiene | UV | 拟茎点霉A123 Phomopsis sp. A123 | 219 mg/L (275.0 folds)[42] |
| 谷胱甘肽Glutathione | UV+NTG | 酿酒酵母Saccharomyces cerevisiae | 230.88 mg/L (3.3 folds)[43] |
| 聚γ-谷氨酸Poly-γ-glutamic acid | UV+LiCl | 枯草芽孢杆菌Bacillus subtilis | 34.3 g/L (18.8 folds)[44] |
| 注:NTG:亚硝基胍;EB:溴化乙锭;DES:硫酸二乙酯;EMS:甲磺酸乙酯;BU:溴尿嘧啶;MW:微波;RE:核糖体工程;ARTP:常压室温等离子体;UV:紫外线;NA:未知. Note: NTG: Nitrosoguanidine; EB: Ethidium bromide; DES: Diethyl sulfate; EMS: Ethyl methanesulfonate; BU: Bromouracil; MW: Microwave; RE: Ribosome engineering; ARTP: Atmospheric and room-temperature plasma; UV: Ultraviolet; NA: Not available. |
|||
生物信息学研究表明,微生物作为活性天然产物的重要来源,其中蕴含着丰富的次级代谢产物生物合成基因簇,但如何激活其中绝大部分沉默的基因簇则是微生物天然产物开发所面临的契机和挑战。通过基因组重排在目标微生物中富集的各种突变很有可能对其基因表达和代谢网络进行多重调控,从而实现对沉默生物合成途径的激活或/和已有生物合成途径的修饰,因此在微生物天然产物开发方面也拥有较大的潜能。例如对从红豆杉中分离的内生真菌瘤座菌TF5 (Tubercularia sp. TF5)进行随机诱变及后续的基因组重排后,从突变菌中分离得到了包括8个倍半萜类化合物、2个二氢异香豆素和1个四氢萘酮在内的11个新化合物和9个已知化合物,其中既有在TF5中已发现天然产物的新衍生物,也有分子结构迥异的新化合物[45]。
3 基因组重排在微生物性状改良上的应用作为菌种选育及其工业化应用的评价标准之一,微生物生理特性上的优化同样尤为重要,并且往往与代谢产物的产量提升相得益彰。在对微生物生理遗传背景缺乏深入了解的前提下,基于表型筛选(一般为条件耐受性筛选)的基因组重排可以富集对生理特性产生影响的基因突变,在微生物性状改良上也得到了广泛的应用(表 2)。以生物乙醇的工业化开发为例,联合应用代谢工程与基因组重排使工业酿酒酵母菌(Saccharomyces cerevisiae)在降低副产物甘油产量的同时提升了对乙醇的耐受性[10];而且基因组重排还能提高活性干酵母在生物乙醇生产中对干燥环境的抗逆性和乙醇发酵性能[46],以及酿酒酵母菌对木质纤维素水解产物中存在的抑制性副产物糠醛和乙酸的耐受性[47]。与此同时,采用电穿孔介导的DNA杂交技术对树干毕赤酵母菌(Pichia stipitis)和酿酒酵母菌进行的基因组重排,成功获得了可直接利用木糖来合成乙醇的杂交重组工程菌ScF2,并大幅提高了其木糖耐受性,为生物转化木质纤维素开发绿色能源创造了条件[48]。
| 生理特性 Physiological feature |
亲本诱变 Parental mutagenesis |
微生物种属 Species |
提升程度 Increased level |
| 葡萄糖耐受性Glucose tolerance | UV+NTG | 禾粟链霉菌Streptomyces graminearus | 70%[11] |
| 耐热性Thermotolerance | UV+DES | 谷氨酸棒状杆菌Corynebacteria glutamicum | ~16%[24] |
| 2-脱氧葡萄糖耐受性2-Deoxyglucose tolerance | UV+NTG+EB | 曲霉NRCF5 Aspergillus sp. NRCF5 | 10 folds[29] |
| 5-羟甲基糠醛耐受性5-Hydroxymethyl-furfural tolerance | NA | 酿酒酵母Saccharomyces cerevisiae | 100%[47] |
| 木糖耐受性Xylose tolerance | NA | 毕赤酵母菌Pichia stipitis酿酒酵母Saccharomyces cerevisiae | 2.5%[48] |
| 正丁醇耐受性n-Butanol tolerance | Fluorescence labeling | 大肠杆菌Escherichia coli | 2.6 folds[49] |
| 脱酸活性Deacidification activity | UV | 粟酒裂殖酵母Schizosaccharomyces pombe | 225.2%[50] |
| 乙醇耐受性Ethanol tolerance | UV | 酿酒酵母Saccharomyces cerevisiae | 7%[51] |
| 异丙醇耐受性Isopropanol tolerance | NTG | 拜氏梭菌Clostridium beijerinckii | 1.4 folds[52] |
| 抗菌活性Antimicrobial activity | UV | 乳酸片球菌Pediococcus acidilactici | 1.43 folds[53] |
| 粘附力Adhesive property | UV+NTG | 植物乳杆菌Lactobacillus plantarum | 10%[54] |
| RDX降解RDX degradation | NA | 嗜麦芽窄食单胞菌Stenotrophomonas maltophilia | 50%[55] |
