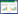扩展功能
文章信息
- 承磊, 郑珍珍, 王聪, 张辉
- CHENG Lei, ZHENG Zhen-Zhen, WANG Cong, ZHANG Hui
- 产甲烷古菌研究进展
- Recent advances in methanogens
- 微生物学通报, 2016, 43(5): 1143-1164
- Microbiology China, 2016, 43(5): 1143-1164
- DOI: 10.13344/j.microbiol.china.150993
-
文章历史
- 收稿日期: 2015-12-01
- 接受日期: 2016-01-28
- 优先数字出版日期(www.cnki.net): 2016-02-03
2. 农业部沼气科学研究所 四川 成都 610041 ;
3. 四川大学轻纺与食品学院 四川 成都 610065
2. Biogas Institute of Ministry of Agriculture, Chengdu, Sichuan 610041, China ;
3. College of Light Industry, Textile and Food Engineering, Sichuan University, Chengdu, Sichuan 610065, China
产甲烷古菌是地球上最古老的生命形式之一,从34.6亿年前就出现在地球上[1-2],它可能还是火星等地外星球上的土著微生物[3]。产甲烷古菌的生长代谢活动与气候变化紧密相关,也许还是2.5亿年前物种大灭绝的元凶[4]。现在每年排放到大气中的甲烷量约500−600 Tg,大约69%是微生物代谢所产生的,产甲烷古菌是其中主要的贡献者[5]。产甲烷古菌分布广泛,存在于湿地[6]、水稻田[7]、淡水和海洋沉积物[8-10]、植物根际[11]、地下油藏和煤藏[12-13]、动物瘤胃和肠道[14-15]及厌氧消化器[16]等自然和人工环境中,甚至存活在干旱的沙漠[17]和高温热泉中[18],在地球主要元素的生物化学循环过程中起着重要作用[19]。产甲烷古菌在有机质厌氧生物降解过程的最后一个环节发挥着关键作用,是生物甲烷形成的直接贡献者,这种可再生能源的开发利用日益受到世界各国的关注[20]。因此,开展产甲烷古菌的研究具有重要的理论和现实意义。
1 产甲烷古菌的生理生化特征 1.1 产甲烷古菌的生理特征产甲烷古菌是严格的厌氧微生物,不能利用氧气作为电子受体,只有少数产甲烷古菌在微量氧气中短时间存活[21-22]。产甲烷古菌的形态学特征与其它微生物差别并不大,但是产甲烷古菌中含有的辅酶F420是参与甲烷代谢途径的关键辅因子,氧化态时吸收420 nm左右的紫外光,激发出约470 nm的蓝绿色荧光[23]。利用这个光谱学特征,可以在荧光/可见光显微镜下区别产甲烷古菌和非产甲烷古菌。产甲烷古菌的生长温度范围非常宽泛[24],最低温度接近0 °C[25-26],最高可达到110 °C[18],在20 mPa的高压条件下产甲烷温度可高达122 °C[27],它也是生长温度最高的微生物之一。pH是影响产甲烷代谢的重要环境因子[28],产甲烷古菌的pH生长范围比较窄,一般在靠近中性条件(pH 6.0−8.0) 生长。只有极少数产甲烷古菌可在pH生长低至4.5左右[29-30],少数产甲烷古菌的最适生长pH值达9.0−9.5[31-32]。此外,产甲烷古菌广泛分布在海洋和盐湖等沉积环境,具有较高的耐盐能力,耐受Na+的最高浓度可达到3.3−3.5 mol/L[31-32]。
1.2 产甲烷古菌的生化特征产甲烷古菌利用的底物种类非常有限,根据底物利用特征主要可分为3种营养类型:氢营养型产甲烷古菌、乙酸营养型产甲烷古菌和甲基营养型产甲烷古菌[33]。
1.2.1 氢营养型产甲烷古菌: 氢营养型产甲烷古菌利用H2、甲酸盐等电子供体还原CO2产生CH4 (表 1),在产甲烷古菌的模式菌株中,3/4以上的产甲烷古菌模式菌株能利用H2/CO2生长。有的氢营养型产甲烷古菌可以利用二元醇、丙酮酸盐作为电子供体,如Methanogenium organophilu m和Methanofollis ethanolicus可以直接利用乙醇、2-丙醇或2-丁醇进行产CH4生长[34-36]。Methanococcus spp.能利用丙酮酸盐作为电子供体(代替H2的功能)还原CO2产生CH4,但是产甲烷速率只有H2/CO2的1%−4%,当H2存在条件下,可以转化丙酮酸盐生成10%−30%的细胞碳,但这不能完全替代其它的碳固定途径[37]。Methanobacterium thermoautotrophicum生长所需要的细胞生物质,最高有80%可以从同化丙酮酸盐中获取[38]。少数氢营养型产甲烷古菌利用CO生长[39]。如Methanothermbacter thermoautotrophicus可以利用低浓度CO (<60%)生长并产CH4,当CO浓度为100%时生长缓慢,其产甲烷速率只有利用H2/CO2的1%[40]。
| 底物利用类型和反应 Substrate utilization and reactions | 标准吉布斯自由能 △G0 (kj/mol CH4)[33] | 产甲烷古菌 Methanogens |
| Hydrogenotrophic | ||
| 4H2+CO2→CH4+2H2O | −135 | Methanobacteriales:Methanobacterium、Methanobrevibacter、Methanothermobacter、Methanothermus Methanococcales:Methanocaldococcus、Methanotorris、Methanococcus、Methanothermococcus Methanocellales:Methanocella Methanomicrobiales:Methanocalculus、Methanocorpusculum、Methanoculleus、Methanofollis、Methanogenium、Methanolacinia、Methanomicrobium、Methanoplanus、Methanolinea、Methanoregula、Methanosphaerula Methanosarcinales:Methanosarcina Methanopyrales:Methanopyrus |
| 4HCOOH→CH4+3CO2+2H2O | −130 | Methanobacteriales:Methanobacterium、Methanobrevibacter、Methanothermobacter Methanococcales:Methanococcus、Methanothermococcus、Methanotorris Methanocellales:Methanocella Methanomicrobiales:Methanocalculus、Methanocorpusculum、Methanoculleus、Methanofollis、Methanogenium、Methanolacinia、Methanolinea、Methanomicrobium、Methanoplanus、Methanoregula、Methanosphaerula、Methanospirillum |
| 2CH3CH2OH+CO2→2CH3COOH+CH4 | −112[59] | Methanomicrobiales:Methanogenium、Methanofollis |
| CO2+4(CH3)2CHOH→CH4+4CH3COCH3+ 2H2O | −37 | Methanobacteriales:Methanobacterium Methanomicrobiales:Methanocorpusculum、Methanoculleus、Methanofollis、Methanogenium、Methanolacinia |
| 4CO+2H2O→CH4+3CO2 | −196 | Methanobacteriales:Methanothermobacter Methanosarcinales:Methanosarcina |
| Methylotrophic | ||
| 4CH3OH→3CH4+CO2+2H2O | −105 | Methanosarcinales:Halomethanococcus、Methanococcoides、Methanohalobium、Methanolobus、Methanomethylovorans、Methanosalsum、Methanosarcina、Methermicoccus |
