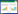扩展功能
文章信息
- 王聪, 曾南方, 刘飔雨, 苗琦, 周磊, 盖新娜, 杨汉春, 郭鑫
- WANG Cong, ZENG Nan-Fang, LIU Si-Yu, MIAO Qi, ZHOU Lei, GE Xin-Na, YANG Han-Chun, GUO Xin
- PIAS1对PRRSV N蛋白SUMO化修饰及病毒复制的影响
- Effects of PIAS1 on sumoylation of PRRSV N protein and viral replication
- 微生物学通报, 2017, 44(12): 2878-2887
- Microbiology China, 2017, 44(12): 2878-2887
- DOI: 10.13344/j.microbiol.china.170476
-
文章历史
- 收稿日期: 2017-07-02
- 接受日期: 2017-09-04
- 优先数字出版日期(www.cnki.net): 2017-09-07
猪繁殖与呼吸综合征(Porcine reproductive and respiratory syndrome,PRRS)是由猪繁殖与呼吸综合征病毒(Porcine reproductive and respiratory syndrome virus,PRRSV)引起的一种以母猪繁殖障碍和仔猪呼吸道疾病为特征的重要猪病[1-2]。2006年,以高热、高发病率和高死亡率及妊娠母猪严重的繁殖障碍为特征的高致病性PRRS (highly pathogenic PRRS,HP-PRRS)在我国流行,给养猪业造成巨大经济损失[3]。PRRSV属套式病毒目(Nidovirales)动脉炎病毒科(Arteriviridae)动脉炎病毒属(Arterivirus)[4],分为以LV (Lelystad virus)为代表的基因Ⅰ型(Type 1)和以VR-2332为代表的基因Ⅱ型(Type 2)[5-6]。PRRSV基因组为单股正链RNA,全长约15 kb,至少包括12个开放阅读框[7-9]。ORF1a和ORF1b编码病毒复制酶多聚蛋白pp1a和pp1ab,其余ORFs编码病毒的结构蛋白[10-12]。N蛋白是PRRSV感染宿主后表达量最高的结构蛋白,免疫原性强并且对PRRSV的生命周期具有重要影响,还可通过自身的共价与非共价相互作用为病毒衣壳的组装提供基础[13]。
SUMO (Small ubiquitin-like modifier)化修饰作为蛋白类泛素化修饰的一种方式,于20世纪90年代被发现,在真核细胞的生命进程中发挥着多种重要作用,如RNA加工、染色质重塑、细胞分裂、核质转移、转录调控等[14]。SUMO化修饰是由E1激活酶、E2结合酶和E3连接酶共同参与完成的一种动态可逆的修饰过程[15-16]。PIAS1属于SP-RING结构域SUMO E3连接酶家族,该家族主要包括人的PIAS (protein inhibitor of activated STAT,有PIAS1、PIAS3、PIASxα/β和PIASy)蛋白[17]及它们的酵母同系物。PIAS1是以STAT1为诱饵利用酵母双杂交方法筛库发现的[18]。PIAS1在转录调控、细胞周期、细胞凋亡及肿瘤发生中发挥着重要的生物学功能。有研究表明,PIAS1可以与A型流感病毒PB2蛋白相互作用抑制病毒的复制[19];PIAS1还可以与人巨细胞病毒IE2蛋白相互作用增强IE2的SUMO化修饰水平[20]。研究人员利用酵母双杂交系统筛选出EB病毒(Epstein-barr virus,EBV)的即刻早期蛋白Rta,发现其可与PIAS1相互作用,且PIAS1可以增强SUMO1与Rta的结合[21]。
目前关于PRRSV的各结构蛋白及非结构蛋白能否被SUMO化修饰的研究还鲜有报道,本实验室前期研究表明PRRSV N蛋白可与Ubc9发生相互作用,并且已证实N蛋白在体外转染和PRRSV感染的情况下皆可发生SUMO化修饰(另文发表)。2012年,Wang等在利用酵母双杂交试验筛选与PRRSV N蛋白相互作用的宿主蛋白时发现PIAS1是候选宿主蛋白之一[22],但对其在调控PRRSV复制及N蛋白SUMO化修饰中的作用并未涉及。研究PIAS1与PRRSV N蛋白的相互作用可为解析PRRSV复制调控和致病分子机制提供科学依据。
1 材料与方法 1.1 主要试剂和仪器MARC-145、HEK 293T和HEK 293FT细胞由农业部动物流行病学重点实验室(以下简称为本实验室)保存;原代猪肺泡巨噬细胞(PAMs)由本实验室灌洗、冻存于液氮;重组质粒pGBKT7-N由本实验室构建、保存;PIAS1 siRNA由上海吉玛制药技术有限公司合成;PRRSV N蛋白单克隆抗体由南京农业大学姜平教授赠送;HA鼠源单克隆抗体(H3663)和Myc兔源多克隆抗体(C3956),Sigma-Aldrich公司;HA兔源单克隆抗体(3724S),CST公司;SUMO2/3兔源多克隆抗体(ab81371),Abcam公司;HRP标记的山羊抗鼠IgG (ZB-2305)和山羊抗兔IgG (ZB-2301),北京中杉金桥生物技术有限公司;KOD FX Neo DNA聚合酶,东洋纺公司;M-MLV反转录酶和FuGENE® HD转染试剂,Promega公司;Trizol™ Reagent、DNA转染试剂(Lipofectamine® LTX and PLUS™ Reagents)和siRNA转染试剂(Lipofectamine™ RNAiMax),Invitrogen公司。2720 Thermal Cycler PCR仪,美国Applied Biosystems公司;Nikon Eclipse Ti-U型倒置荧光显微镜,日本Nikon公司。
1.2 引物设计根据人PIAS1基因序列(GenBank登录号为NM_016166)分别设计重组质粒pCMV-Myc-PIAS1、pGADT7-PIAS1和pWPXL-PIAS1特异性引物,由北京三博远志生物技术有限责任公司合成,引物序列见表 1。
