扩展功能
文章信息
- Miao TIAN, Li-Juan ZHANG, Bo FU, Hong-Bo LIU, Ru-Bing ZHANG, He LIU
- 田淼, 张丽娟, 符波, 刘宏波, 张汝兵, 刘和
- Acetate production from H2/CO2 by mixed cultures from diverse ecosystems and their application for syngas fermentation
- 不同生境微生物转化H2/CO2产乙酸及其在合成气发酵中应用
- Microbiology China, 2017, 44(7): 1563-1578
- 微生物学通报, 2017, 44(7): 1563-1578
- DOI: 10.13344/j.microbiol.china.170084
-
文章历史
- Received: January 31, 2017
- Accepted: April 07, 2017
- Published online(www.cnki.net): April 20, 2017
2. Key Laboratory of Bio-based Material, Chinese Academy of Sciences, Qingdao Institute of Bioenergy and Bioprocess Technology, Qingdao, Shandong 266101, China
2. 中国科学院生物基材料重点实验室 中国科学院青岛生物能源与过程研究所 山东 青岛 266101
Syn(thesis) gas (primarily contains CO, H2 and CO2) has been used as a major feedstock in production of fuels and chemicals. Several laboratory-scale studies focused on acetate or ethanol production from syngas by pure cultures were done[1-2]. Mixed cultures biotechnology could become a more attractive addition or alternative to the traditional pure culture based approach[3] due to its several advantages as no sterilization requirements, more adaptive capacity under various conditions due to microbial diversity, and the possibility of a continuous process implementation[4]. Undefined mixed cultures from raw sludge and anaerobic granular sludge were used for fermentative CO conversion[5]. Singla et al.[6] enriched anaerobic mixed cultures for conversion of syngas to ethanol. H2/CO2 are essential syngas components and are also the products of fermentative CO conversion, however, little attention was paid on the conversion of H2/CO2 by mixed cultures approaches.
H2/CO2 can be biologically converted into chemicals and/or bioenergy such as acetate and ethanol by acetogens[7]. Ryan et al. enriched acetogens by supplying a variety of acetogenic growth substrates to two laboratory-scale high rate upflow anaerobic methanogenic sludge bed reactors operated at 37 ℃ and 55 ℃[8]. Alves et al. reported homoacetogenic communities enriched with a syngas mixture from thermophilic anaerobic sludge[9]. Our previous studies have shown different abundances of acetogens in different natural and engineered environmental samples[10]. However, in these studies, the potential of the undefined mixed cultures from diverse inoculums in bioconversion of H2/CO2 and its application in syngas fermentation was not studied. Furthermore, little information was given about the microbial composition of undefined mixed cultures from diverse inoculum types during the bioconversion of H2/CO2.
In the present study, the undefined mixed cultures from waste activated sludge, freshwater sediment, anaerobic methanogenic sludge and cow manure were used for H2/CO2 bioconversion. Formyltetrahydrofolate synthetase gene (fhs)-specific Real-time quantification PCR (qPCR) assay and Terminal restriction fragment length polymorphism (T-RFLP) analysis, 16S rRNA gene based 454 high-throughput pyrosequencing were also introduced to determine the diversity of the bacterial and acetogenic community during the incubation under H2/CO2. Anaerobic mixed culture obtained from cow manure were applied as inoculum for syngas fermentation.
2 Materials and Methods 2.1 Original samplesWaste activated sludge, freshwater sediment, anaerobic methanogenic sludge and cow manure samples were collected from dewatered sludge of a municipal wastewater treatment plant (Wuxi, China), Lihu Lake (Wuxi, China), a kitchen waste anaerobic digestion reactor (Suzhou, China) and a cattle farm (Wuxi, China), respectively. Basic characteristics of original samples were summarized in Table 1.
| Origin sources初始样品 | pH | TS (%) | VS (%) | COD (mg/L) | Protein (mg/L) | Polysaccharide (mg/L) |
| Waste activated sludge 剩余活性污泥 |
7.7±0.0 | 21.0±0.8 | 53.0±0.4 | 1 977.0±5.0 | 740.0±3.70 | 453.0±2.3 |
| Freshwater sediment 淡水河道底泥 |
6.2±0.0 | 60.0±1.0 | 5.0±0.1 | 108.0±3.5 | 17.0±0.90 | 35.0±0.2 |
| Anaerobic methanogenic sludge 厌氧产甲烷污泥 |
7.2±0.0 | 6.0±0.4 | 42.0±0.6 | 5 229.0±6.4 | 374.0±1.92 | 72.0±1.4 |
| Cow manure 牛粪 |
7.6±0.0 | 18.0±0.6 | 81.0±0.8 | 3 469.0±5.7 | 760.0±3.80 | 308.0±1.5 |
H2/CO2-incubation experiments were conducted in 1 000 mL serum bottles containing 500 mL culture medium[11] and 200 g of original samples. The culture medium consisted of the following basal salts (mmol/L): NH4Cl 9.35, KH2PO4 1.84, K2HPO4∙3H2O 1.10, MgCl2∙6H2O 1.48, FeCl3∙6H2O 0.09, NiSO4∙6H2O 0.06, CaCl2 0.23, ZnCl2 0.08, CoCl∙6H2O 0.04, CuCl2∙2H2O 0.03, and MnCl2∙4H2O 0.08. Oxygen was removed by purging the bottles with nitrogen gas for 5 minutes and 2-bromoethanesulfonate (50 mmol/L) was added to inhibit methanogenesis during incubation. H2/CO2 gas (4:1, V/V) (18.0 mmol/L H2 and 4.4 mmol/L CO2) was purged into headspace every day for substrates. The cultures were incubated at 37±1 ℃ for 30 days. All experiments were carried out in duplicate.
The optimization of trace metal ions and cysteine sulfide for enhanced ethanol production from H2/CO2 was conducted in 100 mL fermentation reactors with 50 mL liquid medium. The gas substrate containing CO2/H2 (80/20, V/V) was flushed daily into the reactor through an inlet steel needle to one standard atmosphere and oxygen was eliminated through an outlet steel needle. The experiments were carried out at 37 ℃. The concentration of metal ions were 0, 20.4, 204 μmol/L Fe2+ and 0, 6.96, 69.60 μmol/L Zn2+, respectively, while the concentration of cysteine sulfide were 0, 0.5, 1.0, 2.0 g/L. The control group was flushed with N2 to one standard atmosphere.
The syngas fermentation experiment was conducted in 250 mL fermentation reactors with 100 mL liquid medium. The gas substrate containing CO2/H2 (80/20, V/V) or CO/CO2/H2 (40/30/30, V/V/V) was flushed daily into the reactor through an inlet steel needle to one standard atmosphere. Oxygen was eliminated through an outlet steel needle. Syngas fermentation experiment was carried out at 37 ℃. 5 mol/L NaOH and 3 mol/L H3PO4 were used to adjust pH at 7.0. The control group was flushed with N2 to one standard atmosphere.
