扩展功能
文章信息
- 解翠晓, 张凯, 孙永革, 吕镇梅
- XIE Cui-xiao, ZHANG Kai, SUN Yong-ge, LU Zhen-mei
- 微宇宙模拟溢油条件下沉积物中蓝细菌多样性
- The impact of oil on the diversity of cyanobacteria in the sediments of a laboratory microcosm
- 微生物学通报, 2017, 44(7): 1579-1588
- Microbiology China, 2017, 44(7): 1579-1588
- DOI: 10.13344/j.microbiol.china.170166
-
文章历史
- 收稿日期: 2017-02-28
- 接受日期: 2017-06-12
- 优先数字出版日期(www.cnki.net): 2017-06-14
2. 浙江大学地球科学系 浙江 杭州 310027
2. Department of Earth Science, Zhejiang University, Hangzhou, Zhejiang 310027, China
在自然环境下,一些湖泊经常由于高温和富营养化等因素出现区域性蓝细菌暴发。有研究发现,溢油环境中也会出现蓝细菌富集,在原油浓度为28.4±1.8 mg/kg wt时沉积物中蓝细菌相对丰度增加[1],经微宇宙模拟实验证明蓝细菌生物量确有很大提升,且产生肉眼可见的蓝细菌富集现象[2]。1991年科威特海湾战争期间海湾受到大批的石油污染,有研究者发现在油膜上出现一层蓝绿色的微生物垫,其中就存在蓝细菌[3]。在自然溢油条件下经常会出现蓝细菌垫,Cohen在苏伊士湾附近受溢油污染的盐沼泽地中发现蓝细菌垫,通过研究表明从中分离出的蓝细菌菌株并不具有原油降解功能[4]。在热带条件下的原油污染位点,无论是土壤表层或水体中经常会出现蓝细菌垫[5]。
目前虽有众多研究发现溢油与蓝细菌垫的生成存在联系,但关于蓝细菌在其中发挥的作用却并无定论。Narro等发现Oscillatoria sp. strain JCM可将萘氧化成1-萘酚[6]。Raghukumar等研究表明Oscillatoria salina、Plectonema terebrans和Aphanocapsa sp.三种蓝细菌混合培养能消除超过40%的原油[7]。一般认为蓝细菌并不直接参与原油降解,而是通过胞外多糖与石油降解菌连接在一起,为石油降解菌提供氧气,此外蓝细菌与石油降解菌的聚合体能起固定作用[8-9],从而进行有效的生物降解。目前关于蓝细菌是否直接参与原油降解仍然具有争议性,但无可否认,一定程度的溢油会刺激蓝细菌生长[10-11]。
许多研究采用显微镜技术或纯培养技术,根据蓝细菌特有的形态来辨别不同蓝细菌[12],此法较为宏观且会错过许多比较小的蓝细菌,从而低估其多样性。根据核糖体数据库(Ribosomal database project)[13]和基因库[14]中蓝细菌16S rRNA基因序列,针对蓝细菌16S rRNA基因的特异性序列设计引物,进而实现蓝细菌序列的特异性扩增[15],此法能避免分离培养等步骤,在分子水平上对蓝细菌进行分类研究。
虽有不少研究发现溢油与蓝细菌的富集存在关联,但这些原油浓度大都处于较低水平,有研究表明溢油事故发生后原油浓度可高达500 mg/g,周边已无植物生长,但此时微生物菌群结构如何尚属未知[16]。本研究通过蓝细菌16S rRNA基因揭示在添加不同高浓度原油条件下蓝细菌多样性和丰度演替过程,旨在对不同种属蓝细菌对原油的响应及耐受性蓝细菌进行挖掘和功能分析。
1 材料与方法 1.1 主要试剂和仪器MoBio强力土壤DNA提取试剂盒,MoBio公司;Gel Extraction kit (200),Omega公司。
BS/BT电子天平,北京赛多利斯仪器系统有限公司;FD-1-50真空冷冻干燥机,北京博医康实验仪器有限公司;真空旋转蒸发仪,IKA公司;安捷伦7890A气相色谱仪,安捷伦公司;MJ MiniTM梯度PCR仪,Bio-Rad公司。
1.2 方法 1.2.1 微宇宙体系的建立:样品取自中国浙江省甬江入海口,该位点属于亚热带与温带气候,非常适宜浮游植物生长,甬江向东流入东海。样品取自水深1 m处表层(0-1.5 cm)沉积物,将样品立即运回实验室,置于4 ℃备用。
微宇宙实验共分为4个处理组:对照组CK、低浓度原油组(25 g/kg干重,LO)、中浓度原油组(125 g/kg干重,MO)及高浓度原油组(250 g/kg干重,HO)。每个处理设置3个重复,容器为透明有机玻璃(20 cm×10 cm×30 cm,长×宽×高),用阻板隔为三等分,每个处理有900 g沉积物。每个组别添加经0.2 μm滤膜过滤的海水,上覆水厚度为2 cm。每个容器上方用可透光有机玻璃覆盖,以减少水分蒸发,并不断添加海水使之与最初条件保持一致。微宇宙实验共持续35 d,按照实验设定的时间取样,取样时间为下午两点。整个微宇宙培养环境温度在(24±6)-(31±8) ℃之间,湿度在45%-69%。
1.2.2 原油降解分析:在微宇宙培养期间,分别在第1、2、5、8、14、20、28及35天取表层1.5 cm的沉积物样品2 g,立即冻存于-80 ℃。实验前将样品取出至冷冻干燥机中处理后经100目过筛,用作后续原油分析。
