扩展功能
文章信息
- 崔丽娇, 王洁, 吕应年, 刘颖文, 江黎明
- CUI Li-Jiao, WANG Jie, LÜ Ying-Nian, LIU Ying-Wen, JIANG Li-Ming
- 湛江沿海潮间带产胞外纤溶活性酶类海洋真菌的生物多样性分析
- Biodiversity analysis of marine fungi producing extracellular fibrinolytic activity enzymes from the intertidal zone along Zhanjiang coast
- 微生物学通报, 2016, 43(7): 1448-1461
- Microbiology China, 2016, 43(7): 1448-1461
- 10.13344/j.microbiol.china.160262
-
文章历史
- 收稿日期: 2016-03-29
- 接受日期: 2016-05-17
- 优先数字出版日期(www.cnki.net): 2016-05-17
2. 漯河医学高等专科学校 医学检验技术教研室 河南 漯河 462002;
3. 广东医科大学 广东省天然药物重点实验室 广东 湛江 524023
2. Department of Medical Laboratory, Luohe Medical College, Luohe, Henan 462002, China;
3. Guangdong Key Laboratory for Research and Development of Natural Drugs, Guangdong Medical University, Zhanjiang, Guangdong 524023, China
国际卫生组织(WHO)报告,2012年因心血管疾病死亡的人数为1 750万,占当年全球因非传染性疾病死亡人数的46.2%,是全球头号死亡原因[1];WHO预计到2030年,全世界因心血管疾病,尤其是因血管内血栓形成所致的急性心肌梗塞、急性缺血性脑卒中和肺动脉栓塞而死亡的年死亡人数将会增加到2 330万[2]。目前常用于血栓性疾病临床治疗和实验研究的溶栓药,可根据其作用机制不同而分为直接酶解血栓中纤维蛋白的纤溶酶样酶(Plasmin-like enzyme)和通过激活纤溶酶原而间接溶解纤维蛋白的纤溶酶原激活物(Plasminogen activator)两大类[3]。前者包括人源纤溶酶及其衍生物、蚓激酶、蛇毒纤溶酶和纳豆激酶[4],后者主要包括人源性的组织纤溶酶原激活物和尿激酶[5]及细菌来源的链激酶和葡激酶[6]。大多数现有的纤溶酶样酶,尤其是纤溶酶经系统给药后会迅速被血浆中的a2-抗纤溶酶和a2-巨球蛋白中和,因此,只有微纤维蛋白溶酶(Microplasmin)和蛇毒纤溶酶被作为溶栓药进行过临床实验,但结果均不理想[4]。迄今被批准用于临床治疗的溶栓药都是纤溶酶原激活物类,包括组织纤溶酶原激活物及其衍生物、尿激酶和链激酶[7]。但这些现有的纤溶酶原激活物对纤维蛋白的特异性较低,在溶解血管中阻塞性血栓的同时,也可使系统中大量的纤维蛋白原和血管损伤部位的止血栓子降解,引起出血的副作用。因此,寻找疗效更好、副作用更小的新型溶栓药仍是目前溶栓药研究中的热门领域之一[8]。
因具有巨大的生物多样性及丰富的代谢产物和酶类,并已发现其中许多物种能产生纤溶酶样酶和纤溶酶原激活物,微生物正逐渐成为溶栓新药研发的重要资源[9]。其中,种类繁多、数量庞大的海洋微生物因长期生活在高盐、高压、低营养、温差大及pH范围广的海洋环境中[10],可产生许多结构和功能与陆栖微生物不同的次级代谢产物和酶类,是至今仍未得到很好挖掘的重要微生物资源[11]。海洋真菌是海洋微生物的重要组成部分,据估计可能有10 000多个种或种系型,包括已被鉴定的500多种专性海洋真菌、300多种兼性海洋真菌和1 500多种海洋酵母等[12]。近年来,海洋微生物中,关于芽孢杆菌属(Bacillus)[13]和希瓦氏菌属(Shewanella)[14]海洋细菌,以及松藻属(Codium)[15]海洋绿藻的纤溶活性酶类的研究已有零星报道,但至今仍未见关于产纤溶活性酶类海洋真菌的研究报道。本研究拟分析研究湛江沿海硇洲岛和徐闻珊瑚礁自然保护区潮间带产胞外纤溶酶样酶和纤溶酶原激活物的海洋真菌的生物多样性,为发掘可用于新型溶栓药物研发的海洋真菌资源奠定基础。
1 材料与方法 1.1 主要试剂和仪器、海洋真菌及其培养基 1.1.1 主要试剂和仪器: 溶细胞酶、酪蛋白、凝血酶和用于配制马铃薯葡萄糖琼脂(PDA)、酵母膏蛋白胨葡萄糖(YPD)、脱脂牛奶马铃薯葡萄糖琼脂(SM-PDA)及纤维蛋白马铃薯葡萄糖琼脂(FN-PDA)等培养基的化学试剂,Sigma-Aldrich公司;真菌基因组DNA提取试剂盒,Bioflux公司;PCR试剂盒,Vazyme公司;真菌rDNA转录间隔区1-5.8S rDNA-转录间隔区2(ITS1-5.8S-ITS2)片段PCR通用引物由上海生工生物工程公司合成;人纤维蛋白原由广东双林生物药业有限公司馈赠;超净工作台,上海蓝季科技;5417R台式冷冻离心机和PCR扩增仪,Eppendorf公司。 1.1.2 海洋真菌: 用于产胞外纤溶酶样酶和纤溶酶原激活物海洋真菌生物多样性分析的海洋真菌共446株,其中178株是研究组之前从硇洲岛(20°52′-20°56′N,110°33′-110°38′E)潮间带分离鉴定的[16],其余268株由本研究组从徐闻珊瑚礁自然保护区(20°10′−20°27′N,109°50′−109°24′E)潮间带采样、分离和鉴定。 