| 引物名称 Primer name |
序列 Sequence (5′→3′) |
| pCMV-Myc-PIAS1 F | CGGAATTCATGGCGGACAGTGCGGAACTAAAGCAAATG |
| pCMV-Myc-PIAS1 R | GGGGTACCGTCCAATGAAATAATGTCTGGTATGATGCCA |
| pGADT7-PIAS1 F | CGGAATTCATGGCGGACAGTGCGGAACTAAAGCAAATG |
| pGADT7-PIAS1 R | CCCTCGAGAGATGGCGGACAGTGCGGAACTAAAGCAAATG |
| pWPXL-PIAS1 F | GGGTTTAAACATGGCGGACAGTGCGGAACTAAAGCAAATG |
| pWPXL-PIAS1 R | CGACGCGTTTAGATGGCGGACAGTGCGGAACTAAAGCAAATG |
| 注:斜体代表保护性碱基;下划线代表限制性酶切位点. Note: Italics represent protective bases; underlines represent restriction sites. |
|
总RNA的提取:按照Trizol™ Reagent试剂盒说明书操作。
反转录:参照M-MLV反转录步骤。建立25 μL反转录体系,42 ℃反转录1−2 h后产物−20 ℃保存备用。
PCR反应体系:2×KOD buffer 25 μL,dNTPs (2 mmol/L) 10 μL,上下游引物(10 pmol/μL)各1.5 μL,KOD FX (1.0 U/μL) 1.0 μL,cDNA 2 μL,无菌去离子水补至50 μL。PCR反应条件:94 ℃ 2 min;98 ℃ 10 s,55 ℃ 30 s,68 ℃ 1 min,35个循环;68 ℃ 7 min。
1.4 酵母回复杂交将重组表达质粒pGBKT7-N和pGADT7-PIAS1各100 ng共转化到制备好的酵母菌株Y2HGold感受态细胞中,同时设立阳性对照组:pGBKT7-53+pGADT7-T;阴性对照组:pGBKT7-N+pGADT7和pGBKT7+pGADT7-PIAS1。酵母感受态制备及具体实验操作参见Clontech说明书。
1.5 体外转染、激光共聚焦和免疫共沉淀根据不同试验要求,采用Lipofectamine® LTX and PLUS™ Reagents转染试剂转染,具体操作按说明书进行,根据实验要求在转染后不同时间对细胞进行处理。
在长有HEK 293T细胞的24孔细胞培养板中共转pCMV-HA-N和pCMV-Myc-PIAS1,同时,用PRRSV (MOI=0.01)感染MARC-145细胞和原代PAMs细胞。在转染或感染36 h后,用预冷的无水乙醇固定20 min,分别孵育相应一抗2 h,PBS洗涤3次后,分别用FITC标记的羊抗鼠IgG和TRITC标记的羊抗兔IgG于37 ℃孵育1 h,DAPI染核,激光共聚焦显微镜下观察。
1.6 siRNA干扰PIAS1对病毒增殖的影响siRNA设计与合成:针对PIAS1基因的3个靶点设计siRNA。siRNA由上海吉玛公司设计和合成,序列见表 2。
| siRNA | 序列Sequence (5′→3′) |
| siRNA-683 | CAGCAAAUCAGUAGUUCCAUGGAUATT |
| UAUCCAUGGAACUACUGAUUUGCUGTT | |
| siRNA-1057 | GAAUCCGGAUCAUUCUAGAGCUUUATT |
| UAAAGCUCUAGAAUGAUCCGGAUUCTT | |
| siRNA-1685 | GCUGUAGACACAAGCUACAUUAAUATT |
| UAUUAAUGUAGCUUGUGUCUACAGCTT |
siRNA转染:将MARC-145细胞接种于6孔细胞培养板中,在37 ℃、5% CO2培养箱中培养,待单层细胞长至70%−80%,按照Lipofectamine™ RNAiMax说明书转染siRNA。用Western blotting方法检测PIAS1基因的沉默效果,确定最佳的干扰浓度。随后,将MARC-145细胞接种于24孔细胞培养板中,选择干扰效果最佳的siRNA进行干扰,48 h后弃掉上清,无菌PBS洗涤3遍,细胞感染PRRSV (MOI=0.01),并于感染后12、24、36、48、60、72 h收集上清及细胞样品,置于−80 ℃反复冻融3次,10 000 r/min离心10 min,收集上清,进行TCID50测定。
1.7 慢病毒包装及转导MARC-145细胞 1.7.1 慢病毒包装质粒转染: 将FuGENE® HD转染试剂和Opti-MEM加入到2 mL无菌EP管中,轻吹混匀,室温静置15 min。将3种质粒(pWPXL-PIAS1 1.18 μg,psPAX2 2.35 μg,pMD2.G 0.47 μg)依次加入到上述混合液中,轻吹混匀,室温静置30 min。静置期间,对HEK 293FT细胞更换10% DMEM新鲜培养液,6 mL/皿。静置后,将配好的混合液均匀加入至每个细胞皿中,混匀,37 ℃、5% CO2培养箱培养,逐日观察荧光和细胞病变。转染后根据GFP荧光和CPE对细胞上清进行收集,并采用超滤的方式对慢病毒液进行浓缩。 1.7.2 慢病毒转导MARC-145细胞: 取生长状态良好的MARC-145细胞接种于24孔板,待细胞长至单层时进行慢病毒转导。转导前更换新鲜的5% DMEM培养基,加入合适比例的慢病毒,转导时需加入终浓度为8 μg/mL Polybrene助转剂,混匀,转导后24 h更换新鲜的10% DMEM培养基。置于37 ℃温箱中培养,转导36 h后,在荧光显微镜下观察蛋白表达效率。 1.8 病毒TCID50测定病毒TCID50测定按照实验室制定的操作程序进行[23]。根据Reed-Müench法计算病毒TCID50值。
