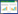扩展功能
文章信息
- 孙旭, 杨柳燕
- SUN Xu, YANG Liu-Yan
- 蓝藻堆积对河蚬N2O释放通量及其肠道细菌群落结构的影响
- Effect of cyanobacterial accumulation on the nitrous oxide emission of Corbicula fluminea and intestinal bacterial community structure
- 微生物学通报, 2018, 45(11): 2376-2386
- Microbiology China, 2018, 45(11): 2376-2386
- DOI: 10.13344/j.microbiol.china.170705
-
文章历史
- 收稿日期: 2017-09-07
- 接受日期: 2018-07-20
- 网络首发日期(www.cnki.net): 2018-08-10
2. 南京工程学院环境工程学院 江苏 南京 211167;
3. 环境保护部南京环境科学研究所 江苏 南京 210042
2. School of Environmental Engineering, Nanjing Institute of Technology, Nanjing, Jiangsu 211167, China;
3. Nanjing Institute of Environmental Sciences, Ministry of Environmental Protection, Nanjing, Jiangsu 210042, China
一氧化二氮(N2O)作为一种温室气体,其在大气中的浓度仅为二氧化碳(CO2)的千分之一,但温室效应是CO2的296倍,所导致的全球性气候变化和生态环境问题已经成为21世纪人类面临的一个严重威胁[1]。随着城市化和工业化水平的提高,人类活动对湖泊的影响也越来越大,大量的污染物输入湖泊水体,引起一系列不可逆的生态环境问题,加速湖泊的富营养化进程。富营养化湖泊硝化反硝化作用强度随之加强,导致湖泊成为继稻田、湿地等传统N2O释放源之外的另一个重要的释放源[2-3]。蓝藻堆积区通常是湖泊N2O释放源的热点区域,水气界面N2O释放通量可达到3.9-85.8 μg N2O/(m·h)[4-6]。
湖泊沉积物中存在大量的底栖动物和氮转化微生物,其中底栖动物在活性氮迁移转化过程中起着重要的媒介作用。近年来研究表明底栖动物不仅能促进沉积物释放N2O,其肠道也能释放N2O,释放通量在0-93.1 nmol/(g·h)[7]。底栖动物自身不能产生N2O,主要是通过两种途径产生N2O,一方面底栖动物肠道内的反硝化微生物经不完全反硝化作用产生N2O,这部分N2O约占底栖动物N2O释放量的30%-68%,是底栖动物N2O释放的主要来源;另一方面底栖动物表面粘附的生物膜也是N2O释放的场所,硝化作用产生的N2O最高可达底栖动物释放量的1/3[8-9]。底栖动物N2O释放能力与其栖息环境密切相关,在4-10 ℃的环境下,摇蚊幼虫(Chironomus plumosus) N2O释放速率与上覆水中硝态氮含量呈显著的正相关[10]。
为了研究富营养化湖泊底栖动物N2O释放规律,本实验以太湖梅梁湾为研究区域,河蚬为研究对象,采用室内微宇宙实验研究蓝藻堆积对底栖动物N2O释放通量的影响,揭示富营养化湖泊底栖动物的环境效应,为湖泊温室气体控制提供理论支撑。
1 材料与方法 1.1 实验材料及设置于2013年9月用彼得森采泥器采集太湖梅梁湾(31.42′65.5′′N,120.20′95.2′′E)沉积物,过0.5 mm筛网后收集筛网上的河蚬;并用64 μm的筛网收集水中的蓝藻,将采集的沉积物、河蚬以及蓝藻带回实验室备用。
将混匀的2 L沉积物装入直径0.20 m、高0.15 m的有机玻璃容器,沿壁采用点滴的方式缓慢加入含NO3--N (2 mg/L)和NH4+-N (2 mg/L)的灭菌自来水(泥水体积比2:1),室温培养2周,每天更换上覆水。预培养结束后,引入3个大小一致的河蚬(密度为500 ind/m2),设置2个实验组,每组5个平行,一组为不加藻的对照组(CK),另一组为添加蓝藻的处理组(T),冻干蓝藻的添加量由梅梁湾上覆水叶绿素a浓度(500 μg/L)换算而来[11],文献记录该区域蓝藻优势种属为Microcystis,相对丰度为95%-98%[12-13]。所有实验组在25 ℃避光培养2周,用0.5 mm的筛网收集存活的河蚬。
1.2 河蚬N2O释放通量的测定各组分别取3个河蚬,用无菌水清洗河蚬表面3次,沥干表面的水分后放入含200 μL NaNO3 (0.5 mmol/L)的50 mL锥形瓶,丁基橡胶密封瓶口后21 ℃避光培养2 h,收集锥形瓶顶空气体测定N2O含量,作为河蚬活体N2O释放通量[8]。
1.3 河蚬肠道细菌基因组的提取分别取培养前(G0)、对照组(GN)和处理组(GA)培养河蚬5个,参照Horn等的方法提取河蚬肠道基因组DNA[14]。用50 μL无菌水洗脱后,0.8%琼脂糖凝胶电泳验证目的条带,并测定230、260、280 nm吸光值检验提取效率和纯度。
1.4 高通量测序及数据分析 1.4.1 454高通量测序将提取的肠道基因组DNA用带有不同TAG标签的通用引物B-27F (5′-AGAGTTTGATCCTGGCT CAG-3′)和A-533R (5′-TTACCGCGGCTGCTGGCA C-3′)对16S rRNA基因进行PCR扩增,A为测序端接头,B为引物共用端接头[15]。PCR反应体系:5×FastPfu缓冲液4 μL,dNTPs (2.5 mmol/L) 2 μL,引物(5 μmol/L)各0.4 μL,FastPfu聚合酶(500 U) 0.4 μL,模板DNA (10 ng/μL) 1 μL,灭菌超纯水补至20 μL。PCR反应条件:95 ℃ 2 min;95 ℃ 30 s,55 ℃ 30 s,72 ℃ 30 s,27个循环;72 ℃ 10 min。扩增产物经2%琼脂糖凝胶电泳检测,切胶回收纯化后送上海美吉生物公司进行454 FLX Titanium测序。
1.4.2 序列处理454测序序列采用Mothur软件进行降噪处理,去掉长度小于300 bp、模糊碱基和引物碱基错配2个以上、质量分数低于25、单碱基重复超过6个不确定碱基的序列,并通过UCHIME的方法去除嵌合体序列,得到的高质量序列用于后续数据分析。