| 注:NTG:亚硝基胍;EB:溴化乙锭;DES:硫酸二乙酯;UV:紫外线;NA:未知. Note: NTG: Nitrosoguanidine; EB: Ethidium bromide; DES: Diethyl sulfate; UV: Ultraviolet; NA: Not available. |
|||
基因组重排富集的多种突变及其突变菌所具备的对应表型为深入解析微生物复杂的基因表达和代谢网络调控创造了条件。例如实时荧光定量PCR (qRT-PCR)和扩增片段长度多态性(Amplified fragment length polymorphism,AFLP)等技术曾被用于挖掘基因组重排突变菌中特定表型的遗传变异。但是,早期许多集中于特定代谢途径的研究仅能进一步说明代谢产物产量提高与其生物合成途径中关键酶的过表达之间本已存在的必然联系,如表面活性肽(Surfactin)与其合成酶基因srfA[36],琥珀酸(Succinic acid)与葡萄糖代谢途径[25],达托霉素(Daptomycin)与其关键合成酶非核糖体肽合成酶(NRPS)[38]等。与此同时,越来越多的研究也开始揭示其它遗传因素对生物合成途径的重要调控,例如基因组重排后普那霉素(Pristinamycin)产量的大幅提升不仅与其生物合成基因snaB、snbA和自我抗性基因ptr的过量表达有关,还受到了AfsR转录调控子和转座酶同源编码基因变异的影响[56];而对纳他霉素(Natamycin)的基因组重排高产突变菌进行的遗传多态性研究发现,54种差异表达的蛋白中仅有葡萄糖激酶调节蛋白直接参与了纳他霉素的生物合成[35]。因此,对微生物遗传和代谢网络的局部研究不能有效地甄别导致相关表型的遗传基础,更无法解析其中潜在的调控机制。
步入后基因组时代,快速发展的各类组学和生物信息分析技术为我们全面了解基因组重排介导的微生物定向进化提供了便利。转录组测序(RNA-Seq)初步揭示了基因组重排引起微生物表型变化的一些潜在遗传基础,如里氏木霉(Trichoderma reesei)合成乙醇能力的增强与一系列糖酵解酶的过表达直接相关[57],酿酒酵母菌抗逆性的提高则主要归因于细胞倍性调节和应激反应基因表达水平的变化[58];而最近对丙酸杆菌(Propionibacterium)突变菌的高通量测序发现,基因组重排主要通过基因转换(而不是长片段基因插入)引起基因组保守区域的单点突变和基因重复等变化[27]。另一方面,蛋白质组学分析表明基因组重排所导致的微生物性能提升(产量提高或/和性状改良)不仅受到其代谢网络全局变化的调控,如蛋白质代谢、细胞膜成分、海藻糖代谢和氧化反应等蛋白表达的改变对活性干酵母抗逆性和乙醇产量的提高[55],还涉及到许多关键生物过程和应激反应过程的交叉影响,如产酸丙酸杆菌(Propionibacterium acidipropionici)对副产物乙酸和终产物丙酸耐受性的提高与包括分泌蛋白3-磷酸甘油醛脱氢酶、ATP合酶α亚基、NADH脱氢酶和丙二酰CoA异构酶等的过表达有关[59];而解淀粉芽孢杆菌(Bacillus amyloliquefaciens)中表面活性肽产量的提升则涉及到包括代谢过程,DNA复制、重组和修复,翻译和翻译后修饰,细胞的分泌和信号转导,表面活性肽合成,能量生产和转换等在内的多个通路的46个差异表达蛋白[60]。这些研究成果表明,基因组重排可以为研究微生物基因转录表达和代谢网络的复杂调控机制提供丰富的素材。
相对而言,多组学分析联合应用可以更有效地研究基因组重排引起的遗传变化与对应表型特征之间的关系。以大肠杆菌产生正丁醇耐受性机制的研究为例,RNA-Seq揭示了各种基因组重排突变菌产生抗性的不同机制,比如影响脂肪酸合成的生物素合成基因bioA以及一系列脂多糖生物合成基因的显著上调表达,有可能通过改变细胞膜的亲水性来增加对正丁醇的耐受性,而与铁离子运输相关的基因表达上调所间接导致的外膜修饰同样有助于增强正丁醇耐受性;与此同时,基因组高通量测序发现了不同突变菌中存在的转座子插入序列迁移和单核苷酸多态性(Single-nucleotide polymorphism,SNP)变化等遗传差异,最终证实大肠杆菌产生正丁醇耐受性的复杂机制涉及到多重基因的参与及潜在的遗传相互作用[48]。类似地,基因组和转录组联合分析发现了酿酒酵母基因组重排突变菌对纤维素水解物抑制剂产生耐受性的关键基因,主要包括与泛素介导蛋白水解有关的去泛素化酶Ubp7p、应激反应转录抑制因子Nrg1p和NADPH依赖性的谷氨酸脱氢酶Gdh1p,并且对去泛素化酶Ubp7p进行的逆向突变可以增加原始酿酒酵母菌对亚硫酸盐废液的耐受性[61]。上述范例充分说明基因组重排介导的微生物定向进化是多个基因共同参与、对代谢网络进行全局调控的结果,而后基因时代飞速发展起来的各类组学分析则成为深入解析相关表型和基因型的有力工具。
5 展望基因组重排作为一种实用且高效的微生物育种技术,在对目标微生物背景缺乏了解、无法有效进行遗传操作等条件下可以人工地加速其定向进化,为许多微生物及其代谢产物的工业化应用奠定了基础。但是,基因组重排同样有其内在局限性和先天不足。比如基因组重排的成功实施需要高质量的原生质体作为基础,因此不适合应用于那些较难培养和不易制备原生质体的微生物。由于绝大部分用于基因组重排的亲本仍然来源于传统的随机诱变,并且其性能的优劣直接影响到最终重排突变菌的质量,所以无法回避传统育种方法固有的效率低下这一局限性。同时,微生物基因表达和代谢网络调控的复杂性使得通过基因组重排富集的多种突变是否一定会产生协同效应仍存在不确定性,也有可能因为相互干扰而无法促进预期的目标菌种定向进化。此外,实现基因组重排突变菌的产业化应用还需要满足一定的条件,作者在早期针对重要微生物天然产物药物雷帕霉素(Rapamycin)的开发中,曾通过基因组重排成功地将雷帕霉素的产量提高了近3倍[62],但在后续应用中发现基因组重排工程菌由于缺乏筛选压力会逐渐退化而导致产量降低,无法应用于工业化生产。因此,拥有特征筛选标记(如耐受性、温敏性、选择性等)对于基因组重排工程菌遗传稳定性的维持和后续的工业化应用是十分必要的,而如何高效地筛选获得具有预期表型的亲本以及遗传稳定的融合菌株都是基因组重排得以有效应用所必须面临的挑战。