1.9 数据统计与分析试验数据用平均值±标准差(x±s)表示,用GraphPad Prism (version 5.0,GraphPad Software,San Diego,CA,USA)软件中的Two-way ANOVA进行差异显著性分析。P > 0.05 (ns)表示差异不显著;P < 0.05 (*)表示差异显著;P < 0.01 (**)和P < 0.001 (***),表示差异极显著。
2 结果与分析 2.1 PIAS1的扩增及重组质粒的构建以PAMs cDNA为模板,扩增得到PIAS1 PCR产物,长度为1 956 bp,与预期一致(图 1)。PCR产物与pGADT7、pCMV-Myc和pWPXL载体经双酶切,连接、转化大肠杆菌Trans10感受态细胞,经PCR鉴定和测序验证,最终获得pGADT7-PIAS1、pCMV-Myc-PIAS1和pWPXL-PIAS1重组质粒。
|
| 图 1 PIAS1基因的扩增及其重组质粒的PCR鉴定 Figure 1 Amplification of PIAS1 gene and PCR identification of recombinant plasmids 注:M:分子量标准DL2000;1:PIAS1;2:pCMV-Myc-PIAS1;3:pGADT7-PIAS1;4:pWPXL-PIAS1. Note: M: Marker DL2000; 1: PIAS1; 2: pCMV-Myc-PIAS1; 3: pGADT7-PIAS1; 4: pWPXL-PIAS1. |
|
|
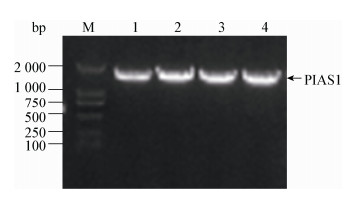
将pGBKT7-N和pGADT7-PIAS1共转化至Y2HGold酵母菌株,酵母回复杂交实验结果显示,PIAS1与N蛋白在酵母细胞中可相互作用(图 2A)。进一步将真核表达质粒pCMV-HA-N和pCMV-Myc-PIAS1共转染HEK 293T细胞,并设置与空载体共转对照,利用Pierce® HA Tag IP/Co-IP Kit进行Co-IP实验,分别用HA和Myc抗体进行Western blotting检测。结果显示,N蛋白和PIAS1之间存在外源性相互作用(图 2B)。通常情况下PIAS1在胞浆和胞核中均有分布。将pCMV-HA-N和pCMV-Myc-PIAS1共转染HEK 293T细胞,分别用HA单抗和Myc多抗进行孵育;同时用PRRSV感染MARC-145细胞和原代PAMs,分别用N单抗和PIAS1多抗进行孵育。激光共聚焦结果显示,PIAS1和N在外源转染HEK 293T细胞中以及在PRRSV感染原代PAMs和MARC-145细胞的情况下均可定位于胞浆中(图 2C)。
|
| 图 2 PIAS1与PRRSV N蛋白相互作用的验证 Figure 2 Confirmation of the interaction of PIAS1 with PRRSV N protein 注:A:酵母回复杂交验证PIAS1与N蛋白相互作用;B:免疫共沉淀验证PIAS1与N蛋白相互作用;C:激光共聚焦验证PIAS1与N蛋白在细胞中共定位. Note: A: Interaction of PIAS1 with N protein by a yeast two-hybrid assay; B: Interaction of PIAS1 with N protein by Co-IP; C: Co-localization of PIAS1 with N protein in cells. |
|
|
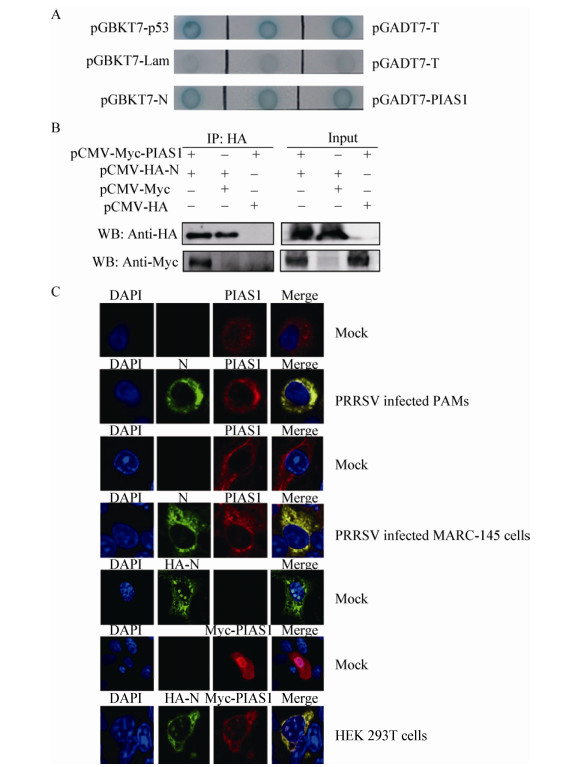
为验证PIAS1是否介导N蛋白的SUMO化修饰,将pCMV-HA-N和pCMV-Myc-PIAS1共转HEK 293T细胞,其中PIAS1设置转染浓度梯度(依次为2.0、1.0、0.5、0 μg),并设置与空载体共转对照,转染30 h后收取蛋白,用N蛋白单克隆抗体进行IP试验,分别利用HA和N单克隆抗体及Myc多克隆抗体进行Western blotting检测。结果表明,外源转染PIAS1不影响N蛋白的SUMO化修饰水平(图 3)。同时,利用siRNA干扰HEK 293T细胞中的PIAS1基因,然后转染pCMV-HA-N,同时设置对照组,结果表明,干扰PIAS1后并不影响N蛋白的SUMO化修饰(图 3B)。
|
| 图 3 外源转染PIAS1对N蛋白SUMO化修饰的影响 Figure 3 Effect of sumoylation of N protein by exogenous transfection PIAS1 Note: A: 1: Myc-PIAS1 (2.0 μg)+HA-N (2.0 μg); 2: Myc-PIAS1 (1.0 μg)+HA-N (2.0 μg)+Myc (1.0 μg); 3: Myc-PIAS1 (0.5 μg)+HA-N (2.0 μg)+Myc (1.5 μg); 4: HA-N (2.0 μg)+Myc (2.0 μg); 5: Myc-PIAS1 (2.0 μg)+HA (2.0 μg). B: No effect on sumoylation of N protein by silencing PIAS1. |
|
|
利用RNA干扰技术分析了干扰MARC-145细胞中PIAS1基因后对PRRSV增殖的影响。结果表明,si-PIAS1-1685能很好地干扰PIAS1基因(图 4A),选取si-PIAS1-1685进行下一步试验。用该siRNA干扰MARC-145细胞中PIAS1基因后,接种PRRSV (MOI=0.01)绘制病毒的生长曲线。结果表明,与对照相比,干扰PIAS1基因的表达可抑制PRRSV复制(图 4B)。
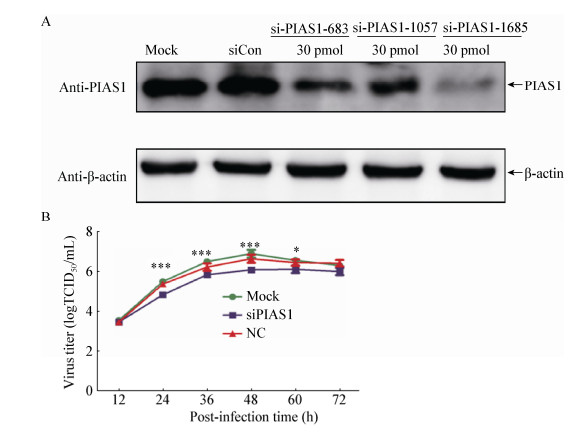
|