1.1.3 分离与发酵培养基: 海水PDA固体培养基(马铃薯200 g,葡萄糖20 g,琼脂16 g,50%自然海水1 l,pH自然,倒平板前加入终浓度为100 U/mL的青链霉素混合液)、海水YPD培养基(胰蛋白胨20 g,酵母提取物10 g,葡萄糖20 g,50%自然海水1 L,pH自然,葡萄糖与其他分别灭菌后再混匀)和用于蛋白酶活性筛选的SM-PDA固体培养基(马铃薯200 g,葡萄糖20 g,琼脂16 g,脱脂速溶奶粉10 g,50%自然海水1 L,pH自然),按文献[16]的方法配制。用于纤溶酶及其激活物筛选的海水FN-PDA固体培养基的配制参照Astrup等的方法[17],并针对海洋真菌的生长特性做出相应的调整。即马铃薯200 g,葡萄糖20 g,琼脂16 g,50%自然海水1 l,1×105 Pa高压灭菌20 min,冷却至55−60 ℃,加入预热的纤维蛋白原缓冲液(0.5 g纤维蛋白原溶于250 ml pH 7.4的磷酸缓冲液),加入3 mL的200 U/ml凝血酶缓冲液(1 000 U凝血酶溶于5 mL pH 7.4的磷酸缓冲液)混匀倒平板。海水纤溶酶活性筛选固体培养基采用Lassen的方法[18],将上述配制的海水FN-PDA固体培养基70 ℃加热60 min而成。
1.2 菌株的分离纯化及保存沉积物样品取2 g悬浮于20 ml灭菌天然海水,其上清液与海水样品经细胞计数后分别适当稀释,取稀释液50 μL涂布在PDA固体培养基上,置28 ℃培养箱培养,一旦发现有菌丝长出后,立即将其转移到另一相同培养基中,经2−3次平板划线纯化后,以海水PDA试管斜面培养基保存于4 ℃。
1.3 DNA的提取及PCR扩增斜面培养基保存的海洋真菌接种到海水PDA培养基,于28 ℃培养14 d。加入灭菌的0.1% Tween 80收集分生孢子,计数后将孢子悬液稀释至约2×108 cells/mL。将孢子悬液接种到3 mL海水YPD培养液中,28 ℃培养48 h。发酵液1 000×g离心3 min,将菌丝体转移到1 ml的Eppendorf管中,加入500 μL PBS(pH 7.0)和100 U溶细胞酶,混匀后37 ℃培养1 h[16]。采用真菌基因组DNA提取试剂盒,按说明书上的方法提取真菌基因组DNA。最后加入100 μl TE缓冲液将DNA溶解,置−20 ℃保存。
采用真菌ITS1-5.8S-ITS2片段特异性通用PCR引物对ITS1(5′-TCCGTAGGTGAACCTGCGG-3′)和ITS4(5′-TCCTCCGCTTATTGATATGC-3′)[16,19],以提取的基因组DNA为模板,PCR扩增各真菌的ITS1-5.8S-ITS2片段。PCR体系为:DNA模板2 μL,2×Taq Master Mix 25 μl,10 µmol/L Primers各2 μl,加ddH2O至50 μl。反应条件为:94 ℃ 5 min;94 ℃ 45 s,52 ℃ 45 s,72 ℃ 1 min,30个循环;72 ℃ 7 min。扩增产物经1.5%琼脂糖凝胶电泳分离后送上海生工生物工程股份有限公司测序。
1.4 系统进化分析ITS1-5.8S-ITS2片段测序结果提交GenBank,并在GenBank网站采用Blast进行相似性分析,采用ClustalW 2两两比对,排除相同序列,用ClustalX进行排列比较,用Phylip软件按照邻接法聚类(选择Bootstrap检验值≥50%,1 000次重复)进行系统树的构建,从而对分离出的微生物进行分类鉴定。
1.5 蛋白酶及纤溶活性菌株的筛选产生蛋白酶的菌株采用之前报道[16]的SM-PDA培养基的方法进行筛选。纤溶活性菌株的筛选采用纤维蛋白培养基方法[17,18]进行,并针对海洋真菌的培养条件做出调整。将分离纯化的单个菌落分别接种于SM-PDA培养基、不经70 ℃加热60 min处理的海水FN-PDA培养基和经70 ℃加热60 min处理的海水FN-PDA培养基,28 ℃恒温培养3−7 d。能在SM-PDA培养基上形成透明圈的菌株为能产生胞外蛋白酶的海洋真菌菌株;能在经过70 ℃加热60 min处理和不处理的海水FN-PDA培养基上都形成透明圈的菌株为能产生胞外纤溶酶样酶的真菌菌株;仅能在未经70 ℃加热60 min处理的海水FN-PDA培养基上形成透明圈的菌株为能产生胞外纤溶酶原激活物的菌株。测量不同阳性菌株的透明圈直径和菌落直径,以透明圈直径与菌落直径的差值(D)表示酶活大小,D值越大,表明活性越强。
2 结果与分析 2.1 海洋真菌生物多样性之前研究组已从硇洲岛潮间带分离鉴定了178株海洋真菌(ITS1-5.8S-ITS2序列GenBank登录号为KM277955−KM278132[16]),本研究从徐闻珊瑚礁自然保护区采集的样品中分离出了268株真菌(ITS1-5.8S-ITS2片段的GenBank序列登录号为KX098015−KX098282),共446株海洋真菌(表 1)。序列比对分析结果表明,这446株海洋真菌包含了真菌的98个种,归属为真菌域2个门的6个纲、18个目、46个科、65个属(表 1)。其中,散囊菌纲(Eurotiomycetes)真菌290株,占分离培养真菌的65.0%;座囊菌纲(Dothideomycetes)真菌66株,约占14.8%;粪壳菌纲(Sordariomycetes)真菌64株,约占14.3%;伞菌纲(Agaricomycetes)真菌19株,约占4.2%,子囊菌纲(Ascomycete)真菌8株,约占1.8%。在446株海洋真菌归类的65个属中,优势属为曲霉属(Aspergillus),含189株,占被分离真菌的42.4%;其次为青霉属(Penicillium),含93株,占被分离真菌的20.8%。