2.3 Chemical analysis methodspH values were measured by a pH meter (METTLER FE 20, Shanghai), chemical oxygen demand (COD), total solids (TS) and volatile solids (VS) were determined according to standard methods described by APHA[12]. Polysaccharides were quantified by phenol-sulfuric acid method using glucose as standard[13] and volatile fatty acids (VFAs) contents were measured every 3 days by a gas chromatograph (GC-2010, Shimadzu, Japan) equipped with an auto injector (AOC-20i, Shimadzu) using a fused-silica capillary column (PEG-20M, 30 m × 0.32 mm×0.5µm, China) as described by Liu et al.[14]. The VFA samples were filtrated through a 0.45 µm membrane before measurement. CO2 and H2 gas samples (0.05 mL) were taken with a gas-tight pressure lock syringe (Shimadzu, Japan) and quantified by gas chromatography (GC-2010, Shimadzu, Japan) equipped with a packed column Porapak Q (50/80 mesh) and a thermal conductivity detector (Shimadzu, Japan)[15].
2.4 DNA extraction2 mL of culture samples were collected every 6 days and centrifuged at 10 000×g for 20 min at 4 ℃. Total DNA was then extracted by MoBio PowerSoil DNA Isolation Kit (MoBio Laboratories, Carlsbad, CA) according to manufacturer's instructions. DNA samples were assayed spectrophotometrically by a NanoDrop-2000 system (Thermo Scientific, USA). DNA samples were stored at -20 ℃ prior to use.
2.5 qPCR assayFormyltetrahydrofolate synthetase fhs gene was chosen as an indicator for the abundance of acetogens during incubation. Copy numbers of fhs gene were measured by qPCR assay according to the description by Xu et al.[10].
Bacteria 16S rRNA genes were amplified by qPCR using forward primer 519F and reverse primer 907R for quantification of total bacteria. Each 25 µL reaction mixture contained 12.5 µL QuantiFast SYBR Green PCR Master Mix (2×), 0.25 µL of each primer (10 μmol/L), 2 µL of DNA sample and 10 µL purified water to complete the final volume. Amplification was carried out with a program consisting of an initial denaturation at 94 ℃ for 5 min; 35 cycles of 94 ℃ for 30 s, 50 ℃ for 30 s, and 72 ℃ for 30 s; and a final elongation cycle at 72 ℃ for 5 min.
qPCR assay was performed using a Rotor-Gene Q system (QIAGEN, Germany). All the PCR assays were carried out using the QuantiFast SYBR Green PCR Kit (QIAGEN, Germany) and reactions were modified based on the instructions of the manufacturer.
2.6 Terminal restriction fragment length polymorphism (T-RFLP) profilingAmplification of the 16S rRNA gene and selection of enzymes for the T-RFLP were performed in accordance with Xu et al.[15]. Bacteria 16S rRNA genes were amplified by qPCR using FAM labeled primer 27F and 1401R. Purified PCR products (10 µL) were digested with 1 µL of restriction enzyme Msp I (TaKaRa, Japan) in a total volume of 20 µL for 3 h at 37 ℃ followed by 10 min at 65 ℃. The digested products were analyzed by Shanghai GeneCore BioTechnologies Co., Ltd. T-RFLP profiles were analyzed on the basis of the peak size and area. Terminal restriction fragments (T-RFs) that differed by ±1 bp in different profiles were considered as identical. Relative abundance of each T-RF was the proportion of that single T-RF's peak area of all T-RFs' total peak area within one sample and the mean of the relative abundances of T-RFs are the results presented.
The Shannon-Wiener index (H′) of TRFLP-based species diversity was calculated by the following equation:

|
Where Pi represents the proportional area of T-RF i in the sum of peak areas in a given T-RFLP and i is the number of T-RFs of each T-RFLP pattern and s is the total number of T-RFs. The evenness (E) of TRFLP-based species diversity was calculated by the following equation:
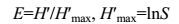
|
t test was performed using Microsoft Excel 2007, and the significance level (p) was determined.
2.7 16S rRNA gene based-454 pyrosequencingThe primers 515F (5′-GTGCCAGCMGCCGCG GTAA-3′) and 926R (5′-CCGTCAATTYYTTTRAG TTT-3′) were used to amplify approximately 412 bp of variable regions V4 to V5 of bacterial 16S rRNA genes. 454 pyrosequencing was accomplished with a Genome Sequencer FLX system (Shanghai Hanyu Bio-Tech Co., Ltd.). All 16S rRNA genes pyrosequencing reads were analyzed using Quantitative Insights Into Microbial Ecology (QIIME) according to protocols described in 454 overview tutorial: de novo OTU picking and diversity analyses using 454 data (http://qiime.org/tutorials/tutorial.html#picking-operational-taxonomic-units-otus-through-making-out-table)[16]. Barcode 16S rRNA gene sequences and information of primers were imported to filter different reads according to Phred quality scores. A minimum sequence length of 300 bp (no "N" was included in a sequence) and a minimum average quality score of 20 were selected as quality criteria. Sequence reads were clustered into operational taxonomic units (OTUs) at 97% sequence similarity using UCLUST and longest sequence in a cluster was used as the representative sequence for that OTU. Then the RDP classifier was applied to assign taxonomic data to each representative sequence[17]. Bacterial taxonomic classifications of our sequences were obtained by BLAST searching against the SILVA database[18]. Sequences were aligned against the Greengenes database[19] using Python Nearest Alignment Space Termination (PyNAST) tool[20].
For all bacterial OTUs at the family level, sequences belonged to the same family were considered as a group, and the sum of percentage of these sequences were considered to represent the relative abundance of that unique group. The 16S rRNA gene sequence analysis was carried out using the BLAST search program[21] against the GenBank and RDPII database[22]. The 16S rRNA gene sequences of OTU#174, OTU#235, OTU#279, OTU#832, OTU#1179, OTU#2347, OTU#2439 and OTU#2848 were submitted to the GenBank and the accession numbers are from KJ125514 to KJ125521. The phylogenetic analysis was conducted using MEGA version 5.0[23].