| 图 4 干扰PIAS1基因的表达抑制病毒的复制 Figure 4 Inhibition of PRRSV replication by silencing PIAS1 gene in MARC-145 cells 注:A:3种siRNA转染对PIAS1表达的影响;B:siPIAS1转染对PRRSV增殖的影响. Notes: A: Silencing efficiency of PIAS1 gene of three siRNAs; B: The effect of siPIAS1 on PRRSV growth. |
|
|
为了分析PIAS1过表达对PRRSV增殖的影响,用表达PIAS1-GFP的慢病毒转导MARC-145细胞,利用免疫荧光和Western blotting检测MARC-145细胞中PIAS1的表达。结果显示,GFP-PIAS1可在转导的MARC-145细胞中表达(图 5A和图 5B)。转导MARC-145细胞24 h后,以MOI=0.01的剂量感染PRRSV进行病毒生长曲线的绘制,分析在MARC-145细胞中过表达PIAS1对PRRSV增殖的影响。结果表明,与对照相比,过表达PIAS1可促进PRRSV的复制(图 5C)。
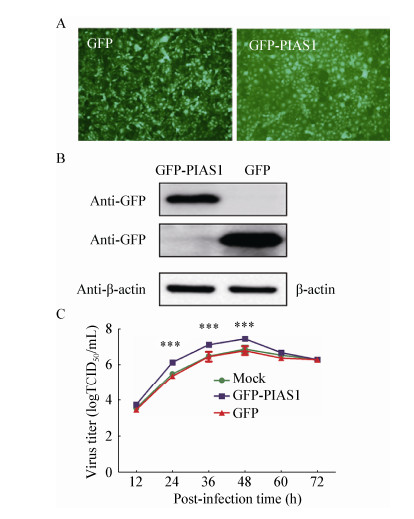
|
| 图 5 过表达PIAS1促进病毒的复制 Figure 5 Enhancement of PRRSV replication by PIAS1 over-expression in MARC-145 cells 注:A:表达GFP-PIAS1的慢病毒在转导MARC-145细胞中的鉴定;B:Western blotting检测GFP-PIAS1的表达;C:过表达PIAS1对PRRSV在MARC-145细胞中滴度的影响. 注: A: The identification of GFP-PIAS1 expression in MARC-145 cells transducted with GFP-PIAS1-expressing lentiviruses; B: Detection of GFP-PIAS1 expression in MARC-145 cells by Western blotting; C: PRRSV titers in MARC-145 cells with PIAS1 over-expression. |
|
|
病毒蛋白与宿主细胞蛋白相互作用及蛋白定位的改变,在调节病毒生命周期甚至病毒复制方面起到重要的作用[24]。本实验室前期的研究表明N蛋白在体外转染和PRRSV感染的情况下皆可发生SUMO化修饰,由此推测属于SP-RING结构域SUMO E3连接酶家族的PIAS1与N蛋白可能存在潜在的相互作用。如同其他RNA病毒一样,PRRSV的复制发生在胞浆中,但在PRRSV的感染过程中N蛋白在胞浆和细胞核中皆有分布[25],N蛋白在10−13 aa和41−47 aa处存在2个核定位信号,分别称为NLS1和NLS2[26]。N蛋白的核定位可能参与多种宿主细胞过程的调节,包括调节细胞周期、细胞凋亡和诱导抗病毒反应。值得注意的是,与野生型病毒相比,具有NLS-2突变的PRRSV可以诱导较低滴度的病毒血症和较高滴度的中和抗体产生[27-29]。此外,N蛋白含有核仁定位序列(NoLS)和核输出信号(NES),所有这些结构域决定了PRRSV N蛋白的核-质穿梭特性[26, 30]。基于以上研究及科学推测,本研究利用酵母回复杂交、免疫共沉淀和激光共聚焦等技术证实了PIAS1与N蛋白间的确存在相互作用,而且无论在瞬时转染的HEK 293T细胞,还是PRRSV感染的MARC-145细胞、原代PAMs细胞,与N蛋白相互作用的PIAS1均从胞核转移到胞浆中,这与Kang等[31]报道的CP2c可以通过与PIAS1相互作用使其亚细胞定位由胞浆转移到核内的研究结果吻合。
SUMO化修饰作为蛋白类泛素化修饰的一种方式在真核细胞的生命进程中发挥着多种重要作用。SUMO共价结合或非共价相互作用可以改变底物稳定性、核定位和功能,此外,一些底物的核定位信号也受SUMO化修饰调节[32-35],因此研究N蛋白SUMO化修饰对深入理解N蛋白生物学功能有着重要意义。有研究表明,宿主蛋白及病毒蛋白均可作为SUMO修饰的靶蛋白受SUMO化修饰系统的调节,此过程对病毒本身的生命周期具有非常重要的作用[36-38]。例如,埃博拉病毒(Ebola virus,EBOV)的基质蛋白VP40可被SUMO化修饰,不仅能维持VP40的稳定性,而且还参与病毒样颗粒(VLPs)的形成[39];甲型流感病毒(Influenza A virus,IAV) NS1的SUMO化增强了NS1的稳定性,最终提高了甲型流感病毒的增殖速度[40];IAV基质蛋白M1的SUMO化有利于形成M1-vRNP复合物,进一步影响病毒的组装和形态发生,而SUMO化修饰的核蛋白对其细胞内运输和病毒生长是必需的[32]。除了Ubc9介导的靶蛋白SUMO化修饰途径外,通过SIM位点介导的病毒蛋白SUMO化修饰也有相关报道,如痘苗病毒(Vaccinia virus,VV)的E3蛋白[41]和人巨细胞病毒(human cytomegalovirus,HCMV) IE2蛋白[42]的SIMs位点对其本身SUMO化修饰水平是必需的。除了上述两种修饰途径外,有学者报道SUMO E3连接酶同样可以介导靶蛋白的SUMO化修饰过程,如E3连接酶Siz1对PCNA的SUMO化修饰起着至关重要的作用[43]。本研究在外源转染PIAS1时,PRRSV N蛋白的SUMO化修饰水平并没有发生改变,表明PIAS1并不参与N蛋白的SUMO化修饰。关于N蛋白SUMO化修饰途径目前可排除通过SCM途径或SIM途径介导,推测N蛋白SUMO化修饰可能由其他SUMO E3连接酶介导完成。
此外,我们分析了PIAS1与N蛋白的相互作用对PRRSV复制的影响。在MARC-145细胞中,沉默PIAS1的表达可以抑制PRRSV的复制,而过表达PIAS1则能促进PRRSV的复制,这表明PIAS1作为一种细胞辅助因子可以正调节病毒的复制。Liu等的研究表明,PIAS1作为一种干扰素信号通路负调节因子,敲除PIAS1可以显著提高机体对病毒或微生物入侵的免疫反应[44],这也与PIAS1促进PRRSV复制的结果相符。