| 类群分布 Distribution groups | 菌株数 Number of strains | 产酶菌株数 Strains producing enzymes | ||||||||||
| 门 Phylum | 纲 Class | 目 Order | 科 Family | 属 Genus | 硇洲岛 NZI | 徐闻 XW | 合计 Total | 蛋白酶 Protease | 纤溶酶样酶 PLE | 纤溶酶原激活物 PA | ||
| Ascomycota | Ascomycete | ? | ? | ? | 2 | 6 | 8 | 6 | 1 | 2 | ||
| Dothideomycetes | Botryosphaeriales | Botryosphaeriaceae | Lasiodiplodia | 8 | 0 | 8 | 0 | 0 | 2 | |||
| Neofusicoccum | 1 | 0 | 1 | 0 | 0 | 0 | ||||||
| Capnodiales | Cladosporiaceae | Cladosporium | 7 | 18 | 25 | 23 | 1 | 2 | ||||
| Toxicocladosporium | 0 | 1 | 1 | 0 | 0 | 0 | ||||||
| Mycosphaerellaceae | Pallidocercospora | 0 | 1 | 1 | 1 | 0 | 0 | |||||
| Teratosphaeriaceae | Devriesia | 0 | 1 | 1 | 1 | 1 | 0 | |||||
| Eupenidiella | 0 | 1 | 1 | 0 | 0 | 0 | ||||||
| Dothideales | Aureobasidiaceae | Aureobasidium | 0 | 1 | 1 | 0 | 0 | 0 | ||||
| ? | Biatriospora | 0 | 1 | 1 | 1 | 0 | 0 | |||||
| Pleosporales | Corynesporascaceae | Corynespora | 1 | 0 | 1 | 0 | 0 | 0 | ||||
| Cucurbitariaceae | Pyrenochaetopsis | 1 | 0 | 1 | 0 | 0 | 0 | |||||
| Didymellaceae | Leptosphaerulina | 0 | 1 | 1 | 1 | 0 | 1 | |||||
| Stagonosporopsis | 0 | 1 | 1 | 1 | 0 | 0 | ||||||
| Didymosphaeriaceae | Paraconiothyrium | 0 | 3 | 3 | 3 | 0 | 3 | |||||
| Sporormiaceae | Westerdykella | 1 | 0 | 1 | 1 | 1 | 0 | |||||
| Montagnulaceae | Paraphaeosphaeria | 7 | 0 | 7 | 6 | 5 | 0 | |||||
| Paraconiothyrium | 2 | 0 | 2 | 2 | 2 | 0 | ||||||
| Phaeosphaeriaceae | Paraphoma | 1 | 0 | 1 | 1 | 0 | 0 | |||||
| Phaeosphaeriopsis | 2 | 0 | 2 | 2 | 0 | 0 | ||||||
| Setophoma | 1 | 0 | 1 | 0 | 0 | 0 | ||||||
| Setophaeosphaeria | 0 | 1 | 1 | 1 | 1 | 0 | ||||||
| Pleosporaceae | Curvularia | 3 | 0 | 3 | 1 | 0 | 0 | |||||
| ? | ? | 1 | 0 | 1 | 0 | 0 | 0 | |||||
| Eurotiomycetes | Eurotiales | Aspergillaceae | Aspergillus | 20 | 169 | 189 | 134 | 34 | 56 | |||
| Thermoascaceae Trichocomaceae Diaporthaceae | Penicillium | 50 | 42 | 92 | 44 | 11 | 11 | |||||
| Paecilomyces | 3 | 0 | 3 | 0 | 0 | 2 | ||||||
| Talaromyces | 3 | 2 | 5 | 1 | 0 | 0 | ||||||
| Sordariomycetes | Diaporthe | 1 | 0 | 1 | 0 | 0 | 0 | |||||
| Glomerellales | Glomerellaceae | Colletotrichum | 1 | 0 | 1 | 1 | 0 | 0 | ||||
| Plectosphaerellaceae | Musicillium | 2 | 0 | 2 | 2 | 1 | 0 | |||||