| CH3OH+H2→CH4+H2O | −113 | Methanobacteriales:Methanobacterium、Methanosphaera Methanomassiliicoccales:Methanomassiliicoccus |
| 2(CH3)2S+3H2O→3CH4+CO2+H2S | −49 | Methanosarcinales:Methanolobus、Methanomethylovorans、Methanosalsum、Methanosarcina |
| 4CH3NH2+2H2O→3CH4+CO2+4NH3 | −75 | Methanosarcinales:Methanococcoides、Methanohalophilus、Methanolobus、Methanomethylovorans、Methanosalsum、Methanosarcina、Methermicoccus |
| 2(CH3)2NH+2H2O→3CH4+CO2+2NH3 | −73 | Methanosarcinales:Methanococcoides、Methanohalophilus、Methanolobus、Methanomethylovorans、Methanosarcina |
| 4(CH3)3N+6H2O→9CH4+3CO2+4NH3 | −74 | Methanosarcinales:Methanococcoides、Methanohalophilus、Methanolobus、Methanomethylovorans、Methanosarcina、Methermicoccus |
| 4(CH3)3N+CH2COO−+2H2O→4(CH3)2NH+CH2COO−+3CH4+CO2 | −721.7[45] | Methanosarcinales:Methanococcoides |
| 4(CH3)3N+CH2CH2OH+6H2O→4H2NCH2CH2OH+9CH4+3CO2+4H+ | −567.6[46] | Methanosarcinales:Methanococcoides |
| 2(CH3)2NCH2CH2OH+2H2O→2H2NCH2CH2OH+3CH4+CO2 | −140.8[46] | Methanosarcinales:Methanococcoides |
| Aciticlasitic | ||
| CH3COOH→CH4+CO2 | −33 | Methanosarcinales:Methanosarcina、Methanothrix |
| 4CH3COCOOH+2H2O→5CH4+7CO2 | −96[47, 60] | Methanosarcinales:Methanosarcina |
1.2.2 甲基营养型产甲烷古菌: 甲基营养型产甲烷古菌能利用甲基类化合物(如甲醇)、甲胺类化合物(如甲胺、二甲胺、三甲胺)和甲基硫化合物(如甲硫醇、二甲基硫)进行产CH4生长(表 1)。甲基营养型产甲烷古菌主要分布在Methanosarcinacea、Methanomassiliicoccus和Methanosphaera中[41-43]。Methanosarcinacea中有8个属都是专性甲基营养型产甲烷古菌,最近研究发现Methanococcoides spp.还可以利用N,N-二甲基乙醇胺、胆碱、甜菜碱等复杂的甲基类化合物生长产CH4[44-46]。Methanosarcina可以利用甲基类化合物生长,有的也利用乙酸、H2/CO2、CO甚至丙酮酸盐[42]。如Methanosarcina barkeri Fusaro可以利用丙酮酸作为碳源和能源产生CH4和CO2,其细胞得率为14 g干重/mol CH4,远高于乙酸发酵产生的3 g干重/mol CH4[47]。M. barkeri利用CO (100%)和甲醇(50 mmol/L)生长,也可以在浓度小于50%的CO中生长产甲烷,并且伴有氢气产生[48]。Methanosphaera只能利用H2/甲醇进行产甲烷代谢,不单独利用甲醇、H2/CO2、乙酸或甲基胺类化合物生长[49-50]。Methanomassiliicoccus luminyensis是从人体粪便分离出的第3个产甲烷古菌新种,只能在H2/甲醇条件下生长产CH4[43]。最近它被归到产甲烷古菌的第7个目中[14, 51-52]。
1.2.3 乙酸营养型产甲烷古菌: 乙酸营养型产甲烷古菌只利用乙酸产生CH4和CO2 (表 1)。目前只有Methanosarcina和Methanothrix (代替之前的Methanosaeta)能利用乙酸产CH4[42, 53]。如上文所述,Methanosarcina能利用多种不同类型底物,如H2/CO2、乙酸和甲基类化合物进行产CH4生长[42]。Methanothrix是专性乙酸营养型产甲烷古菌,包括2个有效种,另外还有1个同名种和1个待有效 种[53]。Methanosarcina利用乙酸的最低极限浓度为0.2−3.0 mmol/L,与此相比,Methanothrix只有7−70 μmol/L[54-56]。这可能与Methanosarcina乙酸激酶对乙酸的亲和力比Methanothrix乙酰辅酶A合成酶的低有关[57]。去年的研究报道发现Methanothrix harudinacea可以直接利用电子还原CO2产生CH4[58]。
2 产甲烷代谢和电子传递产甲烷古菌只能通过产甲烷代谢获取能量来生长繁殖,根据其碳代谢途径差异,可以分为3种(图 1):CO2还原途径、甲基裂解途径和乙酸发酵途径[61]。在碳代谢过程中,产甲烷古菌通过电子呼吸链来推动形成跨膜Na+/H+梯度,再通过A1A0-ATP酶合成ATP[62]。其中铁氧化还原蛋白(Fd)、甲烷吩嗪(MP)、细胞色素(cytochrome)、H2和F420是电子传递的重要载体,介导电子传递的多酶复合体是重要功能单元[63]。
2.1 CO2还原途径CO2首先被甲酰甲烷呋喃脱氢酶还原为甲酰基,并连接在C1载体甲烷呋喃(MFR)上形成CHO-MFR,这是一个吸能反应,需要跨膜离子或质子梯度来驱动。−CHO被转移给另一个C1载体四氢甲烷喋呤(H4MPT),再依次被还原为次甲基(≡CH)、亚甲基(=CH)和甲基(−CH3),催化这些反应需要辅酶F420作为电子载体。在甲基四氢甲烷蝶呤:CoM甲基转移酶(Mtr)的作用下,甲基被转移到第3个C1载体巯基辅酶M(HS-CoM)上,这个跨膜蛋白与Na+跨膜传输耦联,从而在还原CO2过程中推动形成跨膜的Na+梯度(△μNa+)。最后一步是甲基辅酶M还原酶(Mcr)将其转化为甲烷 (图 1)。Mcr是所有产甲烷代谢中的关键功能酶,由3个亚基(α2β2γ2)和辅酶F430[64]组成,编码Mcr α亚基的功能基因(mcrA)可作为研究产甲烷古菌多样性的分子标记物[65],Mcr (或其同系物)可能还是厌氧氧化甲烷的关键功能酶[66]。mcrA不仅存在广古菌门(Euryarchaeota)产甲烷古菌中,还分布在深古菌门(Bathyarchaeota)和其它非Euryarchaeota中[67]。多个C1载体和辅酶(F430和F420)参与了产甲烷代谢过程,一度被认为是产甲烷古菌所特有的辅酶[68],但后来发现它们也存在于非产甲烷古菌中,并执行重要功能[69-71]。
根据产甲烷古菌中是否含有cytochrome,CO2还原产甲烷途径可分为2种,两条途径虽然其碳流向基本一致,但是能量代谢和电子传递方式存在差异(图 2)[73]。含有cytochrome的CO2还原途径存在于Methanosarcina中(图 2a),不含有cytochromes的CO2还原途径主要存在于另外5个产甲烷古菌目(图 2b)[73]。Methanosarcina还原CO2过程中有6个膜结合蛋白复合体与能量储存和电子传递相关(图 2A):MtrA-H、A1A0-ATP合成酶(AhaA-K)、甲烷吩嗪还原氢酶(VhoACG,之前称为甲基紫精还原氢酶或F420-非还原[niFe]氢酶)、甲烷吩嗪依赖型HdrDE、EchA-F和Na+/H+逆向转运体(Nha)。跨膜Mtr从CH3-H4MPT上转移甲基到HS-CoM,同时推动Na+跨膜转运,以用于ATP合成[74]。VhoACG和HdrDE构成了H2:异二硫化物氧化还原酶复合体,Vho是含有细胞色素和氢酶活性的跨膜蛋白,催化H2的氧化,并通过cytochrome b传递电子给MP[75-76]。HdrDE普遍存在Methanosarcina中,也是一种含有cytochrome b的膜结合蛋白[77-78],它接受MP上的电子[79],催化CoM-S-S-CoB还原,同时推动形成的跨膜质子梯度(△μH+)可用于生成ATP。M. barkeri中的Ech是一个膜结合[NiFe]氢酶,与NADH:醌氧化还原酶体系(complex I,真核细胞中的电子传递系统)的序列相似,它借助△μH+推动H2或CO还原Fdox,Fdred可以为甲酰甲烷呋喃脱氢酶催化CO2还原提供电子供体,也可以为丙酮酸合成提供还原力[80]。Nha可能参与形成跨膜Na+/H+梯度[73]。