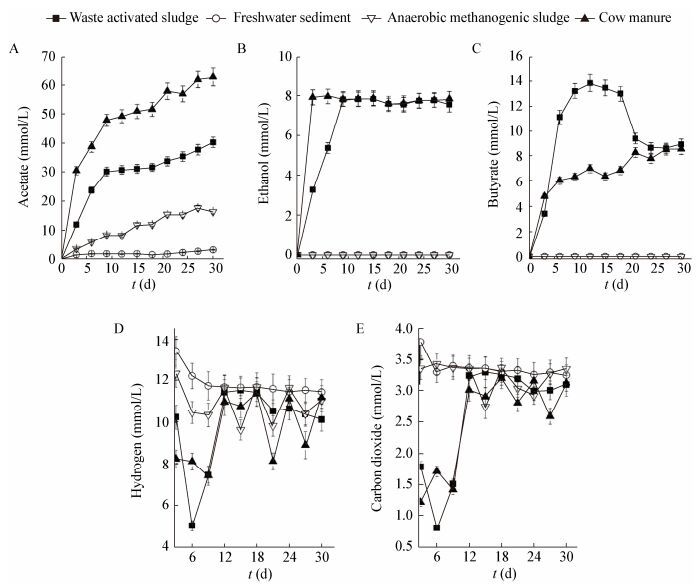
3 Results 3.1 VFAs production during H2/CO2-incubationAcetate, ethanol and butyrate are the main VFAs produced, with the four inocula showing different trends of acetate accumulation during the incubation (Figure 1A). Acetate accumulated in cow manure and waste activated sludge with final concentrations of approximately 63 mmol/L and 40 mmol/L, respectively, which were significantly higher than that of 3 mmol/L and 16 mmol/L in freshwater sediment and anaerobic methanogenic sludge. Acetogenic populations in freshwater sediment and anaerobic methanogenic sludge seemed to poorly develop using H2/CO2 as substrates.
|
| Figure 1 Acetate (A), ethanol (B), butyrate (C) production and H2 (D), CO2 (E) consumption during incubation under H2/CO2 图 1 H2/CO2培养过程中乙酸(A)、乙醇(B)和丁酸(C)生成及底物H2 (D)和CO2 (E)消耗 Note: Initial concentrations of H2, CO2 were 18.0 and 4.4 mmol/L. 注:H2和CO2的初始浓度分别为18.0 mmol/L和4.4 mmol/L. |
|
|
Ethanol productions from cow manure and waste activated sludge samples reached the highest level (around 8 mmol/L) before Day 3, while those from the other two groups were below detection limit (Figure 1B, E). Butyrate accumulations in waste activated sludge samples were concomitant with acetate accumulations. Butyrate concentration reached the highest level of 13 mmol/L on Day 9 and remained constant until Day 21, decreasing to about 8.6 mmol/L afterwards. In the meantime, acetate concentrations increased slightly (Figure 1A, C). In addition, butyrate accumulation or decline was not observed in cow manure samples grown on H2/CO2. The butyrate concentration in cow manure samples maintained at 6-8 mmol/L after 3 days of rapid accumulation (Figure 1C, F).
Substrate consumption during incubation was also monitored. As shown in Figure 1, hydrogen and CO2 consumption (Figure 1D, F) were in accordance with the VFA accumulation profile (Figure 1A-C). Substrates consumption by waste activated sludge and cow manure inocula were much greater than that of the other two groups. Substrates utilization rate increased initially but decreased significantly after 6 days of incubation. Freshwater sediment samples showed faster hydrogen consumption than anaerobic methanogenic sludge, and their CO2 consumption trends were similar. The trends of substrates consumption were highly alike with the products accumulation.
3.2 Abundances of acetogenic fhs gene during H2/CO2-incubationThe copy numbers of fhs genes and 16S rRNA genes were quantified by qPCR approach to measure the abundance of acetogenic bacteria and total bacteria. The initial copy numbers of acetogenic fhs gene in the original samples were (7.39±1.81)×105, (1.28±1.04)×105, (6.44±0.72)×106 and (3.14±0.48)×105 copies/g dry weight (DW) for waste activated sludge, freshwater sediment, anaerobic methanogenic sludge and cow manure, respectively. The quantities of fhs genes in these samples were similar to our previous study[10]. The quantities of fhs gene copy numbers in freshwater sediment and cow manure samples increased about 17 and 29 times at the end of incubation (Figure 2A), while the acetogenic proportions in total bacteria of these two groups reached 5.47%±0.12% and 4.69%±0.32%, which were approximately 5 and 10 times higher than that of the original samples (Figure 2B). The highest copy number of fhs genes occurred in anaerobic methanogenic sludge with (1.95±0.02)×107 copies/g DW (Figure 2A), but only about 3-fold increase of acetogens. The final copy numbers of fhs gene in the waste activated sludge was (5.10±0.58)×106 copies/g DW (Figure 2A). Furthermore, the acetogenic proportions in total bacteria in waste activated sludge and anaerobic methanogenic sludge incubations increased only around 3.2 fold after 30 days (Figure 2B). Considering that acetogenic proportion in total bacteria are about 0.6% to 0.9% in most natural anaerobic environments, this abundance was the upper detection limit of acetogenic detection using Most Probable Numeration as indicated by Harriott and Frazer[24].

|
| Figure 2 Copy numbers of fhs gene and its proportion in total bacteria of cultures using H2/CO2 图 2 H2/CO2培养物中fhs基因拷贝数及其占总细菌的比例 |
|
|
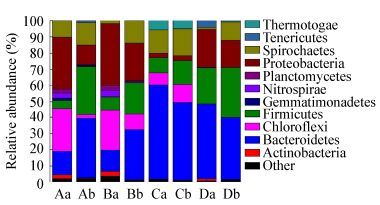
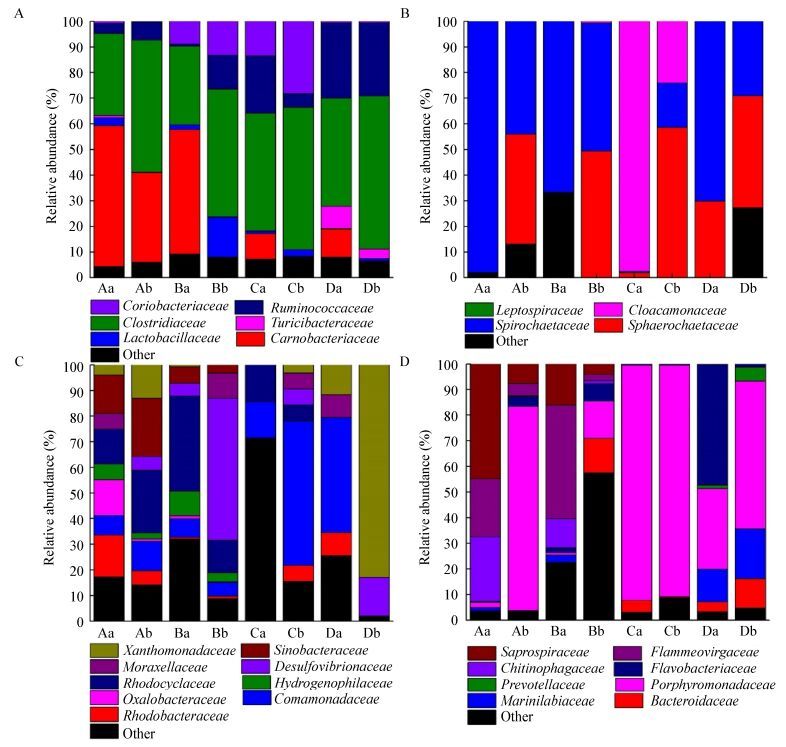
The most abundant sequences in the four samples belonged to the following phyla: Firmicutes, Spirochaetes, Proteobacteria, and Bacteroidetes, accounting for 62.1%-99.1% of the total reads, and other phyla represented smaller fractions (0.9%-37.9%) (Figure 3). The dominant family of Firmicutes shifted from Carnobacteriacea (55% of total Firmicutes) to Clostridiaceae (increase from 32% to 52% of Firmicutes) in waste activated sludge samples (Figure 4A). Similar patterns of community shift were also observed in freshwater sediment. The family of Carnobacteriacea accounted for 48% of total Firmicutes in the initial period, but Clostridiaceae became the dominant family at the end, increasing from 31% to 50% (Figure 4A). For anaerobic methanogenic sludge and cow manure, Clostridiaceae dominated in family Firmicutes throughout the incubation process, but Carnobacteriacea disappeared at the final period. Ruminococcaceae remained stable at about 28% of total Firmicutes in cow manure samples, while Clostridiaceae increased from 42.07% to 59.64% of total Firmicutes (Figure 4A). Bacterial community shift in the family Firmicutes suggested that the cultures were dominated by Clostridiaceae.