1.4.3 OTU聚类注释及多样性分析使用QIIME软件中pick_de_novo_otus.py命令,通过Uclust方法对获得的高质量序列按97%相似度进行OTU (Operational taxonomic unit)聚类,选取每个OTU中数量最多的序列作为代表性序列进行注释,比对数据库为Greengene,获得每个OTU代表序列的物种分类信息。为减少低丰度物种对数据分析的影响,去掉相对丰度小于总序列条数万分之一的OTU,得到的OTU表用于下游数据分析。根据OTU表中的各样品物种丰度情况,应用QIIME软件计算各样品稀释曲线、种群丰富度(Chao1指数)和多样性(Shannon、Simpson指数),并生成不同分类水平上的物种丰度表和多样品物种分布图。本研究中454测序获得的序列在NCBI Sequence Read Archive (SRA)上的登录号为SRX502686。
2 结果与分析 2.1 蓝藻堆积时河蚬N2O释放特征对照组河蚬在引入到微宇宙后逐渐进入沉积物中,处理组河蚬位于泥水界面,外壳呈半张开状态。经2周培养对照组和处理组河蚬总存活数分别为13个和10个,存活率分别达到86%和66%。不同处理组河蚬的N2O释放通量存在显著差异,如图 1所示,2 h内对照组和处理组河蚬N2O释放通量分别为1 335.6 pmol/ind和894.5 pmol/ind (P < 0.01),N2O释放速率分别为667.8 pmol/(ind·h)和447.2 pmol/(ind·h),蓝藻水华颗粒使河蚬N2O释放通量和释放速率分别降低33%和37%,表明蓝藻的堆积减缓了河蚬N2O的释放。

|
| 图 1 蓝藻堆积对河蚬N2O释放的影响 Figure 1 Effect of cyanobacterial accumulation on N2O emission from C. fluminea |
|
|
利用高通量测序技术,经序列过滤和去除嵌合体后,3个肠道样品共得到9 661条高质量序列,长度分布在300-548 bp,其中以序列428 bp居多。如表 1所示,97%的相似度下共获得1 043个OTU,G0、GN、GA 3个样品的OTU数目分别为562、619、65,大多数OTU分布在其中一个或两个样品中(图 2),经2周的培养,对照组河蚬肠道OTU数目有所增加,处理组河蚬肠道OTU数目大幅降低,预示蓝藻水华暴发时河蚬肠道细菌种类急剧减少。从图 3和表 1看出,3个样品的稀释曲线在97%的相似度下趋于平缓并达到饱和,同时样品的覆盖度在96%-99%,说明统一测序深度至2 769时3个样品获取的细菌信息基本能反映河蚬肠道细菌的群落组成。
|
| 图 2 河蚬肠道16S rRNA基因Venn图 Figure 2 Venn diagram of the intestinal 16S rRNA gene from C. fluminea 注:G0:预培养后河蚬;GN:对照组河蚬肠道样品;GA:处理组河蚬肠道样品. Note: G0: Gut before incubation; GN: Gut sample from the control; GA: Gut sample from the treatment. |
|
|

|
| 图 3 稀释曲线 Figure 3 Rarefaction curves |
|
|
| Name | Coverage(%) | OTUs | Chao1 | Shannon | Simpson |
| G0 | 97.03 | 562 | 579.21 | 8.15 | 0.99 |
| GN | 96.59 | 619 | 639.43 | 8.45 | 0.99 |
| GA | 99.06 | 65 | 85.31 | 2.70 | 0.72 |
通过Chao1、香浓指数以及辛普森指数来表征河蚬肠道细菌丰富度和多样性,如表 1所示,处理组河蚬肠道细菌多样性和丰富度均低于对照组,这与稀释曲线的趋势一致,表明蓝藻的堆积能降低河蚬肠道内细菌多样性。
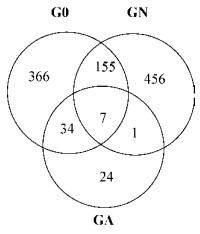
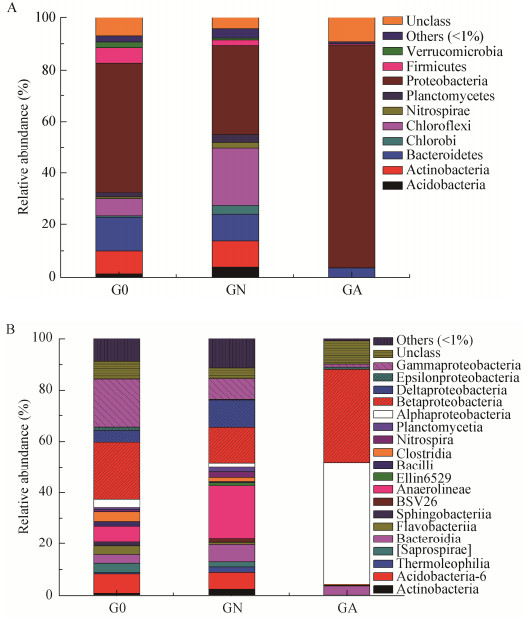
2.3 蓝藻对河蚬肠道细菌群落结构的影响 2.3.1 门和纲水平优势菌分布特征在门分类水平3组河蚬肠道样品细菌群落组成如图 4所示,覆盖了30个细菌门类,包括变形菌门(Proteobacteria)、拟杆菌门(Bacteroidetes)、放线菌门(Actinobacteria)、绿弯菌门(Chloroflexi)、厚壁菌门(Firmicutes)、酸杆菌门(Acidobacteria)等,其中以变形菌门、拟杆菌门、放线菌门、绿弯菌门和厚壁菌门的微生物为主要类群,这5种优势菌群的相对丰度为84.3%-90.1%。不同处理组河蚬肠道细菌群落组成在门分类水平上存在一定的差异,培养前河蚬肠道细菌相对丰度由高到低为变形菌门(50.2%) > 拟杆菌门(13.1%) > 放线菌门(8.64%) > 绿弯菌门(6.5%),经2周培养后对照组(GN)河蚬肠道细菌相对丰度由高到低为变形菌门(34.3%) > 绿弯菌门(22.4%) > 拟杆菌门(10.5%) > 放线菌门(9.9%),而处理组(GA)则主要以变形菌门为优势菌(85.8%)。纵向而言,变形菌门在各组肠道中相对丰度高低为GA > G0 > GN,拟杆菌门为G0 > GA > GN,绿弯菌门为GN > G0 > GA。
|
| 图 4 门(A)和纲(B)分类水平河蚬肠道细菌群落组成 Figure 4 The communities structure of bacterial with phylum (A) and class (B) |
|
|
针对肠道样品中细菌优势门类变形菌门、拟杆菌门、放线菌门、绿弯菌门在纲分类水平进行分析,结果如图 4所示,不同处理的河蚬肠道细菌群落组成存在明显的差异。变形菌门中以β-、α-、γ-变形菌纲为主,相对丰度分别达到13.7%-36.3%、1.4%-37.5%、1.2%-18.9%,而δ-和ε-变形菌纲相对丰度较低,G0组变形菌门以β-和γ-变形菌纲为主,GN组变形菌门以β-和δ-变形菌纲为主,而GA组以α-和β-变形菌纲为主;拟杆菌门中拟杆菌纲、Flavobacteriia、Saprospirae是主要类群,其相对丰度分别为3.4%-6.6%、0.1%-3.2%、0-3.6%,G0组拟杆菌门以上述3种菌为主,GN组和GA组拟杆菌门均是拟杆菌纲为主;放线菌门和绿弯菌门微生物主要在G0和GN组被检出,放线菌门以放线菌纲为主,其在G0和GN组中相对丰度分别为7.4%和6.4%,Anaerolineae是绿弯菌门的优势菌,其在G0和GN组中相对丰度分别为6.1%和20.5%。