| Plectosphaerella | 1 | 0 | 1 | 0 | 0 | 0 | ||||||
| Hypocreales | Bionectriaceae | Gliomastix | 2 | 0 | 2 | 1 | 0 | 0 | ||||
| Geosmithia | 0 | 1 | 1 | 1 | 1 | 0 | ||||||
| ? | ? | 0 | 1 | 1 | 1 | 1 | 1 | |||||
| ? | Acremonium | 0 | 1 | 1 | 1 | 0 | 1 | |||||
| Cordycipitaceae | Beauveria | 1 | 0 | 1 | 0 | 0 | 0 | |||||
| Cordyceps | 0 | 3 | 3 | 1 | 1 | 0 | ||||||
| Lecanicillium | 0 | 2 | 2 | 2 | 0 | 0 | ||||||
| Simplicillium | 0 | 1 | 1 | 1 | 0 | 0 | ||||||
| Hypocreaceae | Trichoderma | 18 | 0 | 18 | 1 | 0 | 0 | |||||
| Nectriaceae | Fusarium | 8 | 0 | 8 | 4 | 0 | 0 | |||||
| Ophiocordycipitaceae | Purpureocillium | 0 | 2 | 2 | 2 | 0 | 0 | |||||
| ? | ? | 1 | 0 | 1 | 0 | 0 | 0 | |||||
| ? | Emericellopsis | 1 | 0 | 1 | 1 | 0 | 0 | |||||
| ? | Stachybotrys | 1 | 0 | 1 | 0 | 0 | 1 | |||||
| Ophiostomatales | Ophiostomataceae | Sporothrix | 0 | 1 | 1 | 0 | 0 | 0 | ||||
| Sordariales | Chaetomiaceae | Humicola | 1 | 0 | 1 | 0 | 0 | 0 | ||||
| Trichosphaeriales | ? | Nigrospora | 1 | 0 | 1 | 0 | 0 | 0 | ||||
| Xylariales | Amphisphaeriaceae | Pestalotiopsis | 1 | 0 | 1 | 1 | 0 | 0 | ||||
| Diatrypaceae | Eutypella | 2 | 0 | 2 | 1 | 0 | 0 | |||||
| Xylariaceae | Daldinia | 2 | 0 | 2 | 0 | 0 | 1 | |||||
| Hypoxylon | 1 | 0 | 1 | 1 | 0 | 0 | ||||||
| ? | ? | Myrmecridium | 2 | 0 | 2 | 2 | 0 | 0 | ||||
| ? | ? | ? | Microsphaeropsis | 5 | 0 | 5 | 4 | 4 | 1 | |||
| Basidiomycota | Agaricomycetes | Agaricales | Amanitaceae | Amanita | 0 | 1 | 1 | 0 | 0 | 0 | ||
| Corticiales | Corticiaceae | Phanerochaete | 2 | 0 | 2 | 0 | 0 | 0 | ||||
| Subulicystidium | 2 | 0 | 2 | 2 | 0 | 0 | ||||||
| Polyporales | Coriolaceae | Leiotrametes | 0 | 1 | 1 | 0 | 0 | 0 | ||||
| Rigidoporus | 1 | 2 | 3 | 2 | 2 | 0 | ||||||
| Trametes | 2 | 3 | 5 | 0 | 0 | 0 | ||||||
| Lentinaceae | Lentinus | 1 | 0 | 1 | 1 | 0 | 0 | |||||
| Flavodon | 2 | 0 | 2 | 1 | 0 | 0 | ||||||
| Phanerochaetaceae | Pseudolagarobasidium | 1 | 0 | 1 | 0 | 0 | 0 | |||||
| Russulales | Peniophoraceae | Peniophora | 1 | 0 | 1 | 1 | 0 | 0 | ||||
| 合计 Total | 178 | 268 | 446 | 265 | 67 | 84 | ||||||
Note: Naozhou island was represented as NZI, Xuwen coral reef nature reserve was represented as XW, plasmin-like enzyme was represented as PLE, plasminogen activator was represented as PA. The closest identified ITS1-5.8S-ITS2 in GenBank has no corresponding class or order or family or genus are represented in ?.