在不含cytochrome的产甲烷古菌中,其能量保存和电子传递方式不同于Methanosarcina,主要是催化CoM-S-S-CoB还原的MvhADG/HdrABC氧化还原酶复合体是细胞质酶,也没有电子载体MP参与电子传递,不能直接推动形成跨膜Na+/H+梯度。但是它可以通过电子歧化作用推动H2还原Fdox[81]。另外一个差异是Eha利用跨膜Na+、而不是H+进行电子传递,来推动H2还原Fdox,生成的Fdred可以补充合成代谢所消耗掉的还原力[82-83]。在Methanococcus maripaludis中,甘油醛-3-磷酸:铁氧还蛋白氧化还原酶(GAPOR)利用甘油醛-3-磷酸、或CO脱氢酶/乙酰辅酶A (ACS/CODH)氧化CO会产生Fdred,这也能回补CO2还原所需要的还原力[84]。鉴于Fd在CO2还原的第一步和最后一步所起到的连接作用,有科学家把CO2还原途径称为“沃夫循环”(the Wolfe cycle)[85]。
2.2 甲基裂解途径首先甲基由甲基转移酶复合体激活和转运,后者包括2个甲基转移酶。甲基转移酶1 (MT1) 将底物上的甲基转移到MT1上与类咕啉蛋白结合,甲基转移酶2 (MT2) 再转移甲基与HS-CoM连接产生CH3-S-CoM。其中1个CH3-S-CoM通过CO2还原逆途径被氧化产生CO2,这个是耗能反应,需要 Na+梯度驱动,产生的6个电子用于还原F420,从而满足另外3个CH3-S-CoM进入与CO2还原代谢途径所需要的F420H2[86-87]。MT1具有底物特异性,MtaB的底物是甲醇,MtmB的是甲胺,MtbB的是二甲胺,MttB的是三甲胺,MtqB是四甲基胺,MtsB的是甲基硫[88-91]。MT2蛋白序列中都含有高度保守的锌指结构,可以与MT1上的类咕啉蛋白结合,完成甲基化合物的激活和甲基的转移[92]。在Methanosarcina基因组中存在多个拷贝的甲基转移酶基因[93-95],不同类型的甲基类化合物诱导Methanosarcina mazei相应甲基转移酶基因mta、mtb、mtt和mtm的表达[96]。有趣的是,编码甲基转移酶的基因阅读框中含有琥珀密码子(UAG),说明其并没有行使终止蛋白翻译的功能,而是编码第22个生物体必需氨基酸——L-吡咯赖氨酸[97-98]。Methanomicrococcus blatticola和Methanosphaera stadtmanae基因组有编码甲基转移酶基因,但是缺少还原CH3-S-CoM代谢产生CO2的基因,这就需要H2作为电子供体来推动HS-CoM和HS-CoB的再生[99-100]。
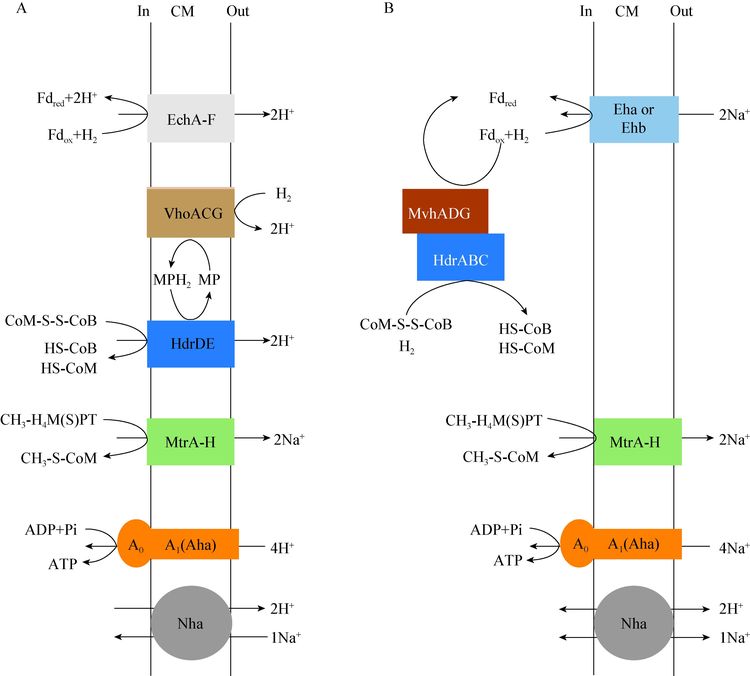
M. mazei有2套膜结合蛋白复合体:H2依赖的异二硫化物氧化还原酶和F420H2依赖的异二硫化物氧化还原酶,分别与HdrDE偶联催化CoM-S-S-CoB的还原,并推动形成跨膜H+梯度[101]。前者在CO2还原(图 2a),后者在甲基合成甲烷过程中起着关键的能量储存作用(图 3a)。在F420H2依赖的异二硫化物氧化还原酶复合体中,F420H2脱氢酶(Fpo)利用F420H2作为电子供体,催化CoM-S-S-CoB还原(图 3a)[102]。Fpo与细菌电子呼吸链中的NADH脱氢酶(NDH-1) 和真核生物中的complex I相似,并通过MP传递F420H2氧化释放的电子[102]。但是并非所有的Methanosarcina都采用这个方式储存能量,M. barkeri通过甲基裂解途径产生的电子,首先被胞内F420H2脱氢酶(Frh)还原为H2,然后扩散到胞外后被膜结合甲烷吩嗪依赖的氢酶(Vht或Vhx)氧化,释放的电子传递到电子呼吸链上,通过MP介导推动HdrDE还原CoM-S-S-CoB,从而转运与Fpo等量的质子,形成相同的△μH+ (图 3a)[103]。
Methanosphaera stadtmanae没有cytochrome,无法像Methanosarcina那样利用膜结合Vho/Hdr或Fpo (或Frh)/Hdr来催化CoM-S-S-CoB还原,并形成△μH+[101]。它利用细胞质MvhADG/HdrABC复合体来催化H2还原CoM-S-S-CoB产生Fdred (图 3B),Fricke等推测MvhADG/HdrABC复合体可能通过HdrB锚定在细胞膜上,从而介导跨膜H+梯度的形成[100]。Thauer等认为跨膜储能氢酶Ehb执行了双重功能:氧化Fdred推动形成跨膜质子梯度,反过来也可以利用跨膜钠离子电势合成ATP[73],这个功能类似Pyrococcus furiosus的能量储存方式[104],但这有待试验验证。第7个产甲烷古菌目中的‘Candidatus Methanoplasma termitum’没有完整的CO2还原途径和cytochrome,无法像M. stadtmanae那样储存能量,可能利用类似Methanosarcina膜结合Fpo/HdrD复合体(没有HdrE亚基,无法氧化F420H2),通过电子歧化催化Fdred的生成和CoM-S-S-CoB的还原[105]。M. luminyensis利用MvhADG/HdrABC氧化2个H2来还原1个CoM-S-S-CoB和1个Fdox,产生的Fdred被Fpo/HdrD用于还原另1个CoM-S-S-CoB,并推动形成△μH+ (图 3c)[106]。
2.3 乙酸发酵途径乙酸发酵产甲烷途径中(图 2),Methanosarcina和Methanothrix激活1个乙酸产生乙酰辅酶A (acetyl CoA)分别需要消耗1和2个ATP[63]。Methanosarcina通过乙酸激酶(AK)和磷酸转乙酰酶(PTA)生成acetyl CoA,Methanothrix利用乙酸辅酶A合成酶(ACS)和焦磷酸酶(PPase)活化乙酸[107]。其中Methanosarcina和Methanothrix激活1个乙酸分别需要消耗1和2个ATP。它们再通过CO脱氢酶/乙酰辅酶A脱羧酶复合体(CODH/ACS)[108-109],氧化乙酸上的羧基为CO2,转移乙酸中的甲基到辅因子四氢八叠喋呤(H4SPT)上,甲基被还原为甲烷的过程与CO2还原途径一致[107, 110]。从M. thermophila中纯化获得水化CO2的碳酸酐酶(CA),可能通过CH3CO2−/HCO3−反向转运系统来促进乙酸的吸收[111]。
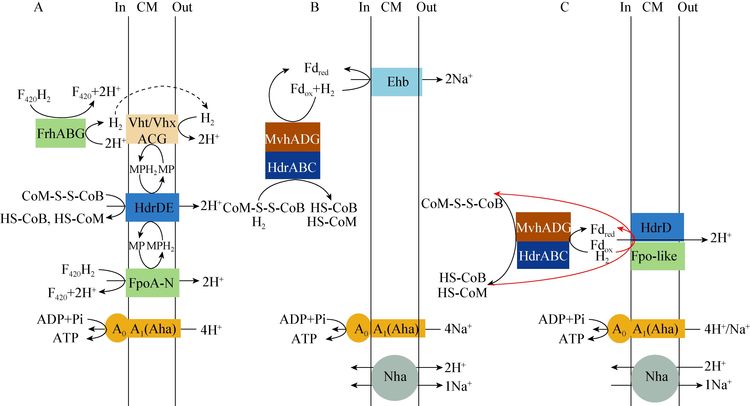
Methanosarcina和Methanothrix的乙酸发酵产甲烷代谢途径类似,都可以利用跨膜Mtr转移甲基(CH3-H4MPT),同时推动Na+跨膜转运[74, 112],这是存在于乙酸发酵途径的第1个与能量储存相关的膜结合蛋白。但是它们催化CoM-S-S-CoB还原和电子传递机制不同[63]。其中M. mazei和M. barkeri利用VhoACG/HdrDE多酶复合体,以H2作为电子供体,在MP介导下传递电子,同时推动形成△μH+ (图 4A),这和Methanosarcina在CO2还原产CH4过程中的电子传递机制一样[63]。另外,它们也利用Ech氧化Fdred产H2,并推动形成△μH+[63]。M. acetivorans不能利用H2/CO2生长,也没有与能量储存相关的Ech,无法氧化Fdred产生H2。但是M. acetivorans含有与Rnf类似的膜结合复合体,具有Fd:MP氧化还原酶活性,能氧化Fdred并推动形成△μNa+,产生的电子通过cytochromec和MP传递给HdrDE,用于还原CoM-S-S-CoB (图 4b)[113-114]。M. thermophila基因组没有ech和rnf相关的基因,它可能通过不完整的F420H2脱氢酶(不含FpoF,不能氧化F420H2)与HdrDE偶联形成多酶复合体,来推动 Na+/H+的跨膜传递,并且可能是直接以Fdred作为电子供体,而不需要其它的电子供体(图 4c)[63]。当然,这还有待于试验验证。
在阳极电极上占绝对优势的Methanobacterium palustre可能直接利用电子还原CO2产生CH4[116]。M. harudinacea和M. barkeri可以直接利用胞外电子来还原CO2产生CH4[58, 117]。这表明不同营养类型的产甲烷古菌都可以直接接受电子产生CH4。 M. acetivorans不利用H2/CO2,但是可以利用CO生长产生乙酸、甲酸和CH4 (甲烷不是主要代谢产物)[118]。M. acetivorans氧化CO产生甲基四氢甲基蝶呤(CH3-THMPT)的代谢途径与CO2还原一样,但是CH3-THMPT进一步还原产甲烷过程涉及到新的甲基转移酶和F420H2:异二硫氧化还原酶来推动形成跨膜质子电势[119]。M. acetivorans通过乙酸裂解产甲烷的反向代谢途径来转化CH3-THMPT产生乙酸,并通过底物磷酸化合成ATP[119]。