|
| Figure 3 Relative abundances of bacterial OTUs based on 454 sequencing of the V4-V5 regions of 16S rRNA genes at phylum level 图 3 基于16S rRNA基因V4-V5区454高通量测序的微生物OTUs门水平的相对丰度 Note: DNA samples were obtained from a: initial inocula; b: final enrichment vials containing H2/CO2; A: waste activated sludge; B: freshwater sediment; C: anaerobic methanogenic sludge; D: cow manure. 注:a:初始样品;b:H2/CO2培养结束时的样品;A:剩余活性污泥;B:淡水河道底泥;C:厌氧产甲烷污泥;D:牛粪. |
|
|
|
| Figure 4 Abundances of the phyla Firmicutes (A), Spirochaetes (B), Proteobacteria (C) and Bacteroidetes (D) 16S rRNA genes at the family level 图 4 厚壁菌门(A)、螺旋体门(B)、变形菌门(C)和拟杆菌门(D) 16S rRNA基因科水平的丰度 Note: DNA samples were obtained from a: initial inocula; b: Final cultures vials containing H2/CO2; A: Waste activated sludge; B: Freshwater sediment; C: Anaerobic methanogenic sludge; D: Cow manure. 注:a:初始DNA样品;b:H2/CO2培养结束时DNA样品;A:活性污泥;B:淡水河道底泥;C:厌氧产甲烷污泥;D:牛粪. |
|
|
Among the families belonged to Spirochaetes phylum, the Spirochaetaceae was dominant in the original waste activated sludge, fresh sediments samples and cow manure with proportions ranging from 67% to 98% of total Spirochaetes (Figure 4B). However, the ratio of this family decreased during the incubation process. The share of the family Sphaerochaetaceae significantly increased from 31.34% to 85.80% in the four cultures. The family of Protebacteria phylum was much richer than the other three phyla. Among the Family composition of Proteobacteria was quite different among the four origins (Figure 4C). Sulfur metabolism-associated OTUs belonging to Betaproteobacteria, Deltaproteobacteria, or Epsilonproteobacteria have been identified from all the samples. Considerable attention should be given to sulfate reducing bacteria populations belonging to Desulfovibrionaceae, which appeared in all the four samples.
75 acetogen-related OTUs were identified to represent significant increases of acetogenic species with the addition of H2/CO2 as substrate. Based on the acetogenic physiology, these OTUs were classified in three physiological groups: H2/CO2 utilizing acetogen, heterotrophic fermentative (often proteolytic) acetogen and autotrophic sulfate reducing acetogen. These physiological acetogenic groups, which are widely distributed at different levels of Firmicutes and Spriochaetes, showed clearly origin-related patterns.
OTU#514 shares 95% identity with heterotropic acetogenic, facultatively sulfur-reducing Sporanaerobacter acetigenes Lup 33[23], which abundance increased in waste activated sludge; while another OTU#666 (94% identical to Caloramator proteoclasticus DSM 12679)[25] showed a relatively higher abundance than OTU#514. OTU#5218 was much less abundant than OTU#514 and shares 92% identity with a fermentative acetogen Sporanaerobacter acetigenes Lup33 (DSM 13106)[26].
In the mixed cultures of anaerobic methanogenic sludge, OTU#5142, 5573, and 216 were 96%-99% identical to Atopobium parvulum DSM 20469[27], which dominated in the acetogenic community at the level of Clostridia class; while they were not detected in other samples. Atopobium parvulum has been reported as a proteolytic, fermentative bacterium associated with sulfur metabolism[27]. The genus Sphaerochaeta in the family Sphaerochaetaceae and the genus Treponema in the family Spirochaetaceae were two dominant populations in the pylum Spirochaetes in three samples, except for anaerobic methanogenic sludge. Instead of using H2/CO2, both Sphaerochaeta and Treponema are sugar utilizing fermentative acetogens.
OTU#5774 from freshwater sediment sample represented the most dominant species. It was probably belonged to Sulfuricurvum kujiense with 98% identity, a facultatively anaerobic, chemolithoautotrophic, sulfur-oxidizing bacterium[28]. These results suggested that sulfur metabolism had been highly activated at the end of incubation. There are two possible explanations for sulfate reducing bacterial growth: (1) Some autotrophic sulfate reducing bacteria could use H2/CO2 as electron donor and carbon source, and use remaining sulfate as electron acceptor for their growth; (2) The addition of 50 mmol/L 2-bromoethanesulfonate inhibits methanogens[15]; However, this significant amount of 2-bromoethanesulfonate also could possibly be used as carbon source/electron donor and electron acceptor to support sulfate reducing bacteria growth.
More importantly, OTU#2347 (84% identical to Acetoanaerobium noterae ATCC 35199)[29] represented common bacterial acetogens in waste activated sludge (Figure 5). In addition, three OTUs were only detected in waste activated sludge. They are OTU#174 and OUT#829, which shared 89% and 92% identity with Sporomusa malonica DSM 5090[30] and Sporomusa ovata strain H1[31], respectively, and represented the highly abundant taxa; and OTU#235, 94% identical with Clostridium carboxidivorans P7[32], which were much less abundant. Besides, OTU#7752 was representative in H2/CO2-utilized waste activated sludge, which was 93% identical with Clostridium methoxybenzovorans SR3[33]. Two acetogenic OTUs (#2848 and #279) were detected from anaerobic methanogenic sludge samples. OTU#2848 was closely related to Eubacterium limosum KIST612[34] with 16S rRNA gene sequence identity of 98%, while OTU#279 was 97% identical to Clostridium magnum. Clostridium magnum related OTU#1179 also was detected in H2/CO2 when freshwater sediment samples were utilized. Besides, OTU#2439, which was 99% identical to Acetobacterium carbinolicum strain VNs25[35], was the representative sequence in H2/CO2 utilized freshwater sediment. It was not detected from the other three kinds of samples. The OTU#6717, #4751 and #6920 were found with highest relative abundances in all the cow manure samples, which had sequence similarities of 90%, 96% and 95% to Clostridium ljungdahlii DSM 13528[36], Clostridium difficile 630[37] and Clostridium mayombei strain SFC-5[38], respectively. OTU#418 with sequence similarity of 92% to Clostridium drakei strain SL1[4] was present in original and H2/CO2-incubated cow manure, and its relative abundance increased. Specially, OTU#5908 and #4900 were close relatives of Treponema azotonutricium ZAS-9[39] and Oxobacter pfennigii strain V5-2[40] with 16S rRNA sequence identity of 89% and 90%, respectively.