基因组重排作为传统诱变和代谢工程之间的连接桥梁,不仅极大地扩展了选育微生物菌种的技术路线,还为通过从表型到基因型的逆向研究来全面解析微生物复杂的代谢网络及调控机制提供了探索的模式。研究者可以联合运用多种组学技术,尤其是新兴的基于液相-质谱分析联用或/和液相-核磁共振分析联用的代谢组学技术[63],从微生物改良的性状出发对基因组重排突变菌及其亲本进行深入剖析,通过系统水平的基因、蛋白和代谢产物的全面网络分析来揭示其不同表型的遗传和物质基础及相互作用关系,并基于生物信息大数据和高效的CRISPR/Cas9基因编辑技术来挖掘潜在的基因靶点,为利用合成生物学技术对微生物进行更为精准的人工调控和实现更高效的定向进化,最终构建面向应用的微生物细胞合成工厂奠定基础。
| [1] |
Liu WS, Jiang RR. Combinatorial and high-throughput screening approaches for strain engineering[J]. Applied Microbiology and Biotechnology, 2015, 99(5): 2093-2104. DOI:10.1007/s00253-015-6400-0 |
| [2] |
Shen B. A new golden age of natural products drug discovery[J]. Cell, 2015, 163(6): 1297-1300. DOI:10.1016/j.cell.2015.11.031 |
| [3] |
Bekker V, Dodd A, Brady D, et al. Tools for metabolic engineering in Streptomyces[J]. Bioengineered, 2014, 5(5): 293-299. DOI:10.4161/bioe.29935 |
| [4] |
Pál C, Papp B, Pósfai G. The dawn of evolutionary genome engineering[J]. Nature Reviews Genetics, 2014, 15(7): 504-512. DOI:10.1038/nrg3746 |
| [5] |
Adrio JL, Demain AL. Genetic improvement of processes yielding microbial products[J]. FEMS Microbiology Reviews, 2006, 30(2): 187-214. DOI:10.1111/j.1574-6976.2005.00009.x |
| [6] |
Zhang YX, Perry K, Vinci VA. Genome shuffling leads to rapid phenotypic improvement in bacteria[J]. Nature, 2002, 415(6872): 644-646. DOI:10.1038/415644a |
| [7] |
Biot-Pelletier D, Martin VJJ. Evolutionary engineering by genome shuffling[J]. Applied Microbiology and Biotechnology, 2014, 98(9): 3877-3887. DOI:10.1007/s00253-014-5616-8 |
| [8] |
Barrick JE, Lenski RE. Genome dynamics during experimental evolution[J]. Nature Reviews Genetics, 2013, 14(12): 827-839. DOI:10.1038/nrg3564 |
| [9] |
Gong JX, Zheng HJ, Wu ZJ, et al. Genome shuffling: Progress and applications for phenotype improvement[J]. Biotechnology Advances, 2009, 27(6): 996-1005. DOI:10.1016/j.biotechadv.2009.05.016 |
| [10] |
Wang PM, Zheng DQ, Liu TZ, et al. The combination of glycerol metabolic engineering and drug resistance marker-aided genome shuffling to improve very-high-gravity fermentation performances of industrial Saccharomyces cerevisiae[J]. Bioresource Technology, 2012, 108: 203-210. DOI:10.1016/j.biortech.2011.12.147 |
| [11] |
Li S, Li F, Chen XS, et al. Genome shuffling enhanced ε-poly-L-lysine production by improving glucose tolerance of Streptomyces graminearus[J]. Applied Biochemistry and Biotechnology, 2012, 166(2): 414-423. DOI:10.1007/s12010-011-9437-2 |