综上,本研究首次证实宿主蛋白PIAS1与PRRSV N蛋白相互作用且共定位于胞浆中;PIAS1可以正调控PRRSV的复制,但不影响PRRSV N蛋白SUMO化修饰。
| [1] |
Holtkamp DJ, Kliebenstein JB, Neumann EJ, et al. Assessment of the economic impact of porcine reproductive and respiratory syndrome virus on United States pork producers[J]. Journal of Swine Health and Production, 2013, 21(2): 72-84. |
| [2] |
Pejsak Z, Stadejek T, Markowska-Daniel I. Clinical signs and economic losses caused by porcine reproductive and respiratory syndrome virus in a large breeding farm[J]. Veterinary Microbiology, 1997, 55(1/4): 317-322. |
| [3] |
Tian KG, Yu XL, Zhao TZ, et al. Emergence of fatal PRRSV variants: unparalleled outbreaks of atypical PRRS in China and molecular dissection of the unique hallmark[J]. PLoS One, 2007, 2(6): e526. DOI:10.1371/journal.pone.0000526 |
| [4] |
Cavanagh D. Nidovirales: a new order comprising Coronaviridae and Arteriviridae[J]. Archives of Virology, 1997, 142(3): 629-633. |
| [5] |
Magar R, Robinson Y, Dubuc C, et al. Evaluation of the persistence of porcine reproductive and respiratory syndrome virus in pig carcases[J]. Veterinary Record, 1995, 137(22): 559-561. DOI:10.1136/vr.137.22.559 |
| [6] |
Murtaugh MP, Elam MR, Kakach LT. Comparison of the structural protein coding sequences of the VR-2332 and Lelystad virus strains of the PRRS virus[J]. Archives of Virology, 1995, 140(8): 1451-1460. DOI:10.1007/BF01322671 |
| [7] |
Conzelmann KK, Visser N, van Woensel P, et al. Molecular characterization of porcine reproductive and respiratory syndrome virus, a member of the arterivirus group[J]. Virology, 1993, 193(1): 329-339. DOI:10.1006/viro.1993.1129 |
| [8] |
Li YH, Tas A, Sun Z, et al. Proteolytic processing of the porcine reproductive and respiratory syndrome virus replicase[J]. Virus Research, 2015, 202: 48-59. DOI:10.1016/j.virusres.2014.12.027 |
| [9] |
Bautista EM, Meulenberg JJM, Choi CS, et al. Structural polypeptides of the American (VR-2332) strain of porcine reproductive and respiratory syndrome virus[J]. Archives of Virology, 1996, 141(7): 1357-1365. DOI:10.1007/BF01718837 |
| [10] |
Johnson CR, Griggs TF, Gnanandarajah J, et al. Novel structural protein in porcine reproductive and respiratory syndrome virus encoded by an alternative ORF5 present in all arteriviruses[J]. The Journal of General Virology, 2011, 92(Pt 5): 1107-1116. |
| [11] |
Meulenberg JJM, Petersen-Den BA, de Kluyver EP, et al. Characterization of proteins encoded by ORFs 2 to 7 of Lelystad virus[J]. Virology, 1995, 206(1): 155-163. DOI:10.1016/S0042-6822(95)80030-1 |