2.3.2 属分类水平优势菌分布特征在属分类水平3个河蚬肠道样品细菌群落组成如图 5所示,获得的1 043个OTU主要集中在311个属,包括110个已知属和201个未知属,相对丰度分别为18.8%和81.2%。311个属中相对丰度大于1%的属有15个,包括Dechloromonas (7.49%)、Acinetobacter (2.8%)、Propionibacterium (2.5%)、Chromobacterium (1.55%)、Bacteroides (1.44%)及未知属10个。

|
| 图 5 河蚬肠道细菌属分类水平群落组成 Figure 5 The bacterial community structure of intestinal bacterial from C. fluminea at genus level |
|
|
对相对丰度大于1%的未知属菌群进行分析发现,立克次氏体目(Rickettsiales,16.1%)、envOPS12 (3.7%)、红环菌科(Rhodocyclaceae,3.5%)、GCA004 (2.4%)、中华杆菌科(Sinobacteraceae,2.2%)等是主要类群。不同样品的未知菌也存在明显差异,G0组未知菌丰度由高到低为丛毛单胞菌科(Comamonadaceae,5.9%) > 嗜甲基菌科(Methylophilaceae,4.5%) > envOPS12 (2.9%) > ACK-M1 (2.7%);GN组主要的优势未知菌为envOPS12 (8.2%)、GCA004 (5.8%)、拟杆菌科(Bacteroidales,5.4%)和中华杆菌科(Sinobacteraceae,5.4%);而GA组主要以立克次氏体目(Rickettsiales,47.2%)和红环菌科(Rhodocyclaceae,7.9%)为主(图 6)。

|
| 图 6 河蚬肠道细菌属分类水平未知菌群落组成 Figure 6 The unclassified community structure of intestinal bacterial from C. fluminea at genus level |
|
|
微生物群落的重要特征之一就是微生物群落的稳定性,主要表现为结构稳定性和功能稳定性,微生物群落结构决定了其微生物功能。
河蚬肠道反硝化细菌组成如表 2所示,3个样品中共获得23种反硝化菌,其相对丰度达到17.7%,主要以脱氮单孢菌属(Dechloromonas,7.5%)、不动杆菌属(Acinetobacter,2.8%)和丙酸杆菌属(Propionibacterium,2.5%)等常见的反硝化菌为主。不同处理的河蚬肠道内反硝化菌组成存在明显的差异,GN组反硝化细菌相对丰度约为10%,以丙酸杆菌属(Propionibacterium,5.3%)、硫化细菌(Thiobacillus,1.2%)和脱氮单孢菌属(Dechloromonas,1.0%)为主;而GA组反硝化菌相对丰度则可达到22.6%,是GN组的2.3倍,脱氮单孢菌属(Dechloromonas,20.9%)是其优势反硝化菌属。
| Denitrify bacterial | Total (%) | G0 (%) | GN (%) | GA (%) |
| Corynebacterium[16] | 0.30 | 0.87 | 0.00 | 0.04 |
| Mycobacterium[17] | 0.10 | 0.29 | 0.00 | 0.00 |
| Propionibacterium[18] | 2.50 | 2.17 | 5.34 | 0.00 |
| Streptomyces[19] | 0.05 | 0.14 | 0.00 | 0.00 |
| Cytophagaceae[20] | 0.07 | 0.22 | 0.58 | 0.00 |
| Flavobacterium[21] | 0.78 | 2.17 | 0.11 | 0.07 |
| Hyphomicrobium[22] | 0.02 | 0.00 | 0.07 | 0.00 |
| Methylosinus[22] | 0.18 | 0.04 | 0.43 | 0.00 |
| Ochrobactrum[23] | 0.05 | 0.04 | 0.00 | 0.11 |
| Rhodoplanes[24] | 0.04 | 0.00 | 0.11 | 0.00 |
| Rhodobacter[23] | 0.10 | 0.18 | 0.00 | 0.00 |
| Rhodospirillaceae[21] | 0.05 | 0.14 | 0.00 | 0.00 |
| Achromobacter[25] | 0.08 | 0.22 | 0.00 | 0.04 |
| Burkholderia[26] | 0.05 | 0.14 | 0.00 | 0.00 |
| Comamonas[21] | 0.25 | 0.76 | 0.00 | 0.00 |
| Hydrogenophaga[27] | 0.24 | 0.58 | 0.07 | 0.00 |
| Rubrivivax[28] | 0.01 | 0.00 | 0.04 | 0.00 |
| Thiobacillus[29] | 0.67 | 0.25 | 1.77 | 0.00 |
| Dechloromonas[21] | 7.50 | 0.51 | 1.01 | 20.91 |
| Zoogloea[21] | 0.29 | 0.79 | 0.07 | 0.00 |
| Arcobacter[21] | 0.61 | 1.05 | 0.00 | 0.79 |
| Acinetobacter[30] | 2.80 | 8.34 | 0.00 | 0.07 |
| Pseudomonas[31] | 0.90 | 2.09 | 0.00 | 0.61 |