3 产甲烷古菌的系统分类产甲烷古菌的生物学特性研究始于20世纪初期,Barker根据细胞形态差异,提出产甲烷古菌可分为八叠球菌、球菌、两种类型的杆菌(根据发酵底物区分杆菌)[120]。但是产甲烷古菌对氧气异常敏感,特别难以纯化和培养,到1947年Schnellen才首次报道了产甲烷古菌Methanobacteriurnformicicum和M. barker的纯培养研究[121-122]。20世纪中期Hungate厌氧操作技术的发明,极大推动了产甲烷古菌纯培养和生理生化特性研究[121, 123],但是依据细胞形态等传统手段对微生物分类的局限性越来越大,甚至有知名微生物学家认为科学分类微生物是一项不可能完成的工作。1977年Woese等提出了基于核糖体rRNA基因序列相似性的系统分类学方法,发现产甲烷古菌不同于细菌和真核细胞,属于独特的一个分支,并命名为古菌[124]。这个“三域学说”彻底改变了人们对微生物多样性的认识,是微生物分子生态学发展的基石。迄今为止生效发表的产甲烷古菌共有6个目(Methanobacteriales、Methanococcales、Methanocellales、Methanomicrobiales、Methanopyrales和Methanosarcinales)、15科、35属,超过150个有效种(表 2),最近发现的Thermoplasmata 代表产甲烷古菌的第7个目[125]。这7个产甲烷古菌目都属于Euryarchaeota。但是最新的研究发现产甲烷古菌还分布在Bathyarchaeota和其它非Euryarchaeota中[67]。下面以目为单元,扼要介绍产甲烷古菌的分类学特征(表 2)。
| 目 Order | 科 Family | 属 Genus | 有效种的 数量 Number of valid published species | 通用底物(特殊底物) common substrates (special substrates) | 最适生长温度的平均值和 范围 mean and range of optimum temperature (oC) | 最适生长pH的平均值和范围 mean and range of optimum pH | 最大NaCl生长的平均值和范围 mean and range of maximum NaCl concentration (%) | DNA G+C mol%的平均值和范围 mean and range of DNA G+C mol% | 分离源 Isolation sources |
| Methano-bacte-riales | Methanobact-eriaceae | Methanobact-erium | 24 | H2/CO2 (H2/M,2P,2B) | 38 (28−65) | 7.1 (5.6−8.6) | 0.69 (0.25−1.40) | 38.1 (31.0−57.0) | 厌氧反应器、水稻田、冻土层、淡水和海洋沉积物 |
| Methanobrev-ibacter | 15 | H2/CO2 (F/CO2) | 36 (30−39) | 7.1 (6.0−7.8) | 0.48 (0.45−0.50) | 30.8 (27.5−38.0) | 哺乳动物粪便、白蚁肠道、厌氧反应器 | ||
| Methanos-phaera | 2 | H2/M (ND) | 37 (37−38) | 6.8 (6.7−6.8) | ND | 24.5 (23.0−26.0) | 人体粪便、兔肠道 | ||
| Methanother-mobacter | 8 | H2/CO2 (F) | 63 (55−70) | 7.3 (6.8−8.1) | 0.56 (0.34−0.68) | 50.4 (41.1−50.4) | 厌氧反应器、油田 | ||
| Methanother-maceae | Methanother-mus | 2 | H2/CO2 (ND) | 86 (83−88) | 6.5 (6.5) | ND | 33 (33.0) | 含硫热泉和泥浆 | |
| Methano-coccales | Methanocal-dococcaceae | Methanocal-dococcus | 7 | H2/CO2 (ND) | 75 (26−85) | 6.4 (6.0−6.5) | 0.93 (0.85−1.0) | 31.5 (30.0−33.0) | 深海热液和沉积物 |
| Methanotor-ris | 2 | H2/CO2 (F) | 82 (75−88) | 6.2 (5.7−6.7) | 1.01 (1.00−1.02) | 32.2 (31.0−33.3) | 深海热液和沉积物 | ||
| Methanococ-caceae | Methanococ-cus | 4 | H2/CO2 (F) | 40 (37−46) | 7.3 (7.0−8.3) | 1 (ND) | 32.8 (32.5−33.0) | 海底沉积物、泥浆 | |
| Methanother-mococcus | 2 | H2/CO2 (F) | 61 (60−63) | 6 (5.5−6.5) | 1.62 (1.60−1.64) | 32.8 (32.0−33.5) | 海底沉积物、热海水 | ||
| Methano-cellales | Methanocell-aceae | Methanocella | 3 | H2/CO2 (F) | 45 (36−45) | 6.9 (6.8−7.0) | 0.13 (0.02−0.30) | 54.7 (52.7−56.6) | 水稻土 |
| Methano-micro-biales | Methanocal-culaceae | Methanocal-culus | 6 | H2/CO2,F ( ) | 36 (35−38) | 7.9 (6.7−9.5) | 1.6 (0.68−2.14) | 51.6 (50.0−55.0) | 油田、盐湖、海水养殖场和垃圾厂 |
| Methanocor-pusculaceae | Methanocor-pusculum | 4 | H2/CO2,F (2P,2B) | 35 (30−37) | 7.0 (7.0) | 0.5 (ND) | 49.6 (47.7−52.0) | 厌氧反应器、沉积物 | |
| Methanomi-crobiaceae | Methanocul-leus | 11 | H2/CO2 (F,2P,2B,CP) | 39 (23−55) | 7.0 (6.5−8.1) | 1.06 (0.54−1.50) | 60.3 (55.2−62.9) | 油田、深部沉积物、湿地和厌氧反应器 | |
| Methanofollis | 5 | H2/CO2,F (E,1P,1B,2P,2B,CP) | 39 (37−43) | 6.8 (6.5−7.0) | 0.55 (0.43−0.63) | 58.3 (54.0−60.9) | 水产养殖场、垃圾处理厂、藕塘、热硫液泥浆 | ||
| Methanoge-nium | 4 | H2/CO2,F (F/CO2,2P,2B,E,1P ) | 24 (15−33) | 7.0 (6.3−7.7) | 0.85 (ND) | 49.4 (46.7−52.0) | 海洋沉积物、南极湖 | ||
| Methanolaci-nia | 2 | H2/CO2 (F,2P) | 39 (37−40) | 7.0 (7.0) | 0.85 (ND) | 47.5 (44.9−50.0) | 油田、海洋沉积物 | ||
| Methanomi-crobium | 1 | H2/CO2,F (ND) | 39 (39) | 6.5 (6.5) | ND | 49 (49.0) | 牛瘤胃 | ||
| Methanopla-nus | 2 | H2/CO2,F (ND) | 36 (32−40) | 7 (7.0−7.1) | ND | 43.1 (38.7−47.5) | 钻井泥浆、 海洋纤毛虫 | ||
| Methanore-gulaceae | Methanolinea | 2 | H2/CO2,F (ND) | 44 (37−50) | 7.0 (7.0) | 0.35 (0.26−0.43) | 55 (53.6−56.4) | 厌氧反应器、水稻田、冻土层、淡水和海洋沉积物 | |