| [12] |
Gao XF, Zhao H, Zhang GH, et al. Genome shuffling of Clostridium acetobutylicum CICC 8012 for improved production of acetone-butanol-ethanol (ABE)[J]. Current Microbiology, 2012, 65(2): 128-132. DOI:10.1007/s00284-012-0134-3 |
| [13] |
Zhang J, Wang XJ, Diao JN, et al. Streptomycin resistance-aided genome shuffling to improve doramectin productivity of Streptomyces avermitilis NEAU1069[J]. Journal of Industrial Microbiology & Biotechnology, 2013, 40(8): 877-889. |
| [14] |
Lv XA, Jin YY, Li YD, et al. Genome shuffling of Streptomyces viridochromogenes for improved production of avilamycin[J]. Applied Microbiology and Biotechnology, 2013, 97(2): 641-648. DOI:10.1007/s00253-012-4322-7 |
| [15] |
Wang QL, Zhang D, Li YD, et al. Genome shuffling and ribosome engineering of Streptomyces actuosus for high-yield nosiheptide production[J]. Applied Biochemistry and Biotechnology, 2014, 173(6): 1553-1563. DOI:10.1007/s12010-014-0948-5 |
| [16] |
Wang L, Chen XS, Wu GY, et al. Genome shuffling and gentamicin-resistance to improve ε-poly-L-lysine productivity of Streptomyces albulus W-156[J]. Applied Biochemistry and Biotechnology, 2016, 180(8): 1601-1617. DOI:10.1007/s12010-016-2190-9 |
| [17] |
Wang HK, Zhang J, Wang XJ, et al. Genome shuffling improves production of the low-temperature alkalophilic lipase by Acinetobacter johnsonii[J]. Biotechnology Letters, 2012, 34(1): 145-151. DOI:10.1007/s10529-011-0749-7 |
| [18] |
El-Gendy MMAA, Al-Zahrani HAA, El-Bondkly AMA. Genome shuffling of mangrove endophytic Aspergillus luchuensis MERV10 for improving the cholesterol-lowering agent lovastatin under solid state fermentation[J]. Mycobiology, 2016, 44(3): 171-179. DOI:10.5941/MYCO.2016.44.3.171 |
| [19] |
Li SB, Qian Y, Liang ZW, et al. Enhanced butanol production from cassava with Clostridium acetobutylicum by genome shuffling[J]. World Journal of Microbiology and Biotechnology, 2016, 32(4): 53. DOI:10.1007/s11274-016-2022-7 |
| [20] |
Li S, Chen XS, Dong CL, et al. Combining genome shuffling and interspecific hybridization among Streptomyces improved ε-poly-L-lysine production[J]. Applied Biochemistry and Biotechnology, 2013, 169(1): 338-350. DOI:10.1007/s12010-012-9969-0 |
| [21] |
Shi J, Zhang M, Zhang LB, et al. Xylose-fermenting Pichia stipitis by genome shuffling for improved ethanol production[J]. Microbial Biotechnology, 2014, 7(2): 90-99. DOI:10.1111/mbt2.2014.7.issue-2 |