|
| Figure 5 Maximum-likelihood phylogenetic tree of 16S rRNA gene sequences from putative acetogens grown on H2/CO2 or formate from waste activated sludge (red), freshwater sediment (blue), anaerobic methanogenic sludge (green) and cow manure (purple) and their closely related species 图 5 不同生境样品中产乙酸菌基于16S rRNA基因序列的系统发生树 Note: Red: Waste activated sludge; Blue: Freshwater sediment; Green: Anaerobic methanogenic sludge; Purple: Cow manure; The GenBank accession numbers appear in brackets; Numbers at internal nodes are the percentage of 100 bootstrap samples; Bar=0.1 represent distance scale. 注:红色:活性污泥;蓝色:淡水河道底泥;绿色:厌氧产甲烷污泥;紫色:牛粪;括号中为菌株序列号;每个节点的数字代表自举值;标尺0.1为距离标尺. |
|
|
Enzymes have an important effect on the metabolism of acetogens, and metal enzymes involves in acetyl-CoA such as carbon monoxide dehydrogenase (CODH) and hydrogenase all contain iron-sulfur protein[41]. Aldehyde:ferredoxin oxidoreductases (AORs) also contain ferredoxin. In addition, acetyl-CoA is catalyzed by alcohol dehydrogenase (ADH) to be reduced to ethanol in solventogenic Clostridiums, and ADH in Clostridiums also contains iron ion or zinc ion. In order to study the effect of trace metal ions on H2/CO2 conversion by mixed culture, different concentration of Fe2+ and Zn2+ were added to test their effects on acetate and ethanol production from H2/CO2 by mixed cultures from cow manure.
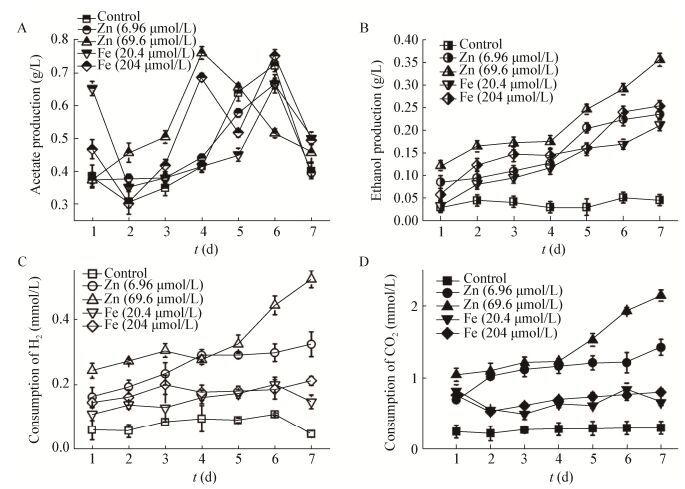
Addition of Zn2+ and Fe2+ promoted the utilization of gas substrate, as well as acetate and ethanol production from the conversion of H2/CO2 (Figure 6), and the increase of acetate yield and ethanol increased with the rise in Zn2+ and Fe2+ concentration. The maximum concentration of acetate and ethanol were 12.83 mmol/L and 7.70 mmol/L, respectively, when the Zn2+ concentration was 6.96 μmol/L. When the concentration of cysteine sulfide increased to 1.0 g/L and 2.0 g/L, the ethanol production increased significantly and the maximum yields were 6.09 mmol/L and 10.65 mmol/L, respectively (Figure 7A, B).
|
| Figure 6 Acetate (A), ethanol (B) accumulation and H2 (C), CO2 (D) consumption under different trace metal ion concentration 图 6 不同痕量金属浓度下乙酸(A)和乙醇(B)生成以及H2(C)和CO2(D)消耗 |
|
|

|
| Figure 7 Acetate (A), ethanol (B) accumulation and H2 (C), CO2 (D) consumption under cysteine sulfide concentration 图 7 不同半胱氨酸硫化物浓度下乙酸(A)和乙醇(B)生成以及H2(C)和CO2(D)消耗 |
|
|
The aim of the syngas fermentation was to examine the selected mixed culture for acetate and ethanol production by utilizing syngas as carbon source. The mixed culture from cow mature produced maximum ethanol and acetic acid during the incubation utilizing H2/CO2 as substrate (Figure 1), and was selected for syngas fermentation. The acetic acid and ethanol accumulations are shown in Figure 8A, B. The acetate and ethanol concentration increased gradually and then remained relatively stable. The final concentrations of acetate were 32.1, 97.1 and 55.4 mmol/L for control, H2/CO2 and syngas fermentation, respectively (Figure 8A), and those of ethanol were 1.78, 8.90 and 11.8 mmol/L respectively (Figure 8B). The average daily consumption of CO2 was 0.12 and 0.14 mmol for H2/CO2 and syngas fermentation, respectively (Figure 8C, D). During the H2/CO2-incubation, the H2 consumption gradually increased and reached 0.41 mmol at Day 7, and then decreased to 0.31 mmol at the end of the fermentation. For syngas fermentation, about 0.35 mmol H2 was consumed every day during the first 16 days, and then decreased to about 0.22 mmol until the end. The CO consumption gradually increased and reached 0.28 mmol at Day 11, and then decreased to 0.12 mmol at the end. The maximum ethanol production during syngas fermentation was 27.3% higher than the one from CO2/H2-incubation. It is suggested that the presence of CO contributes with ethanol production, which is in agreement with the previous results using pure culture as inoculum[42].

|
| Figure 8 Acetate (A) and ethanol (B) accumulation and gas substrates consumption of syngas-incubation (C) and CO2/H2-incubation (D) during syngas fermentation 图 8 合成气发酵过程中乙酸(A)和乙醇(B)的积累以及合成气(C)和H2/CO2(D)的消耗 |
|
|
In mixed culture from cow manure, the diversity indexes H′ for the bacterial communities in the initial samples was 2.98, which decreased to 2.73 and 2.59 after H2/CO2 and syngas fermentation, respectively, showing the reduction in diversity of bacterial community species after the H2/CO2 and syngas fermentation. Bacterial communities involved in syngas fermentation had lower species diversity than H2/CO2 incubation. The Shannon-Wiener index is influenced by both the richness and abundance of species in the community. T-RFLP-based H′ index increases with increasing number of T-RFs or even distribution of individual T-RFs. Most numbers of T-RFs were detected in the initial samples (14 T-RFs) with maximum species evenness (E values of 0.85). 9 T-RFs were detected in the samples of syngas fermentation with E values of 0.80. The samples of H2/CO2 incubation had higher species richness (10 T-RFs) and higher species evenness (E values of 0.83), which resulted in higher species diversity than that of syngas fermentation.