| Methanore-gula | 2 | H2/CO2 (F) | 34 (32−36) | 6.3 (5.1−7.4) | 0.09 (0−0.17) | 55.4 (54.5−56.2) | 泥炭沼泽、 沼气反应器 | ||
| Methanos-phaerula | 1 | H2/CO2,F (ND) | 30 (30) | 5.5 (5.5) | 0.04 (ND) | 58.9 (58.9) | 矿质泥炭沼泽 | ||
| Methanospi-rillaceae | Methanospi-rillum | 4 | H2/CO2 (ND) | 31 (25−45) | 7.4 (7.0−8.0) | 0.2 (0.10−0.30) | 43.2 (40.0−45.3) | 厌氧反应器、 下水污泥、湿地 | |
| Methano-sarcinales | Methanosarc- inaceae | Halomethan-ococcus | 1 | M,Ms (ND) | 36 (36) | 6.9 (6.9) | ND | 43 (43.0) | 盐湖 |
| Methanimicr-ococcus | 1 | H2/M,H2/Ms (ND) | 39 (39) | 7.5 (7.5) | 0.3 (ND) | ND | 蟑螂肠道 | ||
| Methanococ-coides | 4 | TMA (M,Be,Ch,DD) | 27 (23−33) | 7.4 (7.0−7.4) | 0.81 (0.50−1.02) | 41.7 (39.6−43.4) | 海洋沉积物、 南极湖水 | ||
| Methanohalo-bium | 1 | M,Ms (ND) | 48 (48) | 7.3 (.3) | 5.1 (ND) | 36.4 (36.4) | 湖底沉积物 | ||
| Methanohalo-philus | 4 | DMA,TMA (M,) | 35 (31−40) | 7.2 (7.0−7.5) | 2.73 (1.30−3.50) | 45.1 (44.0−48.5) | 沉积物、蓝藻草甸、深层地下水、盐沼 | ||
| Methanolobus | 7 | M (Ms,D,MS) | 36 (25−45) | 7.4 (6.5−8.6) | 1.46 (1.00−2.50) | 41.5 (39.0−45.9) | 海洋和淡水沉积物、煤层水和深层地下水 | ||
| Methanome-thylovo-rans | 3 | M,Ms (D,ME,MT) | 41 (36−50) | 6.5 (6.3−6.8) | 0.23 (0.10−0.40) | 38.4 (37.6−39.2) | 厌氧反应器、湖水沉积物、湿地 | ||
| Methanosal-sum | 2 | M,MMA,D (ND) | 37 (37−38) | 9.3 (9.1−9.5) | 2.8 (2.10−3.50) | 42 (39.2−42.0) | 盐湖、碱水湖 | ||
| Methanosar-cina | 13 | M (H2/CO2,CO,A,Ms MT,D) | 35 (25−50) | 6.9 (6.5−7.8) | 0.85 (0.35−1.20) | 41.1 (36.3−43.5) | 厌氧反应器、深层地下水、海洋和淡水沉积物、冻土 | ||
| Methanotri-chaceae | Methanothrix | 2 | A (ND) | 48 (38-58) | 7.1 (7.0-7.3) | 0.4 (ND) | 57.4 (53.5-61.3) | 厌氧反应器、湖底沉积物 | |
| Methermicoc-caceae | Methermicoc-cus | 1 | M,MMA,TMA (ND) | 65 (65) | 6.3 (6.3) | 1.1 (ND) | 56 (56.0) | 油田 | |
| Methano-pyrales | Methanopyra-ceae | Methanopy-rus | 1 | H2/CO2 (ND) | 98 (98) | 6.5 (6.5) | 0.68 (ND) | 60 (60.0) | 海底热泉沉积物 |
| Methano-massilii-coccales | Methanomas-siliicoccaceae | Methanomas-siliicoc-cus | 1 | H2/M (ND) | 37 (37) | 7.6 (7.6) | ND | 59.9 (59.9) | 人体粪便 |
| 注:通用底物和特殊底物列中的字母所代表的底物如下所示,F:甲酸;M:甲醇;MMA:甲胺;DMA:二甲胺;TMA:三甲胺;Ms:代表甲胺、二甲胺和三甲胺;D:二甲基硫;A:乙酸;MS:甲基硫;E:乙醇;2P:2-丙醇;1P:1-丙醇;1B:1-丁醇;2B:2-丁醇;CP:环戊醇;ME:甲硫醇;Be:甜菜碱Ch:胆碱;DD:N,N-二甲氨基. ND:不存在特殊底物或未检测. | |||||||||
| Note: The letters in lines of “common substrates” and “special substrates” represent the substrates as following,F: Formate; M: Methanol; MMA: Methylamine; DMA: Dimethylamin; TMA: Trimethylamine; Ms: Methylated amines (MMA,DMA,TMA); D: Dimethylsulfide; A: Acetate; MS: Methyl sulfide; E: Ethanol; 2P: 2-Propanol; 1P: 1-Propanol; 1B: 1-Butanol; 2B: 2-Butanol; CP: Cyclopentanol; ME: Methanethiol; Be: Betaine; Ch: Choline; DD: N,N-dimethyl Eamine. ND: Special substrates were or values were not determinated. | |||||||||
Methanobacteriales[126] (甲烷杆菌目)是典型的杆状细胞(长0.6 μm−15.0 μm),有的连接成长丝状。除了Methanosphaera只利用H2/甲醇外,其它都利用H2/CO2生长,有的还能利用甲酸盐、2-丙醇/CO2、2-丁醇/CO2等生长,Methanobacterium lacus和Methanobacteriumveterum也可以利用H2/甲醇[127-128]。Methanobacteriales分为Methanobacteriaceae和Methanothermaceae 2个科,共含有5属53种产甲烷古菌,Methanobacteriaceae共有4个属,其中Methanobacterium、Methanobrevibacter和Methanosphaera都是中温菌,最适生长温度平均在37 °C左右,DNA G+C mol%含量平均是37.8%±9.4%。另外1个Methanothermobacter是嗜热菌,最适生长温度平均是66 °C。Methanothermaceae下只有1属2种(Methanothermus),生理和结构特征明显不同于其它甲烷杆菌目,其最高生长温度达到了97 °C,但是DNA G+C mol%含量只有33%,细胞壁含有假肽聚糖[129]。
3.2 Methanococcales (甲烷球菌目)Methanococcales[130]都是不规则球菌(直径 1 μm-3 μm)、运动,对0.01% (质量体积比) SDS敏感,细胞包裹有蛋白层或覆盖着S-layer,利用H2/CO2,有的可以利用甲酸作为电子供体生长,需要利用硫化物作为硫源,可利用铵作为氮源。Methanococcales都分离自海洋环境,生长需要NaCl,很多也需要酵母膏、硒酸盐和钨酸盐作为生长刺激因子。Methanococcales分为Methanococcaceae和Methanocaldococcaceae 2个科[131-132],共含有4属25种产甲烷古菌。在Methanococcaceae科中,Methanococcus是中温菌,Methanothermococcus是高温菌,最适NaCl生长浓度范围在0.3%−5.0%之间,DNA G+C mol%差别不大,都在32.7%±2.5%左右,它们利用H2/CO2的生长速度非常快,最适条件下的倍增时间分别不超过3 h和1 h[131],因此可以作为产甲烷古菌遗传操作的优良菌种(如Methanococcusmaripaludis)。Methanocaldococcaceae包含Methanocaldococcus和Methanotorris,最适生长温度75−88 °C,最适条件下的倍增时间为25−45 min,它们的生长环境可能与海洋热泉相关[132-133]。