| [22] |
Hou LH, Meng M, Guo L, et al. A comparison of whole cell directed evolution approaches in breeding of industrial strain of Saccharomyces cerevisiae[J]. Biotechnology Letters, 2015, 37(7): 1393-1398. DOI:10.1007/s10529-015-1812-6 |
| [23] |
Gu CK, Wang GY, Mai S, et al. ARTP mutation and genome shuffling of ABE fermentation symbiotic system for improvement of butanol production[J]. Applied Microbiology and Biotechnology, 2017, 101(5): 2189-2199. DOI:10.1007/s00253-017-8093-z |
| [24] |
Zheng P, Liu M, Liu XD, et al. Genome shuffling improves thermotolerance and glutamic acid production of Corynebacteria glutamicum[J]. World Journal of Microbiology and Biotechnology, 2012, 28(3): 1035-1043. DOI:10.1007/s11274-011-0902-4 |
| [25] |
Zheng P, Zhang KK, Yan Q, et al. Enhanced succinic acid production by Actinobacillus succinogenes after genome shuffling[J]. Journal of Industrial Microbiology & Biotechnology, 2013, 40(8): 831-840. |
| [26] |
Zhang GQ, Lin YP, Qi XN, et al. Genome shuffling of the nonconventional yeast Pichia anomala for improved sugar alcohol production[J]. Microbial Cell Factories, 2015, 14: 112. DOI:10.1186/s12934-015-0303-8 |
| [27] |
Luna-Flores CH, Palfreyman RW, Krömer JO, et al. Improved production of propionic acid using genome shuffling[J]. Biotechnology Journal, 2017, 12(2): 1600120. DOI:10.1002/biot.201600120 |
| [28] |
Wang YM, Zhang GY, Zhao X, et al. Genome shuffling improved the nucleosides production in Cordyceps kyushuensis[J]. Journal of Biotechnology, 2017, 260: 42-47. DOI:10.1016/j.jbiotec.2017.08.021 |
| [29] |
El-Bondkly AMA. Molecular identification using ITS sequences and genome shuffling to improve 2-deoxyglucose tolerance and xylanase activity of marine-derived fungus, Aspergillus sp. NRCF5[J]. Applied Biochemistry and Biotechnology, 2012, 167(8): 2160-2173. DOI:10.1007/s12010-012-9763-z |
| [30] |
Wang C, Wu GZ, Li YD, et al. Genome shuffling of Penicillium citrinum for enhanced production of nuclease P1[J]. Applied Biochemistry and Biotechnology, 2013, 170(6): 1533-1545. DOI:10.1007/s12010-013-0297-9 |
| [31] |
Li W, Chen GG, Gu LL, et al. Genome shuffling of Aspergillus niger for improving transglycosylation activity[J]. Applied Biochemistry and Biotechnology, 2014, 172(1): 50-61. DOI:10.1007/s12010-013-0421-x |
| [32] |
Wu P, Zhao XH, Pan SY. Intraspecific protoplast fusion of Brettanomyces anomalus for improved production of an extracellular β-glucosidase[J]. Biotechnology & Biotechnological Equipment, 2014, 28(5): 878-881. |