底栖动物广泛地分布在淡水和海洋沉积物中,是水生态系统的重要组成部分,不仅能反映水质的污染状况,还参与水生态系统的碳、氮、磷、硫等物质循环,研究表明底栖动物也是一个潜在的N2O释放源[7, 32]。本研究以太湖梅梁湾为研究区域,河蚬为研究对象,通过室内微宇宙实验模拟蓝藻堆积情况下底栖动物N2O的释放特征,结果显示上覆水中叶绿素a浓度达到500 μg/L时,河蚬N2O释放速率为447.2 pmol/(ind·h),高于已报道的双壳类平均值300 pmol/(ind·h)[8],若按太湖梅梁湾河蚬最高密度500 ind/m2计算[33],蓝藻暴发时河蚬N2O释放通量可达9.8 μg N2O/(m2·h),约占沉积物释放量的11.4%[11],表明底栖动物也是富营养化湖泊N2O重要的释放源之一,应引起足够重视。如表 3所示,与其他底栖动物N2O释放情况相比,本研究处理组河蚬N2O释放通量处于一个较高的水平,是摇蚊幼虫(Chironomus plumosus)释放量的34.9倍,仅次于触角豆螺(Bithynia tentaculata)的释放水平,这与底栖动物食性和栖息环境有关[7, 28]。底栖动物自身不会释放N2O,但通过吞噬环境中的硝化反硝化微生物在肠道释放N2O,其中肠道释放的N2O占底栖动物释放量的67%-84%。双壳类的河蚬表面通常会形成一层生物膜,大量的硝化细菌附着在生物膜中,硝化过程释放的N2O量约占总释放量的1/3,而肠道反硝化释放量占2/3,本研究将不同处理组河蚬表面清洗后再进行N2O释放通量测定,并采用相应的功能基因amoA引物对肠道AOA (Ammonia-oxidizing archaea)和AOB (Ammonia- oxidizing bacteria)进行扩增,并未成功扩增出目的条带,表明河蚬肠道里不发生或发生很微弱的硝化过程,以致于不能检测到氨氧化菌,因此河蚬N2O释放量基本由肠道产生[8, 35]。
| 底栖动物 Aquatic invertebrate |
食性 Habitat type |
释放量 N2O emission(pmol/(ind·h)) |
| Bithynia tentaculata[7] | Filter-and deposit-feeders | 600-900 |
| Mytilus edulis[7] | 150-426 | |
| Chironomus plumosus[10, 28, 32] | 10-128 | |
| Ephemera danica[34] | 100-351 | |
| Corbicula fluminea (this study) | 447-667 | |
| Anabolia nervosa[7] | Shredders | 100-200 |
| Asellus aquaticus[7] | 20-45 | |
| Ecdyonurus sp.[7] | Grazers | 10-25 |
| Baetis sp.[7] | 2.2-8.1 | |
| Erpobdella octoculata[7] | Predators | 5.1-32.0 |
| Orectochilus sp.[7] | 4.2-18.4 | |
| Sialis lutaria[33] | 2.5-12.2 |
河蚬N2O释放量与其肠道内反硝化底物浓度密切相关,例如NO3--N、DOC及DO等,有研究表明上覆水NO3--N浓度低于15.5 mg/L或DO低于2.9 mg/L时,部分底栖动物肠道内NO3--N和DO浓度与上覆水中相应浓度具有显著的正相关[9-10]。本研究处理组GA组由于堆积蓝藻的降解,释放大量的氨氮及有机酸,上覆水NO3--N和DO浓度分别为3.8 mg/L和1.5 mg/L,明显低于对照组GN的7.6 mg/L和3.7 mg/L,可以推测GA组河蚬肠道内NO3--N和DO均低于对照组GN,而DOC含量高于对照组GN。通常增加可利用碳含量能降低反硝化产物中N2O:N2的比例,且在富含有机质的环境中没有明显的硝氮输入时,会促使更多的N2O转化成N2[36]。
作为自然界氮循环的主要驱动者,微生物在湖泊生态系统氮素的迁移转化过程中发挥着重要作用。利用454高通量测序技术研究了蓝藻堆积时河蚬肠道细菌群落演替规律,结果显示,河蚬肠道内细菌覆盖30门75纲118目237科311属,大多数是未知菌属,比DGGE (Denaturing gradient gel electrophoresis)、T-RFLP (Terminal restriction fragment length polymorphism)、克隆文库等传统的微生物群落分析技术获得更多的细菌门类[37]。经2周的培养处理组河蚬肠道优势菌为变形菌门的α-和β-变形菌,相对丰度高达85%,而对照组河蚬肠道则为变形菌门的β-、γ-变形菌和拟杆菌门,同时处理组河蚬肠道细菌丰富度和多样性均低于对照组。这可能与河蚬在不同栖息环境的习性及肠道对细菌的选择性有关,无蓝藻堆积环境下河蚬在沉积物中掘穴而居,主要以沉积物颗粒为食,导致对照组河蚬肠道细菌与沉积物细菌较为接近;在有蓝藻堆积环境下河蚬栖息在泥水界面之间,双壳敞开,以蓝藻碎屑和沉积物颗粒为食,而在蓝藻暴发时沉积物中以变形菌为主,β-变形菌常常是污染较重湖区沉积物的优势类群[9, 12, 38-39]。进一步对河蚬肠道反硝化细菌进行分析,发现河蚬肠道内反硝化细菌相对丰度为10.0%-22.6%,处理组河蚬肠道反硝化细菌相对丰度是对照组河蚬的2.3倍,主要以Dechloromonas为主,这类反硝化细菌通常在污水处理系统中被检测到,具有完全反硝化能力,可将NO3-转化为N2[40]。这与Svenningsen等[41]研究结果一致,双壳类的斑马贝(Dreissena polymorpha)肠道中nirK型和nirS型反硝化菌多样性较低,nirK型主要以Dechloromonas为主,而nirS型反硝化菌主要是α-变形菌的Rhodobacter和Rhodopseudomonas。值得注意的是基于16S rRNA基因的菌群多样性分析通常会造成对细菌丰度的高估,有研究表明焦磷酸测序的V4-V5区域显示了最低的高估程度(约为3.0%),而V6区域的高估程度最高(约为13%),但也有研究认为V3-V4区域是活性污泥细菌多样性研究的最佳区域,这可能与样本的复杂程度有关,本研究中河蚬肠道细菌多样性明显低于活性污泥,因此采用V3-V4区域进行分析基本能覆盖样品的细菌多样性[42-43]。对于反硝化功能基因而言,由于存在种间转移,且与16S rRNA基因进化速度不同,导致大部分反硝化功能基因与16S rRNA基因的系统发育关系存在较大的差异,因此通过16S rRNA基因对反硝化细菌多样性分析时会引起一定程度的低估,尤其是一些低丰度的反硝化细菌将检测不到[44]。
4 结论通过室内微宇宙实验,模拟蓝藻堆积情况下河蚬N2O释放规律,丰富了N2O释放清单核算,可为全球气候变化控制及湖泊富营养化提供理论支撑,主要结论如下:
(1) 蓝藻的堆积可使河蚬N2O释放通量减少63%;
(2) 蓝藻堆积会降低河蚬肠道内细菌丰富度和多样性,增加肠道内α-和β-变形菌纲相对丰度,降低拟杆菌门和绿弯菌门相对丰度,改变肠道内细菌群落组成;
(3) 蓝藻堆积会增加河蚬肠道内反硝化功能菌的相对丰度,形成特异型的反硝化菌类群(脱氮单孢菌属),相对丰度可达到20.9%左右,进一步强化完全反硝化作用,使更多的N2O转化成N2,减少温室气体N2O的释放,降低湖泊内源氮负荷,是富营养化湖泊生态系统自我调节机制之一。
| [1] |
Jung MY, Well R, Min D, et al. Isotopic signatures of N2O produced by ammonia-oxidizing archaea from soils[J]. The ISME Journal, 2014, 8(5): 1115-1125. DOI:10.1038/ismej.2013.205 |
| [2] |
Wang HJ, Wang WD, Yin CQ, et al. Littoral zones as the "hotspots" of nitrous oxide (N2O) emission in a hyper-eutrophic lake in China[J]. Atmospheric Environment, 2006, 40(28): 5522-5527. DOI:10.1016/j.atmosenv.2006.05.032 |
| [3] |
Kong Q, Wang ZB, Niu PF, et al. Greenhouse gas emission and microbial community dynamics during simultaneous nitrification and denitrification process[J]. Bioresource Technology, 2016, 210: 94-100. DOI:10.1016/j.biortech.2016.02.051 |
| [4] |
Zhong JC, Fan CX, Liu GF, et al. Seasonal variation of potential denitrification rates of surface sediment from Meiliang Bay, Taihu Lake, China[J]. Journal of Environmental Sciences, 2010, 22(7): 961-967. DOI:10.1016/S1001-0742(09)60205-9 |
| [5] |
Fagerstone KD, Quinn JC, Bradley TH, et al. Quantitative measurement of direct nitrous oxide emissions from microalgae cultivation[J]. Environmental Science & Technology, 2011, 45(21): 9449-9456. |
| [6] |
Liu YS, Zhu RB, Ma DW, et al. Temporal and spatial variations of nitrous oxide fluxes from the littoral zones of three alga-rich lakes in coastal Antarctica[J]. Atmospheric Environment, 2011, 45(7): 1464-1475. DOI:10.1016/j.atmosenv.2010.12.017 |
| [7] |
Stief P, Poulsen M, Nielsen LP, et al. Nitrous oxide emission by aquatic macrofauna[J]. Proceedings of the National Academy of Sciences of the United States of America, 2009, 106(11): 4296-4300. DOI:10.1073/pnas.0808228106 |
| [8] |
Heisterkamp IM, Schramm A, Larsen LH, et al. Shell biofilm-associated nitrous oxide production in marine molluscs: processes, precursors and relative importance[J]. Environmental Microbiology, 2013, 15(7): 1943-1955. DOI:10.1111/j.1462-2920.2012.02823.x |
| [9] |
Stief P. Stimulation of microbial nitrogen cycling in aquatic ecosystems by benthic macrofauna: mechanisms and environmental implications[J]. Biogeosciences, 2013, 10(12): 7829-7846. DOI:10.5194/bg-10-7829-2013 |
| [10] |
Stief P, Polerecky L, Poulsen M, et al. Control of nitrous oxide emission from Chironomus plumosus larvae by nitrate and temperature[J]. Limnology and Oceanography, 2010, 55(2): 872-884. DOI:10.4319/lo.2009.55.2.0872 |
| [11] |
Wang HJ, Lu JW, Wang WD, et al. Methane fluxes from the littoral zone of hypereutrophic Taihu Lake, China[J]. Journal of Geophysical Research: Atmospheres, 2006, 111(D17): D17109. DOI:10.1029/2005JD006864 |