称取沉积物样品约1 g分别加入已知浓度的氘代二十四烷烃和氘代菲作为内标,然后用二氯甲烷:甲醇(9:1,体积比)进行3次超声抽提,抽提物经真空旋转蒸发至恒重,取约30 mg抽提有机质进行硅胶柱层析(12.5 cm硅胶+1.5 cm氧化铝)分离成饱和烃、芳烃和非烃沥青质3个组分。
(1) 全油色谱分析。研究所用原油为渤海重质原油。称取原油样品10 mg用正戊烷溶剂稀释100倍,沉淀过夜后取1 μL上样测试,所用仪器为安捷伦7890A气相色谱仪。色谱柱为CP7749弹性石英毛细管柱(50 m×0.32 mm×0.4 μm)。起始温度35 ℃,恒温10 min,以3 ℃/min升至300 ℃,恒温15 min。以氮气为载体,1 mL/min恒速,进样量1 μl。
(2) 饱和烃色谱分析。饱和烃的GC-FID测定采用安捷伦7890A气相色谱仪。色谱柱为DB-1弹性石英毛细管质谱专用柱(60 m×0.32 mm×0.25 μm)。起始温度60 ℃,恒温2 min,2 ℃/min升至290 ℃,恒温30 min。以氮气为载体,1 mL/min恒速,进样量1 μL。
1.2.3 蓝细菌16S rRNA基因克隆文库:对4个组别的第2天和第35天的样品进行克隆文库分析,将基因组DNA作为模板,采用引物CYA395F (5′-GG GGAATYTTCCGCAATGGG-3′)、CYA781(a) (5′-GA CTACTGGGGTATCTAATCCCATT-3′)和CYA781(b) (5′-GACTACAGGGGTATCTAATCCCTTT-3′)[15]进行普通PCR扩增。PCR反应体系为:10×ExTaq缓冲液5 μL,上下游引物各0.5 mmol/L,模板DNA 1 μL,加ddH2O至总体积50 μL。PCR反应条件为:95 ℃ 5 min;95 ℃ 1 min,60 ℃ 45 s,72 ℃ 2 min,共30个循环。PCR产物用凝胶电泳进行验证,切胶后用Gel Extraction Kit进行回收,然后将产物与pMDTM19-T载体进行连接、转化,每个样本挑取80个阳性克隆子。采用通用引物M13F (5′-TGTAAAACGACGGC CAGT-3′)和M13R (5′-GAGCGGATAACAATTTCAC ACAGG-3′)将得到的阳性克隆子进行筛选并测序。
1.2.4 克隆文库多样性指数分析:将得到的全部测序片段进行clustal序列比对,然后通过Phylip package (ver. 3.69) 中dnadist计算出不同序列之间的距离矩阵,最后利用DOTUR软件[17]在97%相似水平上对目的基因序列进行操作分类单元(Operational taxonomic units,OTU)分析。在软件MEGA 5.0上进行系统发育树构建[18],并输出保存为NWK格式,利用在线软件iTOL (http://itol.embl.de/upload.cgi)进行修饰和注释。
将每组样品进行独立的OTU分析,并计算每组样品覆盖率、ACE指数和Chao指数等,从而进行各样品的α多样性分析。
2 结果与分析 2.1 蓝细菌富集现象培养周期共为35 d,在第31天时,LO组表面出现肉眼可见的区域性变绿现象,而在前1天时并未出现此变化。其他处理组包括对照组均未出现变绿现象。
2.2 原油降解结果分析色谱结果导入增强型Chemstation MSD 5977软件(Agilent technologies)进行分析。正构烷烃峰型呈等间距分布的特点,以氘代二十四烷烃出峰时间确定出正构二十四烷烃的出峰时间,以姥鲛烷(Pr)、植烷(Ph)的出峰时间确定出正构C17、C18的出峰时间,然后对C13到C33的正构烷烃进行标注。利用饱和烃峰面积进行统计计算,选取Pr/C17、Pr/Ph、CPI的比值作为重要降解指标。Pr和Ph与其相邻的正构烷烃(nC17,nC18)之比可用作判识有机质中正构烷烃是否发生降解。
由全油色谱结果(图 1A)可知该原油富含轻烃,正构烷烃的碳数分布为nC9-nC36,单驼峰型,主峰碳为nC23,nC36之后无法检出。与常规石油烃优先降解短链烷烃的规律不同,本批次微宇宙培养28 d后正构烷烃高碳数组分的降解程度高于低碳数组分(图 1B),呈现出双峰型构型,主碳峰为nC17,nC31之后组分相对含量降低。

|
| 图 1 全油色谱(a)和LO组原油降解(B)分析图 Figure 1 Chromatography analysis of initial oil (A) and oil degradation in LO group (B) |
|
|
培养周期内饱和烃各组分峰面积积分结果如表 1所示,nC11以下轻烃未检出,而在实验后期有些高碳组分含量有降低趋势。
| Cn | Crude oil (×107) | 0 d (×107) | 2 d (×107) | 8 d (×107) | 20 d (×107) | 35 d (×107) |
| C9 | 0.10 | ND* | ND | ND | ND | ND |
| C10 | 0.15 | ND | ND | ND | ND | ND |
| C11 | 0.13 | ND | ND | ND | ND | ND |