3.3 Methanomicrobiales (甲烷微球目)Methanomicrobiales[134]细胞形态多样,有球状、杆状和鞘杆菌,细胞包裹有单层细胞壁,不含肽聚糖和假肽聚糖。Methanomicrobiales都可以利用H2/CO2,大部分也可以利用甲酸盐作为电子供体来还原CO2,部分利用二级醇/CO2生长。Methanogenium organophilum和Methanofollis ethanolicus利用乙醇/CO2生长产生甲烷和乙酸[34-35]。酵母膏和乙酸盐是大部分Methanomicrobiales的生长必需/刺激因子。Methanomicrobiales分为5个科:Methanocalculaceae、Methanocorpusculaceae、Methanomicrobiaceae、Methanoregulaceae和Methanospirillaceae,在自然界中的分布非常广泛。Methanocalculaceae含1个属(Methanocalculus)和 6个种[32, 135],Methanocalculus都是中温耐盐产甲烷古菌,在中性偏碱条件下生长最快,其中Methanocalculus natronophilus[31]和Methanocalculus alkaliphilus[32]是嗜碱能力最强的产甲烷古菌,其最适pH值也只有9.0−9.5。Methanocalculus的DNA G+C mol%含量较高,一般在50%−56%之间,主要分离自盐湖、油水井、海洋沉积环境中[135]。Methanocorpusculaceae也只含有1个属 (Methanocorpusculum) 4个种,DNA G+C mol%含量比Methanocalculus低,在48%−52%之间。主要分离自厌氧反应器、淡水沉积物等低盐环境,在35 °C、中性pH和低盐条件下生长最佳[136]。Methanomicrobiaceae[137-139]分为6个属26个种:Methanolacinia、Methanomicrobium、Methanofollis、Methanoculleus、Methanogenium和Methanoplanus。利用二级醇/CO2生长的产甲烷古菌也主要集中在Methanomicrobiaceae,它们都是中温菌(生长温度范围:30−40 °C),只有Methanoculleus receptaculi和Methanoculleus thermophilus的最适生长温度在55 °C左右。Methanogenium frigidum最低生长温度可以达到0 °C左右[26]。Methanomicrobiaceae分布比较广泛,先后从水稻田、动物瘤胃、厌氧反应器、湿地、油藏、淡水和海洋沉积环境中分离到了这个科的产甲烷古菌[137]。Methanoregulaceae含有 3个属5个种Methanoregula、Methanolinea和Methanosphaerula[140],这是2012年才建立的新科。它利用H2/CO2生长,乙酸是生长必需因子,DNA G+C mol%含量在53%−59%之间。除Methanolinea tarda是嗜热菌(最适生长温度为50 °C),其它都是中温菌。这个科含有最嗜酸的产甲烷古菌M. boonei,最适pH生长条件为5.5,另外M. palustris也可以在pH 4.8条件下生长[29-30, 141]。Methanospirillaceae通常是长弯曲杆菌,并且可以连接成长的细丝,主要分离自厌氧反应器和湿地,它们生长都不需要NaCl,DNA G+C mol%含量在40%−49%之间[142]。新近分离的2个新种Methanospirillum psychrodurum和Methanospirillum stamsii都可以在4−5 °C的低温条件下生长[143-144]。
3.4 Methanopyrales (甲烷炙热古菌)Methanopyrales自20世纪90年代从海底热泉中分离出以来,迄今只有1个种M. kandleri,这是一个杆状、自养型产甲烷古菌,只利用H2/CO2生长产甲烷,最适生长温度为98 °C[18, 145],在20 MPa的高压条件下,M. kandleri 116最高生长温度可以达到122 °C[27],这也是生长温度最高的产甲烷古菌。M. kandleri的NaCl生长范围较宽(0−0.68 mol/L),最适pH是6.5[18, 145],DNA G+C mol%为61.2%,也是目前已知的G+C含量最高的产甲烷古菌之一[146]。
3.5 Methanosarcinales (甲烷八叠球菌目)Methanosarcinales[147]是细胞形态最多样的一个目,有球状、杆状、鞘状、八叠状和丝状,大部分的细胞壁含有蛋白质层。这也是自然界中分布最广泛的产甲烷古菌之一。Methanosarcinales包括有3个科:Methanotrichaceae、Methermicoccaceae和Methanosarcinaceae。Methanotrichaceae只包括1个有效属Methanothrix,之前曾用名是Methanosaeta,关于哪个是有效属名这个问题,学界争论了很长时间[53, 148]。Methanothrix包括2个有效种Methanothrix concilii和Methanothrix thermoacetophila,和1个同名种Methanothrix harundina cea comb. nov.,另外Methanosaeat pelagica也是专性乙酸营养型产甲烷古菌,但用的是无效属名[148]。Methanothrix[53]只能发酵乙酸产生甲烷,最近研究发现它也可以利用电子来还原CO2产生甲烷[58]。Methanothrix生长缓慢,倍增时间需要1−12 d,培养条件苛刻,很难在固体培养基上形成菌落。Methanothrix可以在中温和高温条件下生长,但最高生长温度低于70 °C。M. harundinacea是杆状细菌,添加群感信号分子Acyl homoserine lactones (AHLs)可以促进形成长丝状细胞[149],这样有利于颗粒污泥的形成,可以提升沼气发酵的稳定性[150]。Methermicoccaceae含 1个种Methermicoccus shengliensis,只能利用甲基类化合物生长产甲烷,最适生长温度为65 °C,是最适生长温度最高的甲基营养型产甲烷古菌,但是从系统发育分析发现它更靠近Methanothrichaceae[151]。Methanosarcinaceae包括9属37种:Halomethanococcus (模式菌株已丢失)、Methani micrococcus、Methanococcoides、Methanohalobium、Methanohalophilus、Methanolobus、Methanomethylovorans、Methanosalsum和Methanosarcina,其多样性仅次于Methanobacteriaceae (49个种)。Methanosarcinaceae[42]是生理生化特征最多样性的产甲烷古菌,其中Methanosarcina可以利用H2/CO2、乙酸和甲醇等不同营养类型的底物生长,并利用不同电子传递方式储存能量(见第1和2部分的论述)。除Methanosarcina的其它Methanosarcinaceae只能利用甲基类化合物生长,并且都是革兰氏阴性菌,很多Methanosarcinaceae生长需要添加Na+和Mg2+。大部分Methanosarcinaceae是中温菌,最适生长温度在23−40 °C之间,最高生长温度不超过60 °C,如Methanolobus zinderi、Methanomethylovorans thermophila和Methanosarcina thermophila的最适生长温度是45−50 °C。有的可以在接近0 °C条件下生长产甲烷,如Methanococcoides alaskense、Methanosarcina lacustris、Methanosarcina spelaei和Methanosarcina soligelidi,以及未有效发表的Methanolobus psychrophilus[9, 25, 152-154],另外M. spelaei和M. soligelidi生长温度非常宽泛,最高生长温度可以达到54 °C。除Methanosalsum zhilinae和M. natronophilum的最适pH在9.0左右外,其它Methanosarcinaceae都在中性pH条件下生长最快。除Methanomethylovorans uponensis[155]、Methanomethylovorans hollandica[156]和Methanolobus chelungpuianus (未有效发表)[157]最适NaCl生长浓度接近0外,大部分Methanosarcinaceae的最适NaCl生长浓度都在0.1−0.5 mol/L。Methanohalophilus部分菌株耐受NaCl浓度可达到2.0−2.2 mol/L。Methanosarcinaceae的平均DNAG+C mol%在42%±3%之间,从厌氧反应器、淡水和海水沉积物、盐湖、湿地等环境都可以分离这个科的产甲烷古菌。