| [12] |
Shao KQ, Zhang L, Wang YP, et al. The responses of the taxa composition of particle-attached bacterial community to the decomposition of Microcystis blooms[J]. Science of the Total Environment, 2014, 488-489: 236-242. DOI:10.1016/j.scitotenv.2014.04.101 |
| [13] |
Chen XF, Jiang HY, Sun X, et al. Nitrification and denitrification by algae-attached and free-living microorganisms during a cyanobacterial bloom in Lake Taihu, a shallow Eutrophic Lake in China[J]. Biogeochemistry, 2016, 131(1/2): 135-146. |
| [14] |
Horn MA, Drake HL, Schramm A. Nitrous oxide reductase genes (nosZ) of denitrifying microbial populations in soil and the earthworm gut are phylogenetically similar[J]. Applied and Environmental Microbiology, 2006, 72(2): 1019-1026. DOI:10.1128/AEM.72.2.1019-1026.2006 |
| [15] |
Wang AH, Li M, Li CQ, et al. Human colorectal mucosal microbiota correlates with its host niche physiology revealed by endomicroscopy[J]. Scientific Reports, 2016, 6: 21952. DOI:10.1038/srep21952 |
| [16] |
Doi Y, Takaya N. A novel A3 group aconitase tolerates oxidation and nitric oxide[J]. Journal of Biological Chemistry, 2015, 290(3): 1412-1421. DOI:10.1074/jbc.M114.614164 |
| [17] |
Darwin KH, Ehrt S, Gutierrez-Ramos JC, et al. The proteasome of Mycobacterium tuberculosis is required for resistance to nitric oxide[J]. Science, 2003, 302(5652): 1963-1966. DOI:10.1126/science.1091176 |
| [18] |
Welsh D, Castadelli G, Bartoli M, et al. Denitrification in an intertidal seagrass meadow, a comparison of 15N-isotope and acetylene-block techniques: dissimilatory nitrate reduction to ammonia as a source of N2O?[J]. Marine Biology, 2001, 139(6): 1029-1036. DOI:10.1007/s002270100672 |
| [19] |
Takaki K, Fushinobu S, Kim SW, et al. Streptomyces griseus enhances denitrification by Ralstonia pickettii K50, which is possibly mediated by histidine produced during co-culture[J]. Bioscience, Biotechnology, and Biochemistry, 2008, 72(1): 163-170. DOI:10.1271/bbb.70528 |
| [20] |
Krustok I, Truu J, Odlare M, et al. Effect of lake water on algal biomass and microbial community structure in municipal wastewater-based lab-scale photobioreactors[J]. Applied Microbiology and Biotechnology, 2015, 99(15): 6537-6549. DOI:10.1007/s00253-015-6580-7 |
| [21] |
Xiao JJ, Guo P, Huo WJ, et al. Application of denitrifying microbes to wastewater denitrification[J]. Environmental Science & Technology, 2009, 32(12): 97-102. (in Chinese) 肖晶晶, 郭萍, 霍炜洁, 等. 反硝化微生物在污水脱氮中的研究及应用进展[J]. 环境科学与技术, 2009, 32(12): 97-102. DOI:10.3969/j.issn.1003-6504.2009.12.022 |
| [22] |
Guo F, Ju F, Cai L, et al. Taxonomic precision of different hypervariable regions of 16S rRNA gene and annotation methods for functional bacterial groups in biological wastewater treatment[J]. PLoS One, 2013, 8(10): e76185. DOI:10.1371/journal.pone.0076185 |