| [33] |
Zhao YP, Jiang CX, Yu HP, et al. Genome shuffling of Aspergillus glaucus HGZ-2 for enhanced cellulase production[J]. Applied Biochemistry and Biotechnology, 2014, 174(4): 1246-1259. DOI:10.1007/s12010-014-1102-0 |
| [34] |
Wang SH, Duan MJ, Liu YL, et al. Enhanced production of fructosyltransferase in Aspergillus oryzae by genome shuffling[J]. Biotechnology Letters, 2017, 39(3): 391-396. DOI:10.1007/s10529-016-2254-5 |
| [35] |
Luo JM, Li JS, Liu D, et al. Genome shuffling of Streptomyces gilvosporeus for improving natamycin production[J]. Journal of Agricultural and Food Chemistry, 2012, 60(23): 6026-6036. DOI:10.1021/jf300663w |
| [36] |
Zhao JF, Li YH, Zhang C, et al. Genome shuffling of Bacillus amyloliquefaciens for improving antimicrobial lipopeptide production and an analysis of relative gene expression using FQ RT-PCR[J]. Journal of Industrial Microbiology & Biotechnology, 2012, 39(6): 889-896. |
| [37] |
Du WJ, Huang D, Xia ML, et al. Improved FK506 production by the precursors and product-tolerant mutant of Streptomyces tsukubaensis based on genome shuffling and dynamic fed-batch strategies[J]. Journal of Industrial Microbiology & Biotechnology, 2014, 41(7): 1131-1143. |
| [38] |
Yu GH, Hu YS, Hui M, et al. Genome shuffling of Streptomyces roseosporus for improving daptomycin production[J]. Applied Biochemistry and Biotechnology, 2014, 172(5): 2661-2669. DOI:10.1007/s12010-013-0687-z |
| [39] |
Zhang YF, Liu SY, Du YH, et al. Genome shuffling of Lactococcus lactis subspecies lactis YF11 for improving nisin Z production and comparative analysis[J]. Journal of Dairy Science, 2014, 97(5): 2528-2541. DOI:10.3168/jds.2013-7238 |
| [40] |
Jin QC, Shen N, Yin H, et al. DNA shuffling of ptr resistance gene leads to improved pristinamycin production in Streptomyces pristinaespiralis[J]. Molecular Biology, 2015, 49(2): 253-259. |
| [41] |
Zhao JF, Zhang C, Lu J, et al. Enhancement of fengycin production in Bacillus amyloliquefaciens by genome shuffling and relative gene expression analysis using RT-PCR[J]. Canadian Journal of Microbiology, 2016, 62(5): 431-436. DOI:10.1139/cjm-2015-0734 |
| [42] |
Wang MZ, Zhang W, Xu WK, et al. Optimization of genome shuffling for high-yield production of the antitumor deacetylmycoepoxydiene in an endophytic fungus of mangrove plants[J]. Applied Microbiology and Biotechnology, 2016, 100(17): 7491-7498. DOI:10.1007/s00253-016-7457-0 |
| [43] |