| C12 | 0.24 | ND | ND | ND | ND | ND |
| C13 | 0.27 | 0.81 | ND | ND | ND | ND |
| C14 | 0.30 | 5.28 | ND | 0.39 | ND | ND |
| C15 | 0.31 | 14.90 | 0.58 | 1.53 | 0.60 | ND |
| C16 | 0.31 | 30.00 | 1.46 | 2.69 | 1.54 | 0.09 |
| C17 | 0.29 | 46.10 | 2.13 | 3.71 | 2.21 | 0.30 |
| Pr | 0.20 | 24.30 | 2.22 | 3.18 | 2.39 | 0.58 |
| C18 | 0.33 | 49.30 | 2.30 | 3.66 | 2.30 | 0.58 |
| Ph | 0.16 | 20.50 | 1.77 | 2.43 | 1.81 | 0.83 |
| C19 | 0.30 | 53.90 | 1.68 | 2.77 | 1.65 | 0.44 |
| C20 | 0.37 | 49.10 | 2.11 | 3.18 | 2.20 | 0.73 |
| C21 | 0.39 | 48.30 | 1.86 | 2.44 | 1.97 | 0.56 |
| C22 | 0.38 | 48.80 | 1.86 | 2.13 | 1.99 | 0.54 |
| C23 | 0.41 | 51.60 | 2.10 | 2.12 | 2.16 | 0.61 |
| C24 | 0.37 | 44.90 | 2.01 | 1.93 | 2.00 | 0.52 |
| C25 | 0.37 | 40.60 | 2.35 | 1.99 | 2.17 | 0.73 |
| C26 | 0.29 | 36.10 | 1.91 | 1.79 | 1.91 | 0.50 |
| C27 | 0.30 | 37.40 | 2.09 | 1.93 | 2.10 | 0.59 |
| C28 | 0.27 | 32.30 | 1.92 | 1.77 | 1.89 | 0.55 |
| C29 | 0.24 | 26.90 | 1.72 | 1.60 | 1.66 | 0.52 |
| C30 | 0.19 | 22.10 | 1.39 | 1.29 | 1.37 | 0.43 |
| C31 | 0.17 | 18.50 | 1.27 | 1.19 | 1.18 | 0.41 |
| C32 | 0.15 | 15.50 | 1.02 | 0.92 | 0.89 | 0.35 |
| C33 | 0.11 | 12.60 | 0.80 | 0.70 | 0.70 | 0.28 |
| C34 | 0.13 | 10.40 | 0.73 | ND | 0.70 | ND |
| C35 | 0.06 | ND | ND | ND | ND | ND |
| 注:ND:未监测到. Note: ND: Non-detection. | ||||||
微生物一般优先利用正构烷烃作为生长繁殖的碳源,因此在生物降解早期阶段或对轻度降解的原油而言,其Pr/Ph值相对保持不变;而Pr和Ph与相邻正构烷烃的比值(Pr/C17和Ph/C18)对于生物降解作用非常敏感,随着生物降解程度的加深,Pr/C17和Ph/C18值均呈现出增加的趋势。随着时间的推移,原油不断被降解,Pr/Ph在35 d约有50%的降低;Pr/C17从0 d开始稳定上升(表 2),表明微生物对正构烷烃的利用,而8 d却作为一个异常点出现了Pr/C17比值的降低,推测可能因为沉积物藻类生物量增加,从而生成C17、C18和C19,造成轻烃富集现象。
| Diagnostic ratio | Crude oil | 0 d | 2 d | 8 d | 20 d | 35 d |
| Pr/Ph | 1.27 | 1.18 | 1.26 | 1.31 | 1.32 | 0.70 |
| Pr/C17 | 0.71 | 0.53 | 1.04 | 0.86 | 1.08 | 1.92 |
| Ph/C18 | 0.48 | 0.42 | 0.77 | 0.66 | 0.79 | 1.44 |
将测序得到的序列进行筛选,去掉双峰序列和载体序列,所有组别第2天及第35天的序列共有596条,大部分序列可归类于蓝细菌,但这些序列中不可培养的占大多数(表 3)。所有处理组中共有11个不同的已知属被监测到,分别为Oscillatoria、Prochlorococcus、Phormidium、Microcystis、Xenococcus、Chroococcidiopsis、Phormidesmis、Cyanothece、Leptolyngbya、Acaryochloris和Synechococcus。丰度最高的属属于Chroococcales (CK-35:17条序列;LO-35:31条序列)。其他丰度较高的属为Oscillatoria (LO-35:25条序列)和Prochlorococcus (MO-35:13条序列)。
| Genus group | CK-2 | LO-2 | MO-2 | HO-2 | CK-35 | LO-35 | MO-35 | HO-35 |