The response of bacterial community in cow manure samples to H2/CO2 and syngas were different. 27, 183 and 190 bp T-RFs accounted for abundance proportions of 22.4%, 17.1% and 27.8% in the populations of initial samples. The three T-RFs were also the main bacterial composition in samples of H2/CO2 incubation with abundance proportions of 25.5%, 15.3% and 28.7% (Figure 9). For cow manure using syngas as substrates, the proportion of 60 bp increased to about 15.1% comparing to 1.9% in the initial samples (Figure 9). Interestingly, T-RFs of 30 bp and 244 bp disappeared throughout the process of syngas fermentation (Figure 9).

|
| Figure 9 Microbial community dynamics as determined by the DNA-based T-RFLP in enrichment of cow manure during syngas fermentation 图 9 牛粪富集物合成气发酵过程中基于T-RFLP技术的微生物群落动态变化 |
|
|
Acetate was the main product of the H2/CO2 bioconversion for each inoculum (Figure 1A-C), which was in accordance with the increase in the number of acetogenic bacteria during H2/CO2-incubation experiments. The copy numbers of fhs genes and the acetogenic proportions in total bacteria increased after incubation under H2/CO2 headspace for each inoculum (Figure 2), indicating the increase of acetogenic number. Acetogens used CO2 as carbon source and H2 as electron donor to synthesize acetyl coenzyme A (acetyl-CoA) through acetyl-CoA pathway and then acetyl-CoA can be converted into acetate or ethanol[43]. 0.3 ATP was generated in the whole process of metabolism when acetyl-CoA was converted into acetate, and thus the energy was positive in this process. 0.1 ATP was needed when acetyl-CoA was converted into ethanol, being the energy balance of this metabolic pathway negative[44]. As a result, acetate production from H2/CO2 was more favorable than ethanol production, and the main product in the present study was acetate. However, ethanol can also be produced by reducing acetate through Aldehyde:ferredoxin oxidoreductases (AOR). Many acetogens such as Clostridium ljungdahlii and Eubacterium limosum KIST612 have been reported to have AORs[44], this maybe the reason for a small amount of ethanol formed in cow manure and waste activated sludge samples.
Relied on high throughput 16S rRNA genes pyrosequencing, the signature OTUs representing acetogenic bacteria from different samples were identified. Different acetogens have been obtained from four different ecosystems after H2/CO2 incubation. Diverse acetogenic bacteria were found in waste activated sludge and cow manure. The major putative acetogenic species belonged to Clostridium spp., Sporomusa malonica and Acetoanaerobium noterae in waste activated sludge, and those were Clostridium spp., Treponema azotonutricium and Oxobacter pfennigii in cow manure. Predominant putative acetogens in methanogenic sludge samples were the species Eubacterium limosum and Clostridium magnum. Acetogenic bacteria were composed also of two species Acetobacterium carbinolicum and Clostridium magnum. The above OTUs sequences provide molecular bioindicators for monitoring key species development and bacterial community dynamics during incubation. The results indicated that the acetogens composition in the studied cultures was decided by the sources of natural inocula under similar incubation condition.
When treated with H2/CO2, anaerobic methanogenic sludge inoculated group possessed the highest number of acetogens, while freshwater sediment inoculated group had the lowest number of that. Besides, cow manure and freshwater sediment inoculated groups achieved the biggest extent of growth (16.8 times and 29.3 times higher). Accordingly, highest proportions of acetogens in total bacteria were detected in these two groups (5.47% and 4.69%). VFAs concentrations, especially acetate, could be indicators to evaluate the capacity of acetogenic bacteria during incubation. Among the four cultures, highest yields of acetate, ethanol, and butyrate were produced in cow manure group after 30 days of incubation. The following was waste activated sludge group. The acetate production in anaerobic methanogenic sludge and freshwater sediment group was much less than in the former two groups. The results showed that the biggest amount of acetogens in mixed culture did not correspond to largest acetate accumulation. Intragenomic heterogeneity of 16S rRNA gene was reported to cause the overestimation of microbial diversity when using 16S rRNA gene-based methods, and overestimation was the least for the V4-V5 region under the dissimilarity level of 3%[45]. In the present study, variable regions V4-V5 of bacterial 16S rRNA genes were amplified through 454 pyrosequencing and sequence reads were clustered into operational taxonomic units (OTUs) at 97% sequence similarity. Although the application of 16S rRNA gene-based methods caused inevitable overestimation of bacterial diversity, this overestimation was minimized in this study, as suggested. Comparatively, acetogenic population of higher diversity with relatively higher content led to higher acetate production from H2/CO2 under the studied situation. Both the richness and number of acetogens in the mixed culture were key factors that regulate the capacity of acetogens to produce acetate.
As ADH contained iron or zinc, the increase of Zn2+ concentration might stimulate the activity of ADH, which catalyzes acetyl-CoA to produce ethanol in solventogenic Clostridiums. Similarly, the addition of Fe2+ also enhanced the yield of acetate and ethanol (Figure 7). The addition of Zn2+ promoted the growth of acetogens. Compared with the control, there was an increase of two orders of magnitude for the Zn2+ concentration of 69.6 μmol/L (Table 2). The existence of cysteine sulfide benefited ethanol and acetate production significantly. Cysteine sulfide reduced the redox potential as reductant[46] and it also provided electrons for acetogens, making the electron flow towards the direction of ethanol production[47]. In addition, cysteine sulfide stimulated the activity of some key enzymes such as CODH involved in acetyl-CoA pathway containing iron-sulfur protein[48]. Cysteine sulfide had no significant effect on the growth of acetogens (Table 2).
| Trace metal ion/ Cysteine sulfide 痕量金属/半胱氨酸硫化物 |
fhs gene copies numbers (copies/g) fhs基因拷贝数(copies/g) |
|
| Trace metal ion | 痕量金属 | |
| Control | 108.03 | |
| Zn2+ | 6.96 μmol/L | 1010.43 |
| 69.6 μmol/L | 108.23 | |
| Fe2+ | 20.4 μmol/L | 108.43 |
| 204 μmol/L | 107.18 | |
| Cysteine sulfide | 半胱氨酸硫化物 | |
| Control | 108.41 | |
| 0.5 g/L | 108.43 | |
| 1.0 g/L | 108.59 | |
| 2.0 g/L | 108.83 | |
The presence of CO is favorable to higher ethanol production and this may be related to the energetics of acetogens in acetyl-CoA pathway. CO can act as a carbon source as well as electron donor to provide reducing equivalents during the syngas fermentation[44]. When using H2/CO2 as energy and carbon source, the reaction is more favorable for the acetate production. 1.5 ATP was generated during converting acetyl-CoA into acetate using CO as electron donor, while 1.7 ATP was generated during converting acetyl-CoA into ethanol. In addition, reducing acetate to form ethanol through AOR generated 2.1 ATP[44].