Yin H, Ma YL, Deng Y, et al. Genome shuffling of Saccharomyces cerevisiae for enhanced glutathione yield and relative gene expression analysis using fluorescent quantitation reverse transcription polymerase chain reaction[J]. Journal of Microbiological Methods, 2016, 127: 188-192. DOI:10.1016/j.mimet.2016.06.012 |
| [44] |
Zeng W, Chen GG, Wu H, et al. Improvement of Bacillus subtilis for poly-γ-glutamic acid production by genome shuffling[J]. Microbial Biotechnology, 2016, 9(6): 824-833. DOI:10.1111/mbt2.2016.9.issue-6 |
| [45] |
Wang MZ, Liu SS, Li YY, et al. Protoplast mutation and genome shuffling induce the endophytic fungus Tubercularia sp. TF5 to produce new compounds[J]. Current Microbiology, 2010, 61(4): 254-260. DOI:10.1007/s00284-010-9604-7 |
| [46] |
Zheng DQ, Zhang K, Gao KH, et al. Construction of novel Saccharomyces cerevisiae strains for bioethanol active dry yeast (ADY) production[J]. PLoS One, 2013, 8(12): e85022. DOI:10.1371/journal.pone.0085022 |
| [47] |
Cheng C, Almario MP, Kao KC. Genome shuffling to generate recombinant yeasts for tolerance to inhibitors present in lignocellulosic hydrolysates[J]. Biotechnology Letters, 2015, 37(11): 2193-2200. DOI:10.1007/s10529-015-1895-0 |
| [48] |
Zhang W, Geng AL. Improved ethanol production by a xylose-fermenting recombinant yeast strain constructed through a modified genome shuffling method[J]. Biotechnology for Biofuels, 2012, 5(1): 46. |
| [49] |
Reyes LH, Almario MP, Winkler J, et al. Visualizing evolution in real time to determine the molecular mechanisms of n-butanol tolerance in Escherichia coli[J]. Metabolic Engineering, 2012, 14(5): 579-590. DOI:10.1016/j.ymben.2012.05.002 |
| [50] |
Ding S, Zhang Y, Zhang J, et al. Enhanced deacidification activity in Schizosaccharomyces pombe by genome shuffling[J]. Yeast, 2015, 32(2): 317-325. DOI:10.1002/yea.v32.2 |
| [51] |
Snoek T, Nicolino MP, van den Bremt S, et al. Large-scale robot-assisted genome shuffling yields industrial Saccharomyces cerevisiae yeasts with increased ethanol tolerance[J]. Biotechnology for Biofuels, 2015, 8: 32. DOI:10.1186/s13068-015-0216-0 |
| [52] |
de Gérando HM, Fayolle-Guichard F, Rudant L, et al. Improving isopropanol tolerance and production of Clostridium beijerinckii DSM 6423 by random mutagenesis and genome shuffling[J]. Applied Microbiology and Biotechnology, 2016, 100(12): 5427-5436. DOI:10.1007/s00253-016-7302-5 |
| [53] |
Han GG, Song AA, Kim EB, et al. Improved antimicrobial activity of Pediococcus acidilactici against Salmonella gallinarum by UV mutagenesis and genome shuffling[J]. Applied Microbiology and Biotechnology, 2017, 101(13): 5353-5363. DOI:10.1007/s00253-017-8293-6 |