3.6 Methanocellales (甲烷胞菌目)Methanocellales[158]有1属(Methanocella) 3个种,它们呈不规则杆菌,有的在生长后期会变成拟球菌。Methanocella能利用H2/CO2生长产甲烷,有的还可以利用甲酸盐作为电子供体。Methanocella全部分离自水稻土,是水稻根际甲烷排放的主要贡献者[11]。有趣的是,水稻田温度一般在15−30 °C[159],3个属Methanocella模式菌株的最适生长温度分别是35、45和55 °C[158],明显高于水稻土的原位温度,难道是因为水稻根际活跃的微生物代谢活动导致根际原位温度上升,而Methanocella较其它产甲烷古菌能更好的适应这种温度变化/胁迫么?Methanocella广泛分布在全球不同的水稻田[7],可能与其对低浓度H2的高亲和力有关[160]。Methanocella基因组中存在编码超氧化物歧化酶等抗氧基因,可能与它应对氧胁迫机制有关[161-162]。最近研究发现一类未培养的Methanocella (属于Rice cluster II)是永久冻土层解冻过程甲烷排放的主要功能菌[163]。采用宏基因组技术获得了它的近全长基因组序列,发现它具有CO2还原产甲烷代谢的基因,暂命名为‘Candidatus Methanoflorens stordalenmirensis’,它可能代表了Methanocellales中的另一个新科[164]。
3.7 ‘Methanomassiliicoccales’ (甲烷马赛球 菌目)‘Methanomassiliicoccales’是近年来发现的一个的甲烷古菌新目。这个目中只有1个纯培养物M. luminyensisB10T,它是从人体粪便中分离到的一个产甲烷古菌新菌,只能利用H2/甲醇进行产甲烷生长,而不能利用甲酸盐、乙酸盐、三甲胺、乙醇和二元醇[43]。分析发现它与Thermoplasmatales中的‘Candidatus Aciduliprofundum boonei’的16S rRNA基因相似性最高,但是只有83%,与产甲烷古菌中的Methanobrevibacter smithii相似性最高(只有76%)[43]。Paul等发现海洋沉积物、水稻土、白蚁、蟑螂和哺乳动物肠道中的未培养环境基因序列能与M. luminyensis聚成一类,处于Euryarchaeota的顶部,并远离其它产甲烷古菌,培养实验也证实了它可 以利用H2/甲醇生长产甲烷。因此,作者提出了第7个产甲烷古菌新目‘Methanoplasmatales’[14]。Borrel等从老年人粪便中富集获得了‘Candidatus Methanomethylophilus alvus’的基因组序列,发现它不仅可以利用H2/甲醇生长产甲烷,可能还利用甲胺类化合物,从而提出它也属于第7个产甲烷古菌新目[165]。Iino等发现从厌氧反应器中富集到的‘Candidatus Methanogranum caenicola’可以利用H2/甲醇生长,根据细菌命名准则把‘Methanoplasmatales’改 为‘Methanomassiliicoccales’[125]。比较‘Ca. Methanomethylophilus alvus’、‘Candidatus Methanomassiliicoccus intestinalis’和M. luminyensis的基因组序列,发现它们都采用相同的基因来储存能量[166]。‘Ca. Methanoplasma termitum’的能量储存机制不同于M. stadtmanae,可能利用Fpo/HdrD复合体(没有HdrE亚基,无法氧化F420H2),以电子歧化的方式来推动Fdred的生成和CoM-S-S-CoB的还原[105]。M. luminyensis利用MvhADG/HdrABC和Fpo/HdrD复合体,以Fd作为电子载体来分别催化CoM-S-S-CoB还原和形成跨膜质子电势[106]。
目前分离获得的产甲烷古菌都属于Euryarchaeota,但是最新的研究表明产甲烷古菌还分布在非Euryarchaeota中。Evans等[67]采用宏基因组技术,从地下煤层水构建获得了2个微生物的全基因组,系统发育分析表明它们属于Bathyarchaeota,但是编码产甲烷代谢相关的功能基因。其中BA1中还含有mtsA、mtbA、mtaA、mttBC、mtbBC和mtrH等与甲基裂解途径相关的基因,这些基因与Methanomassiliicoccales的相似,表明它可能利用H2/甲基化合物产甲烷。BA2也编码有甲基裂解途径相关的基因,但是其能量储存和电子传递机制可能不同于BA1。此外,还发现不属于Euryarchaeota的mcrA基因广泛分布在自然环境中,表明这类非Euryarchaeota产甲烷古菌可能在全球甲烷循环中也起着重要作用。
4 产甲烷古菌的生态学功能复杂有机质厌氧降解产生短链脂肪酸(VFA:乙、丙、丁酸)、H2/CO2和各种醇类化合物,当O2、Fe(III)、NO3−和SO42−等外源电子受体供给有限的时候,细菌需要与产甲烷古菌耦联进行产甲烷代谢,才能完全厌氧降解有机质[167]。这是因为在标准热力学条件下,中间代谢产物(如丙酸、丁酸等)降解产乙酸和H2是一个吸热反应,无法自发进行[167]。而产甲烷古菌可以利用H2/CO2生长产CH4,使得原本不利的吸热反应变成了放能反应,从而保证有机质的厌氧降解可以持续稳定进行[167]。产甲烷古菌通过种间氢和/或甲酸转移来推动有机质厌氧降解产甲烷过程[168-169]。最近的研究发现产甲烷古菌还可以直接接受胞外电子还原CO2并产生CH4[58, 117]。在湿地、水稻田、海洋沉积物、白蚁和瘤胃动物、厌氧反应器等缺氧环境中,产甲烷古菌虽然不是复杂有机质起始降解的参与者,但是有机质持续降解的重要推动者。
产甲烷古菌在自然界中分布非常广泛,在全球碳生物地球化学循环过程中起着重要作用。每年排放到大气中的甲烷量约500−600 Tg,大约69%是产甲烷古菌代谢产生的[5]。基于微生物分子生态学技术研究发现很多未培养产甲烷古菌是环境中甲烷排放的主要参与者。Lu等采用稳定同位素示踪技术证实Rice cluster I是水稻田根际甲烷排放的主要贡献者[11]。后续的分离培养研究证实了这类微生物代表产甲烷古菌的第6个目(Methanocella)[158]。在北环极永久冻土层蕴藏的碳汇超过全球地下有机碳的50%[170],全球气候变暖导致永久冻土层解冻,其中一类未培养的Methanocella(属于Rice cluster II)是永久冻土层解冻过程甲烷排放的主要功能菌[163]。采用宏基因组技术获得了未培养产甲烷古菌‘Ca. Methanoflorens stordalenmirensis’的基因组草图,它可能通过CO2还原来进行产甲烷代谢[164]。从地下煤层水发现了2个属于Bathyarchaeota的未培养产甲烷古菌,可能是通过甲基裂解途径参与地下煤层甲烷的形成[67]。在动物瘤胃和昆虫肠道等生境中发现的第7个产甲烷古菌新目‘Methanoplasmatales’,可以利用H2/甲醇生长产甲烷,是动物甲烷排放的一个重要贡献者[14]。当然,产甲烷古菌可能不仅参与了甲烷的产生,可能还是特殊环境条件下甲烷的消耗者[66],限于篇幅,不在此讨论。
产甲烷古菌既是全球碳素生物地球化学的重要参与者和推动者,也可以作为可再生能源的生产者,与发酵细菌和互营菌一起合作,利用畜禽粪便和秸秆等农业废弃物,为人类社会提供清洁干净的可再生能源——甲烷。以中国为例,截止2014年底,共建有4.15千万口农村户用沼气池,年产沼气136.7亿m3,大中型沼气工程接近10万座,年产沼气21亿m3[171]。在沼气发酵反应器中,水解细菌降解非水溶性复杂有机物产生可溶性化合物,发酵细菌再降解产生H2/CO2、短链脂肪酸和醇等小分子化合物,互营细菌和产甲烷古菌通过互营代谢产生CH4。其中主要是通过乙酸发酵和CO2还原途径来介导甲烷的产生。沼气发酵的物料成分复杂,参与沼气发酵的微生物群落多变,产甲烷古菌的生长条件比较苛刻,容易受到环境因子(如pH、O2、温度和铵)的干扰[172],因此控制和优化沼气发酵过程,提升产甲烷古菌在内的沼气发酵微生物的代谢活性,提高物料的利用率和转化率,一直是沼气发酵微生物研究的重点。