| [12] |
Kappes MA, Miller CL, Faaberg KS. Highly divergent strains of porcine reproductive and respiratory syndrome virus incorporate multiple isoforms of nonstructural protein 2 into virions[J]. Journal of Virology, 2013, 87(24): 13456-13465. DOI:10.1128/JVI.02435-13 |
| [13] |
Wootton SK, Yoo D. Homo-oligomerization of the porcine reproductive and respiratory syndrome virus nucleocapsid protein and the role of disulfide linkages[J]. Journal of Virology, 2003, 77(8): 4546-4557. DOI:10.1128/JVI.77.8.4546-4557.2003 |
| [14] |
Flotho A, Melchior F. Sumoylation: A regulatory protein modification in health and disease[J]. Annual Review of Biochemistry, 2013, 82(1): 357-385. DOI:10.1146/annurev-biochem-061909-093311 |
| [15] |
Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins[J]. Annual Review of Cell and Developmental Biology, 2006, 22(1): 159-180. DOI:10.1146/annurev.cellbio.22.010605.093503 |
| [16] |
Capili AD, Lima CD. Taking it step by step: mechanistic insights from structural studies of ubiquitin/ubiquitin-like protein modification pathways[J]. Current Opinion in Structural Biology, 2007, 17(6): 726-735. DOI:10.1016/j.sbi.2007.08.018 |
| [17] |
Schmidt D, Müller S. Members of the PIAS family Act as SUMO ligases for c-Jun and p53 and repress p53 activity[J]. Proceedings of the National Academy of Sciences of the United States of America, 2002, 99(5): 2872-2877. DOI:10.1073/pnas.052559499 |
| [18] |
Liu B, Liao JY, Rao XP, et al. Inhibition of Stat1-mediated gene activation by PIAS1[J]. Proceedings of the National Academy of Sciences of the United States of America, 1998, 95(18): 10626-10631. DOI:10.1073/pnas.95.18.10626 |
| [19] |
Tsai JS. The interaction between Influenza A viral PB2 protein and cellular PIAS1 protein[D]. Taiwan, China: Master's Thesis of Taiwan University, 2012 (in Chinese) 蔡季书. A型流行性感冒病毒PB2蛋白质与细胞蛋白质PIAS1之间的交互作用[D]. 中国台湾: 台湾大学硕士学位论文, 2012 |
| [20] |
Lee JM, Kang HJ, Lee HR, et al. PIAS1 enhances SUMO-1 modification and the transactivation activity of the major immediate–early IE2 protein of human cytomegalovirus[J]. FEBS Letters, 2003, 555(2): 322-328. DOI:10.1016/S0014-5793(03)01268-7 |
| [21] |
Chang L, Lee Y, Cheng T, et al. Post-translational modification of Rta of Epstein-Barr Virus by SUMO-1[J]. Journal of Biological Chemistry, 2004, 279(37): 38803-38812. DOI:10.1074/jbc.M405470200 |
| [22] |
Wang XY, Bai J, Zhang LL, et al. Poly(A)-binding protein interacts with the nucleocapsid protein of porcine reproductive and respiratory syndrome virus and participates in viral replication[J]. Antiviral Research, 2012, 96(3): 315-323. DOI:10.1016/j.antiviral.2012.09.004 |