| [23] |
Nguyen VK, Park Y, Yang H, et al. Effect of the cathode potential and sulfate ions on nitrate reduction in a microbial electrochemical denitrification system[J]. Journal of Industrial Microbiology & Biotechnology, 2016, 43(6): 783-793. |
| [24] |
Hiraishi A, Ueda Y. Rhodoplanes gen. nov., a new genus of phototrophic bacteria including Rhodopseudomonas rosea as Rhodoplanes roseus comb. nov. and Rhodoplanes elegans sp. nov.[J]. International Journal of Systematic Bacteriology, 1994, 44(4): 665-673. DOI:10.1099/00207713-44-4-665 |
| [25] |
Gui MY, Chen Q, Ni JR. Effect of NaCl on aerobic denitrification by strain Achromobacter sp. GAD-3[J]. Applied Microbiology and Biotechnology, 2017, 101(12): 5139-5147. DOI:10.1007/s00253-017-8191-y |
| [26] |
Nie YX, Li L, Wang MC, et al. Nitrous oxide emission potentials of Burkholderia species isolated from the leaves of a boreal peat moss Sphagnum fuscum[J]. Bioscience, Biotechnology, and Biochemistry, 2015, 79(12): 2086-2095. DOI:10.1080/09168451.2015.1061420 |
| [27] |
Yang J, Zhang GZ, Yang XN, et al. Microbial community structure and diversity in cellar water by 16S rRNA high-throughput sequencing[J]. Environmental Science, 2017, 38(4): 1704-1716. (in Chinese) 杨洁, 张国珍, 杨晓妮, 等. 16S rRNA高通量测序研究集雨窖水中微生物群落结构及多样性[J]. 环境科学, 2017, 38(4): 1704-1716. |
| [28] |
Sun X, Hu ZX, Jia W, et al. Decaying Cyanobacteria decrease N2O emissions related to diversity of intestinal denitrifiers of Chironomus plumosus[J]. Journal of Limnology, 2015, 74(2): 261-271. |
| [29] |
Li R, Feng CP, Hu WE, et al. Woodchip-sulfur based heterotrophic and autotrophic denitrification (WSHAD) process for nitrate contaminated water remediation[J]. Water Research, 2016, 89: 171-179. DOI:10.1016/j.watres.2015.11.044 |
| [30] |
Huang XF, Li WG, Zhang DY, et al. Ammonium removal by a novel oligotrophic Acinetobacter sp. Y16 capable of heterotrophic nitrification-aerobic denitrification at low temperature[J]. Bioresource Technology, 2013, 146(10): 44-50. |
| [31] |
He TX, Li ZL, Sun Q, et al. Heterotrophic nitrification and aerobic denitrification by Pseudomonas tolaasii Y-11 without nitrite accumulation during nitrogen conversion[J]. Bioresource Technology, 2016, 200: 493-499. DOI:10.1016/j.biortech.2015.10.064 |
| [32] |
Poulsen M, Kofoed MVW, Larsen LH, et al. Chironomus plumosus larvae increase fluxes of denitrification products and diversity of nitrate-reducing bacteria in freshwater sediment[J]. Systematic and Applied Microbiology, 2014, 37(1): 51-59. DOI:10.1016/j.syapm.2013.07.006 |
| [33] |
Cai YJ, Gong ZJ, Qin BQ. Influences of habitat type and environmental variables on benthic macroinvertebrate communities in a large shallow subtropical lake (Lake Taihu, China)[J]. Annales de Limnologie-International Journal of Limnology, 2011, 47(1): 85-95. DOI:10.1051/limn/2010028 |