| Chroococcales | 2 | 3 | 2 | 3 | 17 | 31 | 2 | 6 |
| Oscillatoria | 1 | 1 | 25 | |||||
| Prochlorococcus | 3 | 4 | 5 | 1 | 13 | 6 | ||
| Phormidium | 4 | 1 | 4 | 4 | 1 | 2 | ||
| Microcystis | 4 | |||||||
| Xenococcus | 5 | |||||||
| Chroococcidiopsis | 1 | 2 | ||||||
| Phormidesmis | 3 | |||||||
| Oscillatoriales | 1 | 1 | 1 | |||||
| Cyanothece | 2 | |||||||
| Leptolyngbya | 2 | |||||||
| Acaryochloris | 1 | |||||||
| Synechococcus | 1 | 5 | 3 | 4 | 2 | 1 | ||
| Uncultured | 60 | 56 | 54 | 54 | 58 | 19 | 56 | 59 |
| Total sequences | 75 | 75 | 72 | 75 | 75 | 75 | 74 | 75 |
| Simpson | 0.014 8 | 0.024 1 | 0.052 3 | 0.068 5 | 0.289 4 | 0.450 5 | 0.077 4 | 0.117 1 |
| Shannon | 3.77 | 3.60 | 3.20 | 2.99 | 1.45 | 0.96 | 2.74 | 2.61 |
| ACE | 113.32 (77.32-196.72) | 162.13 (97.18-311.16) | 102.32 (62.87-201.97) | 77.52 (48.13-160.47) | 15.00 (10.77-29.36) | 5.60 (5.097-8.72) | 57.86 (35.88-124.31) | 69.88 (41.70-148.65) |
| Coverage | 76.0% | 73.0% | 84.0% | 86.7% | 100% | 100% | 94.6% | 94.6% |
| 注:CK-2、LO-2、MO-2和HO-2分别为对照组、低浓度、中浓度和高浓度处理组第2天样品蓝细菌16S rRNA基因克隆文库结果;CK-35、LO-35、MO-35和HO-35分别为对照组、低浓度、中浓度和高浓度处理组第35天样品蓝细菌16S rRNA基因克隆文库结果. Note: CK-2, LO-2, MO-2 and HO-2 were derived from the control group, low oil-contaminated group, medium oil-contaminated group, and high oil-contaminated group on day 2, respectively; CK-35, LO-35, MO-35 and HO-35 were derived from the control group, low oil-contaminated group, medium oil-contaminated group, and high oil-contaminated group on day 35, respectively. | ||||||||
稀释曲线(图 2)表明所得序列很大程度上能够满足每个处理组中蓝细菌种属的分布情况,α多样性分析表明LO-35组具有最低的香农指数值(0.96) 和最高的辛普森指数值(0.45)。整体上看,第35天所有处理组与第2天相比,其香农指数下降而辛普森指数上升,其中变化最大的当属LO组。丰度(Richness)可用ACE指数进行说明,CK-35和LO-35有最低的ACE值,而后者更低。
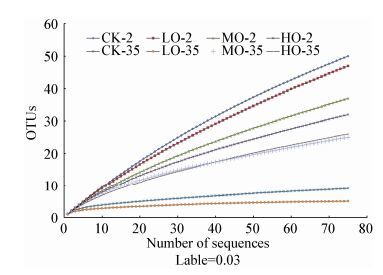
|
| 图 2 各组样品16S rRNA基因克隆文库稀释曲线 Figure 2 Rarefaction curve generated for 16S rRNA gene clone libraries from all groups 注:CK-2、LO-2、MO-2和HO-2分别为对照组、低浓度、中浓度和高浓度处理组第2天样品;CK-35、LO-35、MO-35和HO-35分别为对照组、低浓度、中浓度和高浓度处理组第35天样品. Note: CK-2, LO-2, MO-2 and HO-2 were derived from the control group, low oil-contaminated group, medium oil-contaminated group, and high oil-contaminated group on day 2, respectively; CK-35, LO-35, MO-35 and HO-35 were derived from the control group, low oil-contaminated group, medium oil-contaminated group, and high oil-contaminated group on day 35, respectively. |