| [23] |
Zhao SC, Ge XN, Wang XL, et al. The DEAD-box RNA helicase 5 positively regulates the replication of porcine reproductive and respiratory syndrome virus by interacting with viral Nsp9 in vitro[J]. Virus Research, 2015, 195: 217-224. DOI:10.1016/j.virusres.2014.10.021 |
| [24] |
Brass AL, Dykxhoorn DM, Benita Y, et al. Identification of host proteins required for HIV infection through a functional genomic screen[J]. Science, 2008, 319(5865): 921-926. DOI:10.1126/science.1152725 |
| [25] |
Rowland RR, Kervin R, Kuckleburg C, et al. The localization of porcine reproductive and respiratory syndrome virus nucleocapsid protein to the nucleolus of infected cells and identification of a potential nucleolar localization signal sequence[J]. Virus Research, 1999, 64(1): 1-12. DOI:10.1016/S0168-1702(99)00048-9 |
| [26] |
Rowland RRR, Schneider P, Fang Y, et al. Peptide domains involved in the localization of the porcine reproductive and respiratory syndrome virus nucleocapsid protein to the nucleolus[J]. Virology, 2003, 316(1): 135-145. DOI:10.1016/S0042-6822(03)00482-3 |
| [27] |
Lee C, Hodgins D, Calvert JG, et al. Mutations within the nuclear localization signal of the porcine reproductive and respiratory syndrome virus nucleocapsid protein attenuate virus replication[J]. Virology, 2006, 346(1): 238-250. DOI:10.1016/j.virol.2005.11.005 |
| [28] |
Lee C, Hodgins DC, Calvert JG, et al. The nuclear localization signal of the PRRS virus nucleocapsid protein modulates viral replication in vitro and antibody response in vivo[J]. Advances in Experimental Medicine and Biology, 2006, 581: 145-148. DOI:10.1007/978-0-387-33012-9 |
| [29] |
Pei YL, Hodgins DC, Lee C, et al. Functional mapping of the porcine reproductive and respiratory syndrome virus capsid protein nuclear localization signal and its pathogenic association[J]. Virus Research, 2008, 135(1): 107-114. DOI:10.1016/j.virusres.2008.02.012 |
| [30] |
Rowland RRR, Yoo D. Nucleolar-cytoplasmic shuttling of PRRSV nucleocapsid protein: a simple case of molecular mimicry or the complex regulation by nuclear import, nucleolar localization and nuclear export signal sequences[J]. Virus Research, 2003, 95(1/2): 23-33. |
| [31] |
Kang HC, Chae JH, Jeon J, et al. PIAS1 regulates CP2c localization and active promoter complex formation in erythroid cell-specific α-globin expression[J]. Nucleic Acids Research, 2010, 38(16): 5456-5471. DOI:10.1093/nar/gkq286 |
| [32] |