| [34] |
Stief P, Schramm A. Regulation of nitrous oxide emission associated with benthic invertebrates[J]. Freshwater Biology, 2010, 55(8): 1647-1657. |
| [35] |
Monteiro M, Séneca J, Magélhaes C. The history of aerobic ammonia oxidizers: from the first discoveries to today[J]. Journal of Microbiology, 2014, 52(7): 537-547. DOI:10.1007/s12275-014-4114-0 |
| [36] |
Saggar S, Jha N, Deslippe J, et al. Denitrification and N2O:N2 production in temperate grasslands: processes, measurements, modelling and mitigating negative impacts[J]. Science of the Total Environment, 2013, 465: 173-195. DOI:10.1016/j.scitotenv.2012.11.050 |
| [37] |
Wu TT, Sheng JP, Shen L. Application of microbial molecular ecology techniques in microbial diversity of lake[J]. Biotechnology Bulletin, 2010(3): 62-66. (in Chinese) 武婷婷, 生吉萍, 申琳. 微生物分子生态学技术在湖泊微生物多样性研究中的应用[J]. 生物技术通报, 2010(3): 62-66. |
| [38] |
Shao KQ, Gao G, Qin BQ, et al. Comparing sediment bacterial communities in the macrophyte-dominated and algae-dominated areas of eutrophic Lake Taihu, China[J]. Canadian Journal of Microbiology, 2011, 57(4): 263-272. DOI:10.1139/w11-003 |
| [39] |
Wang S, Xu Q, Zhang GS, et al. Operational performance and microbial community structure in a completely mixed aeration system[J]. Environmental Science, 2017, 38(2): 665-671. (in Chinese) 王硕, 徐巧, 张光生, 等. 完全混合式曝气系统运行特性及微生物群落结构解析[J]. 环境科学, 2017, 38(2): 665-671. |
| [40] |
Wen DH, Zhang N, Yu C, et al. Community structure and contaminant degradation function of biofilm in environmental engineering systems[J]. Microbiology China, 2014, 41(7): 1394-1401. (in Chinese) 温东辉, 张楠, 于聪, 等. 环境中生物膜的菌群结构与污染物降解特性[J]. 微生物学通报, 2014, 41(7): 1394-1401. |
| [41] |
Svenningsen NB, Heisterkamp IM, Sigby-Clausen M, et al. Shell biofilm nitrification and gut denitrification contribute to emission of nitrous oxide by the invasive freshwater mussel Dreissena polymorpha (Zebra mussel)[J]. Applied and Environmental Microbiology, 2012, 78(12): 4505-4509. DOI:10.1128/AEM.00401-12 |
| [42] |
Sun DL, Jiang X, Wu QL, et al. Intragenomic heterogeneity of 16S rRNA genes causes overestimation of prokaryotic diversity[J]. Applied and Environmental Microbiology, 2013, 79(19): 5962-5969. DOI:10.1128/AEM.01282-13 |
| [43] |
Cai L, Ye L, Tong AHY, et al. Biased diversity metrics revealed by bacterial 16S pyrotags derived from different primer sets[J]. PLoS One, 2013, 8(1): e53649. DOI:10.1371/journal.pone.0053649 |
| [44] |
Liang LH, Zuo JE. Denitrifying functional genes-the molecular marker for detection of denitrifying community structure[J]. Microbiology China, 2009, 36(4): 627-633. (in Chinese) 梁丽华, 左剑恶. 反硝化功能基因-检测反硝化菌种群结构的分子标记[J]. 微生物学通报, 2009, 36(4): 627-633. |
 2018, Vol. 45
2018, Vol. 45