| [54] |
Zhao YJ, Duan CC, Gao L, et al. Genome shuffling of Lactobacillus plantarum C88 improves adhesion[J]. Bioscience, Biotechnology, and Biochemistry, 2017, 81(1): 184-193. DOI:10.1080/09168451.2016.1224637 |
| [55] |
Lee BU, Choi MS, Kim DM, et al. Genome shuffling of Stenotrophomonas maltophilia OK-5 for improving the degradation of explosive RDX (Hexahydro-1, 3, 5-trinitro-1, 3, 5-triazine)[J]. Current Microbiology, 2017, 74(2): 268-276. DOI:10.1007/s00284-016-1179-5 |
| [56] |
Jin QC, Jin ZH, Zhang LJ, et al. Probing the molecular mechanisms for pristinamycin yield enhancement in Streptomyces pristinaespiralis[J]. Current Microbiology, 2012, 65(6): 792-798. DOI:10.1007/s00284-012-0233-1 |
| [57] |
Huang J, Wu RZ, Chen D, et al. Transcriptional profiling of the Trichoderma reesei recombinant strain HJ48 by RNA-seq[J]. Journal of Microbiology and Biotechnology, 2016, 26(7): 1242-1251. DOI:10.4014/jmb.1602.02003 |
| [58] |
Zheng DQ, Chen J, Zhang K, et al. Genomic structural variations contribute to trait improvement during whole-genome shuffling of yeast[J]. Applied Microbiology and Biotechnology, 2014, 98(7): 3059-3070. DOI:10.1007/s00253-013-5423-7 |
| [59] |
Guan NZ, Shin HD, Chen RR, et al. Understanding of how Propionibacterium acidipropionici respond to propionic acid stress at the level of proteomics[J]. Scientific Reports, 2014, 4: 6951. |
| [60] |
Zhao JF, Cao L, Zhang C, et al. Differential proteomics analysis of Bacillus amyloliquefaciens and its genome-shuffled mutant for improving surfactin production[J]. International Journal of Molecular Sciences, 2014, 15(11): 19847-19869. DOI:10.3390/ijms151119847 |
| [61] |
Pinel D, Colatriano D, Jiang H, et al. Deconstructing the genetic basis of spent sulphite liquor tolerance using deep sequencing of genome-shuffled yeast[J]. Biotechnology for Biofuels, 2015, 8: 53. DOI:10.1186/s13068-015-0241-z |
| [62] |
Chen XY, Wei PL, Fan LM, et al. Generation of high-yield rapamycin-producing strains through protoplasts-related techniques[J]. Applied Microbiology and Biotechnology, 2009, 83(3): 507-512. DOI:10.1007/s00253-009-1918-7 |
| [63] |
Watrous J, Roach P, Alexandrov T, et al. Mass spectral molecular networking of living microbial colonies[J]. Proceedings of the National Academy of Sciences of the United States of America, 2012, 109(26): E1743-E1752. DOI:10.1073/pnas.1203689109 |
 2018, Vol. 45
2018, Vol. 45