采用SM-PDA培养基培养的方法,对获得的海洋真菌进行胞外蛋白酶活性分析的结果显示,446株海洋真菌中有265株真菌可产生胞外蛋白酶(表 1),占分离培养真菌的59.4%。其中优势菌群为曲霉属(Aspergillus),含海洋真菌134株,占可产生胞外蛋白酶真菌的50.6%;其次为青霉属(Penicillium),含海洋真菌44株,占可产生胞外蛋白酶真菌的16.6%。265株可产生胞外蛋白酶的真菌含61个种,分布于真菌域的41个属、32个科、13个目、6个纲的2个门。
2.3 产胞外纤溶活性酶类海洋真菌的生物多样性采用经70 ℃加热60 min处理和不处理的两种海水FN-PDA培养基培养的方法,对获得的海洋真菌的胞外纤溶酶样酶和纤溶酶原激活物进行分析的结果(图 1)显示:在446株分离培养的海洋真菌中,能在经70 ℃加热60 min处理和不处理的海水FN-PDA培养基上形成透明圈的,即产生胞外纤溶酶样酶活性的菌株有67株;仅能在未经70 ℃加热60 min处理的海水FN-PDA培养基上形成透明圈的,即产生胞外纤溶酶原激活物的菌株有84株(表 1)。

|
|
图 1
采用海水纤维蛋白PDA培养基筛选产胞外纤溶酶样酶及纤溶酶原激活物海洋真菌
Figure 1
Screening for the marine fungi producing plasmin-like enzymes and plasminogen activators with sea water FN-PDA culture plates
注: A1、B1和C1是接种了不同菌株的未经加热处理的海水FN-PDA培养基;A2、B2和C2是接种了不同菌株的经加热处理的海水FN-PDA培养基. Note: A1, B1 and C1 were the unheated sea water FN-PDA plates innoculated with different strains; A2, B2 and C2 were the unheated sea water FN-PDA plates innoculated with different strains. |
这些具有胞外纤溶活性菌株的类群分布(表 1)及ITS1-5.8S-ITS2序列系统进化树(图 2−5)显示:67株产胞外纤溶酶样酶真菌菌株含22个种,分布在14个属、13个科、8个目、5个纲的2个门,即子囊菌门(Ascomycota)和担子菌门(Basidiomycota);优势属为曲霉属(Aspergillus),含海洋真菌34株,占产胞外纤溶酶样酶活性真菌的50.7%;其次为青霉属(Penicillium),含海洋真菌11株,占产胞外纤溶酶样酶活性真菌的16.4%。84株产胞外纤溶酶原激活物的真菌菌株含23个种,分布于13个属、11个科、8个目、4个纲的1个门,即子囊菌门(Ascomycota);优势属也是曲霉属(Aspergillus),含海洋真菌56株,占产胞外纤溶酶原激活物活性真菌的66.7%;其次为青霉属(Penicillium),含海洋真菌11株,占产胞外纤溶酶原激活物活性真菌的13.1%。

|
|
图 2
产纤溶酶样酶菌株(硇洲岛)与GenBank数据库中相应真菌菌种的ITS1-5.8S-ITS2序列系统进化树
Figure 2
Phylogenetic tree of ITS1-5.8S-ITS2 sequence of the isolated fungal strains (Naozhou island) with plasmin-like enzymes and appropriate fungal species in GenBank database
注:分离的真菌菌株号用粗线表示;括号里的数字为GenBank登录号;节点上的数字为Bootstrap值(1 000次重复抽样的百分比);比例尺代表每1 000个核苷酸中有0.5个核苷酸发生替代. Note: The number of fungal strains isolated in this study are indicated in bold; Numbers in parentheses are GenBank accession numbers; Bootstrap values (expressed as percentages of 1 000 replications) are given at nodes; The scale bar represents 0.5 substitutions per 1 000 nucleotide positions. |
硇洲岛和徐闻珊瑚礁自然保护区是湛江沿海目前环境保护较好的海岛和珊瑚礁自然保护区。本研究组先后分别从硇洲岛和徐闻珊瑚礁自然保护区潮间带的海水和沉积物中分离、鉴定了海洋真菌178株[16]和268株,合计共446株,98个种,基本都是兼性海洋真菌。其中优势属为Aspergillus,共189株,占分离培养菌株的42.4%;其次为Penicillium,共93株,占分离培养菌株的20.8%(表 1),与研究组之前的报道[16]一致。
除蛋白酶、纤维素酶、淀粉酶和脂肪酶等重要的工业用酶[20]外,微生物也是产纤溶酶样酶和纤溶酶原激活物的重要资源[11]。已发现可产生和释放纤溶酶样酶和纤溶酶原激活物的陆栖微生物包括细菌[9]、放线菌[21]和真菌[8]。细菌来源的纤溶活性酶类有最早从溶血性链球菌(Streptococcus hemolyticus)发现、目前常用于血栓性疾病临床治疗的链激酶,以及从金黄色葡萄球菌(Staphylococcus aureus)获得的葡激酶[9]和从纳豆芽孢杆菌(Bacillus natto)获得的纳豆激酶[22];已发现可产生具有显著纤溶活性蛋白酶的放线菌主要是链霉菌属(Streptomyces)的放线菌[21];可产生具有显著纤溶活性蛋白酶的真菌有赭曲霉(Asperigillus ochraceus) KSK-3、产黄青霉(Penicillum chrysogenum)H9、多孢木霉(Tolypocladium inflatum)k1、镰胞菌(Fusarium oxysporum)和中华根霉(Rhizopus chinesis)12等[8,9,23]。海洋微生物中,已发现可产生纤溶活性酶类的有细菌和绿藻[13,14,15],但至今仍未见关于产纤溶活性酶类海洋真菌的研究报道。