|
|
从时间序列上看,从第2天到第35天,所有组别蓝细菌多样性都在下降,其中变化最明显的是对照组和LO组,虽然对照组中并未出现蓝细菌富集现象,但其对应的克隆文库结果显示LO组中已知种属多样性减少,似乎与LO组有相同的演替趋势。个别属如Oscillatoria丰度在LO组中增幅较大,预示该种属的蓝细菌在富集蓝细菌中可能占据主要地位。另外,在第35天时对照组多样性也大幅下降,但其中不可培养蓝细菌仍占大多数,而且没有监测到Oscillatoria的存在。
所有组别的序列共得到135个OTU,在第35天时各处理组包括对照组OTU个数相比第2天有很大减少。根据这些序列建立系统发育树(图 3),共可分为Uncultured cyanobacteria、Prochlorales、Chroococcales、Oscillatoriales和Pleurocapsales 5个部分。其中,不可培养种属占很大部分,在596条序列中,其比例可达69.8%。OTU1 (64条序列)和OTU2 (59条序列)序列条数最多,OTU7 (25条序列)独在LO-35中存在。由于OTU数目过多,部分OTU被折叠,其中,OTU1位于折叠部分。

|
| 图 3 在第2天和第35天4个组别中蓝细菌特异序列系统发育树 Figure 3 Phylogenetic tree of cyanobacterial specific sequences for four samples on day 2 and 35 注:CK-2、LO-2、MO-2和HO-2分别为对照组、低浓度、中浓度和高浓度处理组第2天样品;CK-35、LO-35、MO-35和HO-35分别为对照组、低浓度、中浓度和高浓度处理组第35天样品.系统树四周不同颜色柱及其对应的数字表示每个处理组中该OTU所占个数. Note: CK-2, LO-2, MO-2 and HO-2 were derived from the control group, low oil-contaminated group, medium oil-contaminated group, and high oil-contaminated group on day 2, respectively; CK-35, LO-35, MO-35 and HO-35 were derived from the control group, low oil-contaminated group, medium oil-contaminated group, and high oil-contaminated group on day 35, respectively. The numbers followed by colored columns indicate the numbers of OTU in each treatment. |
|
|
烷烃(n-alkanes)是原油的主要组分,在降解过程中一般遵循短链优先降解原则,短链烷烃(C11-C12)首先几乎降解完全时,长链烷烃依次被降解[19-20]。本研究所得结果与以往研究有所不同,在微宇宙培养后期,低碳数组分似乎降解程度不明显,此时沉积物表面蓝细菌富集,有研究表明蓝细菌暴发过程中会产生许多有机碳类物质,被异养微生物进一步降解[21],产生的有机物可能是一些轻烃含量增加的原因。
目前关于原油与蓝细菌的研究多停留在溢油环境中蓝细菌富集现象的揭示,而原油对蓝细菌是否具有抑制或刺激作用仍无定论,本研究结果表明有些蓝细菌种属可能对原油有很高的耐受性。本研究设置的原油浓度整体偏高,最低浓度(25 g/kg)也比一般研究浮游植物影响的浓度如Chronopoulou等[22](300 mL/m2)要高很多,但其中仍然有蓝细菌暴发。由此可见,蓝细菌对原油有较高的耐受性。但除LO组之外的其他组中蓝细菌丰度均低于对照组,且处于监测水平之下,由此可见过高的原油浓度对蓝细菌的毒性作用会大于生长促进作用[23]。Huang等研究认为,高浓度原油对浮游植物有毒害作用[24]。本研究发现在原油最低浓度下蓝细菌富集,而更高浓度原油处理组却未监测到,说明蓝细菌的暴发需要合适的原油浓度。
溢油会降低蓝细菌的多样性[25],本研究中对照组和原油添加组多样性均减少,推测可能是由于实验室微宇宙和原位条件的差异引起,但结果也表明原油的存在会引起部分蓝细菌种属丰度增加。Prochlorococcus和Synechococcus,尤其是前者,在第35天时,MO-35和HO-35组中其序列与第2天时相比增加很多,反而在LO-35组中没有监测到,表明Prochlorococcus对原油具有较高耐受性,这与Rigonato等通过16S rRNA基因分析所得Prochlorococcus和Synechococcus对溢油很敏感的结果相反[26],而González等研究结果也表明低浓度有机烃污染会对Prochlorococcus和Synechococcus的生长产生抑制作用[27],这可能与溢油时间长短及原油的种类不同有关,本实验周期相较于Rigonato等的要短很多,只有一个月,后者为25年,而González等[27]的实验周期只有72 h。沉积物表面发生蓝细菌富集的LO组中监测到最多的序列与Oscillatoria有较高相似性,Oscillatoria是典型的具有固氮功能的蓝细菌[28],也经常在溢油环境中被监测到,预示着这一种属可能在原油存在条件下发挥重要固氮作用,但仍需后续研究进一步确认。