5 ConclusionThe studied cow manure and waste activated sludge possess an ability to convert H2/CO2 to acetate, ethanol and butyrate. The major putative acetogens belonged to Clostridium spp., Sporomusa malonica and Acetoanaerobium noterae in waste activated sludge, and those were Clostridium spp., Treponema azotonutricium and Oxobacter pfennigii in cow manure. Diverse acetogens were found in waste activated sludge and cow manure. Both richness and number of acetogens in mixed cultures are important for regulating acetogenesis during the bioconversion of H2/CO2. The addition of trace metal ions (Fe2+ and Zn2+) and cysteine sulfide effectively improved ethanol production during the conversion of H2/CO2 by mixed culture from cow manure. Mixed culture obtained from cow manure had potential for acetate and ethanol production from syngas fermentation.
| [1] | Mohammadi M, Younesi H, Najafpour G, et al. Sustainable ethanol fermentation from synthesis gas by Clostridium ljungdahlii in a continuous stirred tank bioreactor[J]. Journal of Chemical Technology and Biotechnology, 2012, 87(6): 837–843. DOI:10.1002/jctb.3712 |
| [2] | Abubackar HN, Veiga MC, Kennes C. Biological conversion of carbon monoxide: rich syngas or waste gases to bioethanol[J]. Biofuels, Bioproducts and Biorefining, 2011, 5(1): 93–114. DOI:10.1002/bbb.256 |
| [3] | Ni BJ, Rittmann BE, Yu HQ. Soluble microbial products and their implications in mixed culture biotechnology[J]. Trends in biotechnology, 2011, 29(9): 454–463. DOI:10.1016/j.tibtech.2011.04.006 |
| [4] | Kö pke M, Mihalcea C, Bromley JC, et al. Fermentative production of ethanol from carbon monoxide[J]. Current opinion in biotechnology, 2011, 22(3): 320–325. DOI:10.1016/j.copbio.2011.01.005 |
| [5] | Liu YF, Wan JJ, Han S, et al. Selective conversion of carbon monoxide to hydrogen by anaerobic mixed culture[J]. Bioresource Technology, 2016, 202: 1–7. DOI:10.1016/j.biortech.2015.11.071 |
| [6] | Singla A, Verma D, Lal B, et al. Enrichment and optimization of anaerobic bacterial mixed culture for conversion of syngas to ethanol[J]. Bioresource Technology, 2014, 172: 41–49. DOI:10.1016/j.biortech.2014.08.083 |
| [7] | Fast AG, Schmidt ED, Jones SW, et al. Acetogenic mixotrophy: novel options for yield improvement in biofuels and biochemicals production[J]. Current Opinion in Biotechnology, 2015, 33: 60–72. DOI:10.1016/j.copbio.2014.11.014 |
| [8] | Ryan P, Forbes C, McHugh S, et al. Enrichment of acetogenic bacteria in high rate anaerobic reactors under mesophilic and thermophilic conditions[J]. Water Research, 2010, 44(14): 4261–4269. DOI:10.1016/j.watres.2010.05.033 |
| [9] | Alves JI, Stams AJM, Plugge CM, et al. Enrichment of anaerobic syngas-converting bacteria from thermophilic bioreactor sludge[J]. FEMS Microbiology Ecology, 2013, 86(3): 590–597. DOI:10.1111/fem.2013.86.issue-3 |
| [10] | Xu KW, Liu H, Du GC, et al. Real-time PCR assays targeting formyltetrahydrofolate synthetase gene to enumerate acetogens in natural and engineered environments[J]. Anaerobe, 2009, 15(5): 204–213. DOI:10.1016/j.anaerobe.2009.03.005 |
| [11] | Oh SE, van Ginkel S, Logan BE. The relative effectiveness of pH control and heat treatment for enhancing biohydrogen gas production[J]. Environmental Science & Technology, 2003, 37(22): 5186–5190. |
| [12] | American Public Health Association. Standard Methods for the Examination of Water and Wastewater[M]. Washington DC, USA: American Public Health Association (APHA), 2005 American Public Health Association. Standard Methods for the Examination of Water and Wastewater[M]. Washington DC, USA: American Public Health Association (APHA), 2005 |
| [13] | DuBois M, Gilles KA, Hamilton JK, et al. Colorimetric method for determination of sugars and related substances[J]. Analytical Chemistry, 1956, 28(3): 350–356. DOI:10.1021/ac60111a017 |
| [14] | Liu XL, Liu H, Chen YY, et al. Effects of organic matter and initial carbon-nitrogen ratio on the bioconversion of volatile fatty acids from sewage sludge[J]. Journal of Chemical Technology and Biotechnology, 2008, 83(7): 1049–1055. DOI:10.1002/(ISSN)1097-4660 |
| [15] | Xu KW, Liu H, Chen J. Effect of classic methanogenic inhibitors on the quantity and diversity of archaeal community and the reductive homoacetogenic activity during the process of anaerobic sludge digestion[J]. Bioresource Technology, 2010, 101(8): 2600–2607. DOI:10.1016/j.biortech.2009.10.059 |
| [16] | Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data[J]. Nature Methods, 2010, 7(5): 335–336. DOI:10.1038/nmeth.f.303 |
| [17] | Cole JR, Wang Q, Cardenas E, et al. The ribosomal database project: improved alignments and new tools for rRNA analysis[J]. Nucleic Acids Research, 2009, 37(S1): D141–D145. |
| [18] | Russell JA, Moreau CS, Goldman-Huertas B, et al. Bacterial gut symbionts are tightly linked with the evolution of herbivory in ants[J]. Proceedings of the National Academy of Sciences of the United States of America, 2009, 106(50): 21236–21241. DOI:10.1073/pnas.0907926106 |
| [19] | DeSantis TZ, Hugenholtz P, Larsen N, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB[J]. Applied and Environmental Microbiology, 2006, 72(7): 5069–5072. DOI:10.1128/AEM.03006-05 |
| [20] | Caporaso JG, Bittinger K, Bushman FD, et al. PyNAST: a flexible tool for aligning sequences to a template alignment[J]. Bioinformatics, 2010, 26(2): 266–267. DOI:10.1093/bioinformatics/btp636 |
| [21] | Altschul SF, Madden TL, Schä ffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs[J]. Nucleic Acids Research, 1997, 25(17): 3389–3402. DOI:10.1093/nar/25.17.3389 |
| [22] | Wang Q, Garrity GM, Tiedje JM, et al. Naï ve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy[J]. Applied and Environmental Microbiology, 2007, 73(16): 5261–5267. DOI:10.1128/AEM.00062-07 |
| [23] | Tamura K, Peterson D, Peterson N, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods[J]. Molecular Biology and Evolution, 2011, 28(10): 2731–2739. DOI:10.1093/molbev/msr121 |