本研究从硇洲岛和徐闻珊瑚礁自然保护区潮间带的海水和沉积物中分离到了67株产胞外纤溶酶样酶的海洋真菌,含22个种,分布于Aspergillus (曲霉属)、Penicillium (青霉属)、Cladosporium (芽枝霉属)、Cordyceps (虫草属)、Rigidoporus (硬孔菌属)、Devriesia、Geosmithia、Microsphaeropsis、Musicillium、Paraphaeosphaeria、Paraconiothyrium、Setophaeosphaeria、Westerdykella等14个属,优势属为曲霉属(Aspergillus),其次为青霉属(Penicillium)(表 1,图 2、3);分离到了84株产胞外纤溶酶原激活物的海洋真菌,含23个种,分布于Acremonium (枝顶孢属)、Cladosporium (芽枝霉属)、Daldinia (轮层炭菌属)、Lasiodiplodia (二孢属)、Leptosphaerulina (小光壳属)、Paecilomyces (拟青霉属)、Stachybotrys (葡萄穗霉属)、Paraconiothyrium、Microsphaeropsis等13个属的海洋真菌,优势属也是曲霉属(Aspergillus),其次为青霉属(Penicillium)(表 1,图 4、5)。本研究还分离到了8株镰胞菌属(Fusarium)的海洋真菌,其中4株有蛋白酶活性,但都没有纤溶酶样酶和纤溶酶原激活物的活性。

|
|
图 3
产纤溶酶样酶菌株(徐闻珊瑚礁自然保护区)及GenBank数据库中相应真菌菌种的ITS1-5.8S-ITS2序列系统进化树
Figure 3
Phylogenetic tree of ITS1-5.8S-ITS2 sequence of the isolated fungal strains (Xuwen coral reef nature reserve) with plasmin-like enzymes and appropriate fungal species in GenBank database
注:分离的真菌菌株号用粗线表示;括号里的数字为GenBank登录号;节点上的数字为Bootstrap值(1 000次重复抽样的百分比);比例尺代表每100个核苷酸中有0.5个核苷酸发生替代. Note: The number of fungal strains isolated in this study are indicated in bold; Numbers in parentheses are GenBank accession numbers; Bootstrap values (expressed as percentages of 1 000 replications) are given at nodes; The scale bar represents 0.5 substitutions per 100 nucleotide positions. |

|
|
图 4
产纤溶酶原激活物菌株(硇洲岛)与GenBank数据库中相应真菌菌种的ITS1-5.8S-ITS2序列系统进化树
Figure 4
Phylogenetic tree of ITS1-5.8S-ITS2 sequence of the isolated fungal strains (Naozhou island) with plasminogen activators and appropriate fungal species in GenBank database
注:分离的真菌菌株号用粗线表示;括号里的数字为GenBank登录号;节点上的数字为Bootstrap值(1 000次重复抽样的百分比);比例尺代表每100个核苷酸中有0.5个核苷酸发生替代. Note: The number of fungal strains isolated in this study are indicated in bold; Numbers in parentheses are GenBank accession numbers; Bootstrap values (expressed as percentages of 1 000 replications) are given at nodes; The scale bar represents 0.5 substitutions per 100 nucleotide positions. |
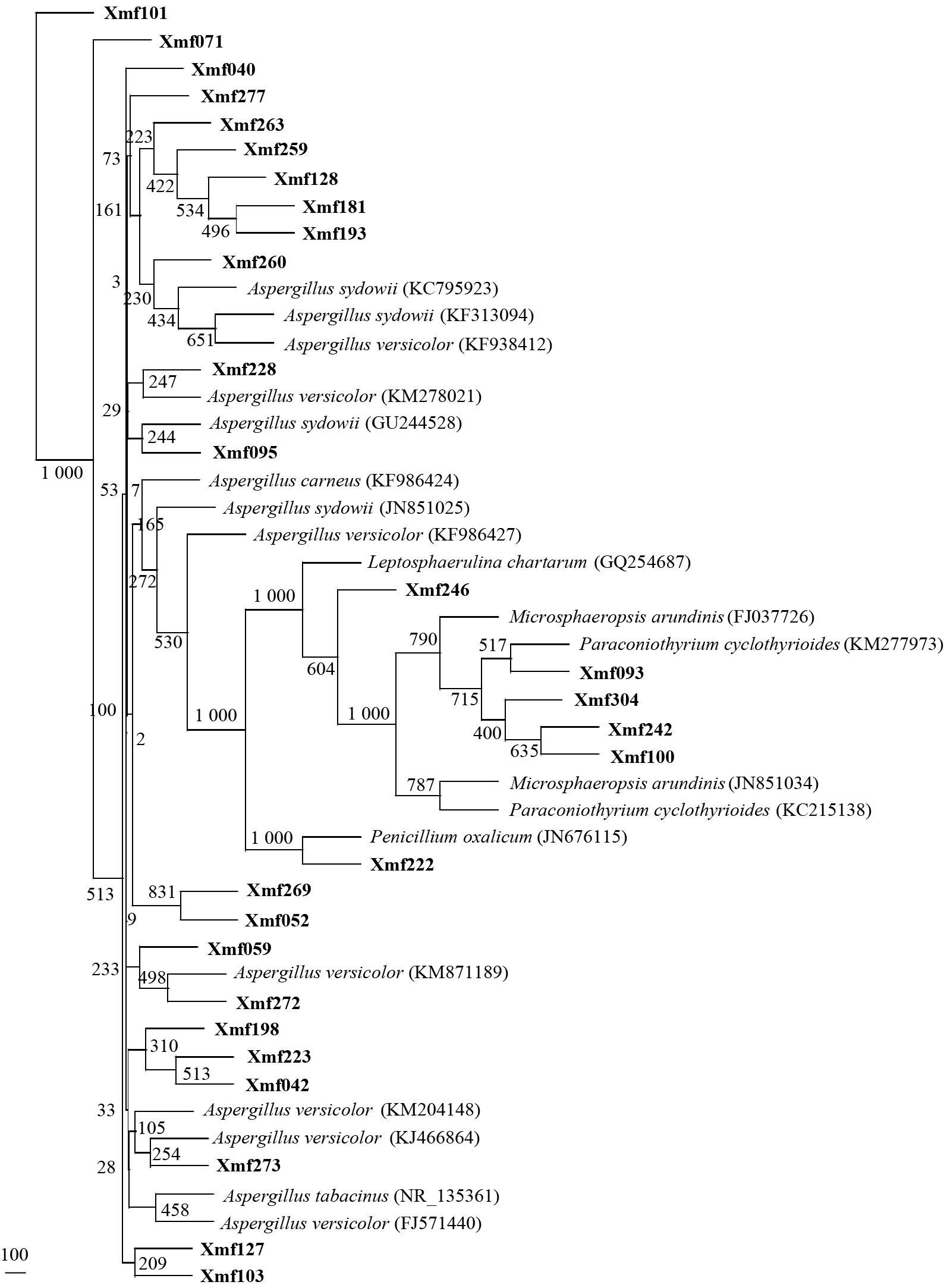
|
|
图 5
产纤溶酶原激活物菌株(徐闻珊瑚礁自然保护区)与GenBank数据库中相应真菌菌种的 ITS1-5.8S-ITS2序列系统进化树
Figure 5
Phylogenetic tree of ITS1-5.8S-ITS2 sequence of the isolated fungal strains (Xuwen coral reef nature reserve) with plasminogen activators and appropriate fungal species in GenBank database