本研究中通过微宇宙模拟实验发现蓝细菌对原油具有高耐受性,采用蓝细菌16S rRNA基因克隆文库分析表明,蓝细菌多样性在原油存在条件下降低,但也有不少种属丰度大幅增加,表明这些蓝细菌如Oscillatoria可能是组成富集蓝细菌的重要部分,也可能在自然环境溢油条件下发挥着重要功能。原核生物的单个基因组内常有16S rRNA基因的多样性[29],蓝细菌也不例外,在该文报道的2 013条细菌和古菌系列中共有44条蓝细菌基因序列,大部分菌株具有16S rRNA基因多拷贝,且各拷贝并不完全一样的有9个菌株。采用蓝细菌16S rRNA基因克隆文库的方法可能在一定程度上高估蓝细菌多样性和相对丰度。本研究选择16S rRNA V3-V4区来评估蓝细菌多样性,相对来说可降低对其基因多样性的高估[29]。
| [1] | Coulon F, Chronopoulou PM, Fahy A, et al. Hydrocarbon biodegradation in coastal mudflats: the central role of dynamic tidal biofilms dominated by aerobic hydrocarbonoclastic bacteria and diatoms[J]. Applied and Environmental Microbiology, 2012, 78(10) : 3638–3648. DOI:10.1128/AEM.00072-12 |
| [2] | Sumaila UR, Cisneros-Montemayor AM, Dyck A, et al. Impact of the Deepwater Horizon well blowout on the economics of US Gulf fisheries[J]. Canadian Journal of Fisheries and Aquatic Sciences, 2012, 69(3) : 499–510. DOI:10.1139/f2011-171 |
| [3] | Sorkhoh N, Al-Hasan R, Radwan S, et al. Self-cleaning of the Gulf[J]. Nature, 1992, 359(6391) : 109. |
| [4] | Cohen Y. Bioremediation of oil by marine microbial mats[J]. International Microbiology, 2002, 5(4) : 189–193. DOI:10.1007/s10123-002-0089-5 |
| [5] | Chaillan F, Gugger M, Saliot A, et al. Role of cyanobacteria in the biodegradation of crude oil by a tropical cyanobacterial mat[J]. Chemosphere, 2006, 62(10) : 1574–1582. DOI:10.1016/j.chemosphere.2005.06.050 |
| [6] | Narro ML, Cerniglia CE, van Baalen C, et al. Evidence for an NIH shift in oxidation of naphthalene by the marine cyanobacterium Oscillatoria sp. strain JCM[J]. Applied and Environmental Microbiolog, 1992, 58(4) : 1360–1363. |
| [7] | Raghukumar C, Vipparty V, David J, et al. Degradation of crude oil by marine cyanobacteria[J]. Applied Microbiology and Biotechnology, 2001, 57(3) : 433–436. DOI:10.1007/s002530100784 |
| [8] | Radwan SS, Al-Hasan RH. Oil pollution and cyanobacteria[A]//Whitton BA, Potts M. The Ecology of Cyanobacteria: Their Diversity in Time and Space[M]. Netherlands: Springer, 2002: 307-319 |