| [24] | Harriott OT, Frazer AC. Enumeration of acetogens by a colorimetric most-probable-number assay[J]. Applied and Environmental Microbiology, 1997, 63(1): 296–300. |
| [25] | Tarlera S, Muxí L, Soubes M, et al. Caloramator proteoclasticus sp. nov., a new moderately thermophilic anaerobic proteolytic bacterium[J]. International Journal of Systematic and Evolutionary Microbiology, 1997, 47(3): 651–656. |
| [26] | Hernandez-Eugenio G, Fardeau ML, Cayol JL, et al. Sporanaerobacter acetigenes gen. nov., sp. nov., a novel acetogenic, facultatively sulfur-reducing bacterium[J]. International Journal of Systematic and Evolutionary Microbiology, 2002, 52(4): 1217–1223. |
| [27] | Copeland A, Sikorski J, Lapidus A, et al. Complete genome sequence of Atopobium parvulum type strain (IPP 1246T)[J]. Standards in Genomic Sciences, 2009, 1(2): 166–173. DOI:10.4056/sigs.29547 |
| [28] | Harms C, Schleicher A, Collins MD, et al. Tissierella creatinophila sp. nov., a Gram-positive, anaerobic, non-spore-forming, creatinine-fermenting organism[J]. International Journal of Systematic and Evolutionary Microbiology, 1998, 48(3): 983–993. |
| [29] | Sleat R, Mah RA, Robinson R. Acetoanaerobium noterae gen. nov., sp. nov.: an anaerobic bacterium that forms acetate from H2 and CO2[J]. International Journal of Systematic and Evolutionary Microbiology, 1985, 35(1): 10–15. |
| [30] | Biebl H, Schwab-Hanisch H, Sprö er C, et al. Propionispora vibrioides, nov. gen., nov. sp., a new gram-negative, spore-forming anaerobe that ferments sugar alcohols[J]. Archives of Microbiology, 2000, 174(4): 239–247. DOI:10.1007/s002030000198 |
| [31] | Stupperich E, Konle R. Corrinoid-dependent methyl transfer reactions are involved in methanol and 3, 4-dimethoxybenzoate metabolism by Sporomusa ovata[J]. Applied and Environmental Microbiology, 1993, 59(9): 3110–3116. |
| [32] | Ahmed A, Cateni BG, Huhnke RL, et al. Effects of biomass-generated producer gas constituents on cell growth, product distribution and hydrogenase activity of Clostridium carboxidivorans P7T[J]. Biomass and Bioenergy, 2006, 30(7): 665–672. DOI:10.1016/j.biombioe.2006.01.007 |
| [33] | Mechichi T, Labat M, Patel BKC, et al. Clostridium methoxybenzovorans sp. nov., a new aromatic o-demethylating homoacetogen from an olive mill wastewater treatment digester[J]. International Journal of Systematic and Evolutionary Microbiology, 1999, 49(3): 1201–1209. |
| [34] | Roh H, Ko HJ, Kim D, et al. Complete genome sequence of a carbon monoxide-utilizing acetogen, Eubacterium limosum KIST612[J]. Journal of Bacteriology, 2011, 193(1): 307–308. DOI:10.1128/JB.01217-10 |
| [35] | Basso O, Lascourreges JF, Le Borgne F, et al. Characterization by culture and molecular analysis of the microbial diversity of a deep subsurface gas storage aquifer[J]. Research in Microbiology, 2009, 160(2): 107–116. DOI:10.1016/j.resmic.2008.10.010 |
| [36] | Nevin KP, Hensley SA, Franks AE, et al. Electrosynthesis of organic compounds from carbon dioxide is catalyzed by a diversity of acetogenic microorganisms[J]. Applied and Environmental Microbiology, 2011, 77(9): 2882–2886. DOI:10.1128/AEM.02642-10 |
| [37] | Farrow KA, Lyras D, Rood JI. The macrolide-lincosamide-streptogramin B resistance determinant from Clostridium difficile 630 contains two erm (B) genes[J]. Antimicrobial Agents and Chemotherapy, 2000, 44(2): 411–413. DOI:10.1128/AAC.44.2.411-413.2000 |
| [38] | Kane MD, Brauman A, Breznak JA. Clostridium mayombei sp. nov., an H2/CO2 acetogenic bacterium from the gut of the African soil-feeding termite, Cubitermes speciosus[J]. Archives of Microbiology, 1991, 156(2): 99–104. DOI:10.1007/BF00290980 |
| [39] | Drö ge S, Rachel R, Radek R, et al. Treponema isoptericolens sp. nov., a novel spirochaete from the hindgut of the termite Incisitermes tabogae[J]. International Journal of Systematic and Evolutionary Microbiology, 2008, 58(5): 1079–1083. DOI:10.1099/ijs.0.64699-0 |
| [40] | Sipma J, Henstra AM, Parshina SN, et al. Microbial CO conversions with applications in synthesis gas purification and bio-desulfurization[J]. Critical Reviews in Biotechnology, 2006, 26(1): 41–65. DOI:10.1080/07388550500513974 |
| [41] | Pandelia ME, Ogata H, Currell LJ, et al. Inhibition of the[NiFe] hydrogenase from Desulfovibrio vulgaris Miyazaki F by carbon monoxide: an FTIR and EPR spectroscopic study[J]. Biochimica et Biophysica Acta (BBA)-Bioenergetics, 2010, 1797(2): 304–313. DOI:10.1016/j.bbabio.2009.11.002 |
| [42] | Yun SI, Gang SJ, Ro HM, et al. Tracing carbon monoxide uptake by Clostridium ljungdahlii during ethanol fermentation using 13C-enrichment technique[J]. Bioprocess and Biosystems Engineering, 2013, 36(5): 591–595. DOI:10.1007/s00449-012-0815-0 |
| [43] | Diender M, Stams AJM, Sousa DZ. Pathways and bioenergetics of anaerobic carbon monoxide fermentation[J]. Frontiers in Microbiology, 2015, 6: 1275. |
| [44] | Bertsch J, Müller V. Bioenergetic constraints for conversion of syngas to biofuels in acetogenic bacteria[J]. Biotechnology for Biofuels, 2015, 8(1): 210. DOI:10.1186/s13068-015-0393-x |
| [45] | Sun DL, Jiang X, Wu QL, et al. Intragenomic heterogeneity of 16S rRNA genes causes overestimation of prokaryotic diversity[J]. Applied and Environmental Microbiology, 2013, 79(19): 5962–5969. DOI:10.1128/AEM.01282-13 |
| [46] | Sim JH, Kamaruddin AH. Optimization of acetic acid production from synthesis gas by chemolithotrophic bacterium — Clostridium aceticum using statistical approach[J]. Bioresource Technology, 2008, 99(8): 2724–2735. DOI:10.1016/j.biortech.2007.07.004 |
| [47] | Abubackar HN, Veiga MC, Kennes C. Biological conversion of carbon monoxide to ethanol: effect of pH, gas pressure, reducing agent and yeast extract[J]. Bioresource Technology, 2012, 114: 518–522. DOI:10.1016/j.biortech.2012.03.027 |
| [48] | Gao J, Atiyeh HK, Phillips JR, et al. Development of low cost medium for ethanol production from syngas by Clostridium ragsdalei[J]. Bioresource Technology, 2013, 147: 508–515. DOI:10.1016/j.biortech.2013.08.075 |
 2017, Vol. 44
2017, Vol. 44