注:分离的真菌菌株号用粗线表示;括号里的数字为GenBank登录号;节点上的数字为Bootstrap值(1 000次重复抽样的百分比);比例尺代表每100个核苷酸中有0.5个核苷酸发生替代. Note: The number of fungal strains isolated in this study are indicated in bold; Numbers in parentheses are GenBank accession numbers; Bootstrap values (expressed as percentages of 1 000 replications) are given at nodes; The scale bar represents 0.5 substitutions per 100 nucleotide positions. |
纤溶酶和大多数纤溶酶原激活物都是蛋白酶。本研究首先采用SM-PDA培养基培养的方法[16],筛选可产生胞外蛋白酶的海洋真菌,然后用Permin于1947首次建立并用于纤溶酶原激活物检测,后经Astrup等和Lassen改进后用于血浆纤溶酶和纤溶酶原激活物检测的纤维蛋白培养基培养法[17,18]来筛选产胞外纤溶酶和纤溶酶原激活物的海洋真菌。该方法中用来形成底物的纤维蛋白原,在其分离纯化的过程中至今仍无法将其中少量的纤溶酶原去除掉,所以制备的FN-PDA培养基中均有少量的纤溶酶原,因此不论是纤溶酶还是纤溶酶原激活物都能在FN-PDA培养基上形成纤维蛋白溶解圈;纤溶酶可直接酶解培养基中纤维蛋白形成纤维蛋白溶解圈,纤溶酶原激活物则可将培养基中纤溶酶原激活为纤溶酶进而间接使培养基中的纤维蛋白酶解,形成纤维蛋白溶解圈。Lassen发现[18],可采用70 ℃加热60 min的方法使FN-PDA培养基中纤溶酶原变性失活而用于纤溶酶活性的特异性检测;同时,采用未经70 ℃加热60 min处理和经70 ℃加热60 min处理的两种FN-PDA培养基分别检测纤溶活性的样品则可区分纤溶酶和纤溶酶原激活物的活性,前者可分别在两种不同的FN-PDA培养基上形成纤维蛋白溶解圈,后者只能在未经70 ℃加热60 min处理的FN-PDA培养基上形成纤维蛋白溶解圈。针对海洋真菌的生长特征,本研究改用50%自然海水来配制FN-PDA等培养基,以利于海洋真菌的生长。
| [1] | WHO. Global status report on noncommunicable diseases 2014[R]. Geneva: World Health Organization, 2014 |
| [2] | WHO. Global atlas on cardiovascular disease prevention and control[R]. Geneva: World Health Organization, 2011 |
| [3] | Urano T, Suzuki Y. Accelerated fibrinolysis and its propagation on vascular endothelial cells by secreted and retained tPA[J]. Journal of Biomedicine & Biotechnology, 2012, 2012: 208108 |
| [4] | Marder VJ, Novokhatny V. Direct fibrinolytic agents: biochemical attributes, preclinical foundation and clinical potential[J]. Journal of Thrombosis and Haemostasis, 2009, 8(3): 433-444 |
| [5] | Kunamneni A, Ravuri BD, Ellaiah P, et al. Urokinase - A strong plasminogen activator[J]. Biotechnology and Molecular Biology Reviews, 2008, 3: 58-70 |
| [6] | Aisina RB, Mukhametova LI, Gulin DA, et al. Streptokinase and staphylokinase: differences in the kinetics and mechanism of their interaction with plasminogen, inhibitors, and fibrin[J]. Russian Journal of Bioorganic Chemistry, 2015, 41(5): 506-517 |
| [7] | Thelwell C. Biological standards for potency assignment to fibrinolytic agents used in thrombolytic therapy[J]. Seminars in Thrombosis and Hemostasis, 2014, 40(2): 205-213 |
| [8] | Sharkova TS, Kurakov AV, Osmolovskiy AA, et al. Screening of producers of proteinases with fibrinolytic and collagenolytic activities among micromycetes[J]. Microbiology, 2015, 84(3): 359-364 |
| [9] | Raju EVN, Divakar G. An overview on microbial fibrinolytic proteases[J]. International Journal of Pharmaceutical Sciences and Research, 2014, 5(3): 643-656 |
| [10] | Mondol MA, Shin HJ, Islam MT. Diversity of secondary metabolites from marine Bacillus species: chemistry and biological activity[J]. Marine Drugs, 2013, 11(8): 2846-2872 |