| [9] | de Philippis R, Vincenzini M. Exocellular polysaccharides from cyanobacteria and their possible applications[J]. FEMS Microbiology Reviews, 1998, 22(3) : 151–175. DOI:10.1111/j.1574-6976.1998.tb00365.x |
| [10] | Parsons ML, Morrison W, Rabalais NN, et al. Phytoplankton and the Macondo oil spill: a comparison of the 2010 phytoplankton assemblage to baseline conditions on the Louisiana shelf[J]. Environmental Pollution, 2015, 207 : 152–160. DOI:10.1016/j.envpol.2015.09.019 |
| [11] | Gilde K, Pinckney JL. Sublethal effects of crude oil on the community structure of estuarine phytoplankton[J]. Estuaries and Coasts, 2012, 35(3) : 853–861. DOI:10.1007/s12237-011-9473-8 |
| [12] | Abed RMM, Safi NMD, K ster J, et al. Microbial diversity of a heavily polluted microbial mat and its community changes following degradation of petroleum compounds[J]. Applied and Environmental Microbiology, 2002, 68(4) : 1674–1683. DOI:10.1128/AEM.68.4.1674-1683.2002 |
| [13] | Maidak BL, Larsen N, McCaughey MJ, et al. The ribosomal database project[J]. Nucleic Acids Research, 1994, 22(17) : 3485–3487. DOI:10.1093/nar/22.17.3485 |
| [14] | Benson DA, Boguski MS, Lipman DJ, et al. GenBank[J]. Nucleic Acids Research, 1999, 27(1) : 12–17. DOI:10.1093/nar/27.1.12 |
| [15] | Nübel U, Garcia-Pichel F, Muyzer G. PCR primers to amplify 16S rRNA genes from cyanobacteria[J]. Applied and Environmental Microbiology, 1997, 63(8) : 3327–3332. |
| [16] | Lin QX, Mendelssohn IA. Impacts and recovery of the Deepwater Horizon oil spill on vegetation structure and function of coastal salt marshes in the northern Gulf of Mexico[J]. Environmental Science & Technology, 2012, 46(7) : 3737–3743. |
| [17] | Schloss PD, Handelsman J. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness[J]. Applied and Environmental Microbiology, 2005, 71(3) : 1501–1506. DOI:10.1128/AEM.71.3.1501-1506.2005 |
| [18] | Tamura K, Peterson D, Peterson N, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods[J]. Molecular Biology and Evolution, 2011, 28(10) : 2731–2739. DOI:10.1093/molbev/msr121 |
| [19] | Harayama S, Kishira H, Kasai Y, et al. Petroleum biodegradation in marine environments[J]. Journal of Molecular Microbiology and Biotechnology, 1999, 1(1) : 63–70. |