Han QL, Chang C, Li L, et al. Sumoylation of influenza A virus nucleoprotein is essential for intracellular trafficking and virus growth[J]. Journal of Virology, 2014, 88(16): 9379-9390. DOI:10.1128/JVI.00509-14 |
| [33] |
Mahajan R, Delphin C, Guan TL, et al. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2[J]. Cell, 1997, 88(1): 97-107. DOI:10.1016/S0092-8674(00)81862-0 |
| [34] |
Müller S, Matunis MJ, Dejean A. Conjugation with the ubiquitin-related modifier SUMO-1 regulates the partitioning of PML within the nucleus[J]. The EMBO Journal, 1998, 17(1): 61-70. DOI:10.1093/emboj/17.1.61 |
| [35] |
Salinas S, Brian on-Marjollet A, Bossis G, et al. SUMOylation regulates nucleo-cytoplasmic shuttling of Elk-1[J]. The Journal of Cell Biology, 2004, 165(6): 767-773. DOI:10.1083/jcb.200310136 |
| [36] |
Everett RD, Boutell C, Hale BG. Interplay between viruses and host sumoylation pathways[J]. Nature Reviews Microbiology, 2013, 11(6): 400-411. DOI:10.1038/nrmicro3015 |
| [37] |
Mattoscio D, Segré CV, Chiocca S. Viral manipulation of cellular protein conjugation pathways: the SUMO lesson[J]. World Journal of Virology, 2013, 2(2): 79-90. DOI:10.5501/wjv.v2.i2.79 |
| [38] |
Wimmer P, Schreiner S, Dobner T. Human pathogens and the host cell SUMOylation system[J]. Journal of Virology, 2012, 86(2): 642-654. DOI:10.1128/JVI.06227-11 |
| [39] |
Baz-Martínez M, El Motiam A, Ruibal P, et al. Regulation of Ebola virus VP40 matrix protein by SUMO[J]. Scientific Reports, 2016, 6: 37258. DOI:10.1038/srep37258 |
| [40] |
Xu K, Klenk C, Liu B, et al. Modification of nonstructural protein 1 of influenza A virus by SUMO1[J]. Journal of Virology, 2011, 85(2): 1086-1098. DOI:10.1128/JVI.00877-10 |
| [41] |
González-Santamaría J, Campagna M, García MA, et al. Regulation of vaccinia virus E3 protein by small ubiquitin-like modifier proteins[J]. Journal of Virology, 2011, 85(24): 12890-12900. DOI:10.1128/JVI.05628-11 |
| [42] |
Berndt A, Hofmann-Winkler H, Tavalai N, et al. Importance of covalent and noncovalent SUMO interactions with the major human cytomegalovirus transactivator IE2p86 for viral infection[J]. Journal of Virology, 2009, 83(24): 12881-12894. DOI:10.1128/JVI.01525-09 |
| [43] |
Yunus AA, Lima CD. Structure of the Siz/PIAS SUMO E3 Ligase Siz1 and determinants required for SUMO modification of PCNA[J]. Molecular Cell, 2009, 35(5): 669-682. DOI:10.1016/j.molcel.2009.07.013 |
| [44] |
Liu B, Mink S, Wong KA, et al. PIAS1 selectively inhibits interferon-inducible genes and is important in innate immunity[J]. Nature Immunology, 2004, 5(9): 891-898. DOI:10.1038/ni1104 |
 2017, Vol. 44
2017, Vol. 44