| [11] | Mahajan PM, Nayak S, Lele SS. Fibrinolytic enzyme from newly isolated marine bacterium Bacillus subtilis ICTF-1: media optimization, purification and characterization[J]. Journal of Bioscience and Bioengineering, 2012, 113(3): 307-314 |
| [12] | Jones EBG. Are there more marine fungi to be described?[J]. Botanica Marina, 2011, 54(4): 343-354 |
| [13] | Huang SH, Pan SH, Chen GG, et al. Biochemical characteristics of a fibrinolytic enzyme purified from a marine bacterium, Bacillus subtilis HQS-3[J]. International Journal of Biological Macromolecules, 2013, 62: 124-130 |
| [14] | Vijayaraghavan P, Prakash Vincent SG. A low cost fermentation medium for potential fibrinolytic enzyme production by a newly isolated marine bacterium, Shewanella sp. IND20[J]. Biotechnology Reports, 2015, 7: 135-142 |
| [15] | Matsubara K, Hori K, Matsuura Y, et al. Purification and characterization of a fibrinolytic enzyme and identification of fibrinogen clotting enzyme in a marine green alga, Codium divaricatum[J]. Comparative Biochemistry and Physiology Part B: Biochemistry and Molecular Biology, 2000, 125(1): 137-143 |
| [16] | Wang J, Cui LJ, Lan LB, et al. Diversity of culturable extracellular proteases producing marine fungi isolated from the intertidal zone of Naozhou island in South China Sea[J]. Microbiology China, 2015, 42(2): 238-253 |
| [17] | Astrup T, Müllertz S. The fibrin plate method for estimating fibrinolytic activity[J]. Archives of Biochemistry and Biophysics, 1952, 40(2): 346-351 |
| [18] | Lassen M. Heat denaturation of plasminogen in the fibrin plate method[J]. Acta Physiologica Scandinavica, 1952, 27(4): 371-376 |
| [19] | White TJ, Bruns T, Lee S, et al. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics[A]//PCR Protocol—a Guide to Methods and Applications[M]. San Diego: Academic Press, 1990: 315-322 |
| [20] | Gurung N, Ray S, Bose S, et al. A broader view: microbial enzymes and their relevance in industries, medicine, and beyond[J]. Biomed Research International, 2013, 2013: 329121 |
| [21] | Sasirekha C, Ramya S, Balagurunathan AR. Fibrinolytic enzymes from actinomycetes[J]. Journal of Pharmacy Research, 2012, 5(12): 5457-5463 |
| [22] | Jia Y, Cao XH, Deng Y, et al. Four residues of propeptide are essential for precursor folding of nattokinase[J]. Acta Biochimica et Biophysica Sinica, 2014, 46(11): 957-964 |
| [23] | Shirasaka N, Naitou M, Okamura K, et al. Purification and characterization of a fibrinolytic protease from Aspergillus oryzae KSK-3[J]. Mycoscience, 2012, 53(5): 354-364 |
 2016, Vol. 43
2016, Vol. 43