| [20] | Sei K, Sugimoto Y, Mori K, et al. Monitoring of alkane-degrading bacteria in a sea-water microcosm during crude oil degradation by polymerase chain reaction based on alkane-catabolic genes[J]. Environmental Microbiology, 2003, 5(6) : 517–522. DOI:10.1046/j.1462-2920.2003.00447.x |
| [21] | Eiler A, Bertilsson S. Composition of freshwater bacterial communities associated with cyanobacterial blooms in four Swedish lakes[J]. Environmental Microbiology, 2004, 6(12) : 1228–1243. DOI:10.1111/emi.2004.6.issue-12 |
| [22] | Chronopoulou PM, Fahy A, Coulon F, et al. Impact of a simulated oil spill on benthic phototrophs and nitrogen-fixing bacteria in mudflat mesocosms[J]. Environmental Microbiology, 2013, 15(1) : 242–252. DOI:10.1111/emi.2013.15.issue-1 |
| [23] | D'souza NA, Subramaniam A, Juhl AR, et al. Elevated surface chlorophyll associated with natural oil seeps in the Gulf of Mexico[J]. Nature Geoscience, 2016, 9(3) : 215–218. DOI:10.1038/ngeo2631 |
| [24] | Huang YJ, Jiang ZB, Zeng JN, et al. The chronic effects of oil pollution on marine phytoplankton in a subtropical bay, China[J]. Environmental Monitoring and Assessment, 2011, 176(1/4) : 517–530. |
| [25] | Llirós M, Gaju N, de Oteyza TG, et al. Microcosm experiments of oil degradation by microbial mats. Ⅱ. The changes in microbial species[J]. Science of the Total Environment, 2008, 393(1) : 39–49. DOI:10.1016/j.scitotenv.2007.11.034 |
| [26] | Rigonato J, Kent AD, Alvarenga DO, et al. Drivers of cyanobacterial diversity and community composition in mangrove soils in south-east Brazil[J]. Environmental Microbiology, 2013, 15(4) : 1103–1114. DOI:10.1111/emi.2013.15.issue-4 |
| [27] | González J, Figueiras FG, Aranguren-Gassis M, et al. Effect of a simulated oil spill on natural assemblages of marine phytoplankton enclosed in microcosms[J]. Estuarine, Coastal and Shelf Science, 2009, 83(3) : 265–276. DOI:10.1016/j.ecss.2009.04.001 |
| [28] | Severin I, Stal LJ. NifH expression by five groups of phototrophs compared with nitrogenase activity in coastal microbial mats[J]. FEMS Microbiology Ecology, 2010, 73(1) : 55–67. |
| [29] | Sun DL, Jiang X, Wu QL, et al. Intragenomic heterogeneity of 16S rRNA genes causes overestimation of Prokaryotic diversity[J]. Applied and Environmental Microbiology, 2013, 79(19) : 5962–5969. DOI:10.1128/AEM.01282-13 |
 2017, Vol. 44
2017, Vol. 44