5 展望近年来,基于高通量测序的组学技术,揭示了大量未培养产甲烷古菌在地球碳循环中起着重要的生态学功能,但是缺少纯培养物及其遗传操作系统限制了人们深入研究它们的生理生化和遗传学特性。因此,如何应用新的厌氧培养思路和方法,从复杂环境中定向分离生长条件苛刻、但是具有特殊生态学功能的产甲烷古菌,将是微生物家必须面对和解决的一个关键问题。产甲烷代谢直接与产甲烷古菌的能量储存相结合,是产甲烷古菌获取能量进行生长的直接代谢途径,不同营养类型的产甲烷古菌可能采用的不同电子传递机制来推动能量储存,因此,结合比较基因组学、遗传突变和转录本分析,研究产甲烷古菌的能量储存和电子传递过程,将为我们深入阐明产甲烷古菌的生长特性奠定基础。产甲烷古菌在生物圈分布广泛,具有重要的生态学功能,但是自然环境因子(如温度、pH、溶氧、竞争性电子受体)的波动和变化会影响产甲烷古菌的种群结构和功能,因此,在面对这些环境胁迫时,原位产甲烷古菌(系)响应和应答机制研究将是人们深入了解产甲烷古菌生态学功能的重要途径。
致谢: 感谢中国农业科学院农业资源与农业区划研究所阮志勇博士为本文成稿提供的帮助。| [1] | Ueno Y, Yamada K, Yoshida N, et al. Evidence from fluid inclusions for microbial methanogenesis in the early Archaean era[J]. Nature, 2006, 440 (7083): 516–519. |
| [2] | Battistuzzi FU, Feijao A, Hedges SB. A genomic timescale of prokaryote evolution: insights into the origin of methanogenesis, phototrophy, and the colonization of land[J]. BMC Evolutionary Biology, 2004, 4 (1): 44. |
| [3] | Tung HC, Bramall NE, Price PB. Microbial origin of excess methane in glacial ice and implications for life on Mars[J]. Proceedings of the National Academy of Sciences of the United States of America, 2005, 102 (51): 18292–18296. |
| [4] | Rothman DH, Fournier GP, French KL, et al. Methanogenic burst in the end-Permian carbon cycle[J]. Proceedings of the National Academy of Sciences of the United States of America, 2014, 111 (15): 5462–5467. |
| [5] | Conrad R. The global methane cycle: recent advances in understanding the microbial processes involved[J]. Environmental Microbiology Reports, 2009, 1 (5): 285–292. |
| [6] | Zhang GS, Tian JQ, Jiang N, et al. Methanogen community in Zoige wetland of Tibetan plateau and phenotypic characterization of a dominant uncultured methanogen cluster ZC-I[J]. Environmental Microbiology, 2008, 10 (7): 1850–1860. |
| [7] | Conrad R, Erkel C, Liesack W. Rice Cluster I methanogens, an important group of Archaea producing greenhouse gas in soil[J]. Current Opinion in Biotechnology, 2006, 17 (3): 262–267. |
| [8] | Newberry CJ, Webster G, Cragg BA, et al. Diversity of prokaryotes and methanogenesis in deep subsurface sediments from the Nankai Trough, Ocean Drilling Program Leg 190[J]. Environmental Microbiology, 2004, 6 (3): 274–287. |
| [9] | Simankova MV, Parshina SN, Tourova TP, et al. Methanosarcina lacustris sp. nov., a new psychrotolerant methanogenic archaeon from anoxic lake sediments[J]. Systematic and Applied Microbiology, 2001, 24 (3): 362–367. |
| [10] | Ferry JG, Lessner DJ. Methanogenesis in marine sediments[J]. Annals of the New York Academy of Sciences, 2008, 1125 (1): 147–157. |
| [11] | Lu YH, Conrad R. In situ stable isotope probing of methanogenic archaea in the rice rhizosphere[J]. Science, 2005, 309 (5737): 1088–1090. |
| [12] | Strąpoć D, Mastalerz M, Dawson K, et al. Biogeochemistry of microbial coal-bed methane[J]. Annual Review of Earth and Planetary Sciences, 2011, 39 (1): 617–656. |
| [13] | Cheng L, Qiu TL, Deng Y, et al. Recent advances in anaerobic microbiology of petroleum reservoirs[J]. Chinese Journal of Applied & Environmental Biology, 2006, 12 (5): 740–744. (in chinese) 承磊, 仇天雷, 邓宇, 等. 油藏厌氧微生物研究进展[J]. 应用与环境生物学报, 2006, 12 (5):740–744. |
| [14] | Paul K, Nonoh JO, Mikulski L, et al. “Methanoplasmatales,” Thermoplasmatales-related archaea in termite guts and other environments, are the seventh order of methanogens[J]. Applied and Environmental Microbiology, 2012, 78 (23): 8245–8253. |
| [15] | Schulz K, Hunger S, Brown GG, et al. Methanogenic food web in the gut contents of methane-emitting earthworm Eudrilus eugeniae from Brazil[J]. The ISME Journal, 2015, 9 (8): 1778–1792. |
| [16] | Karakashev D, Batstone DJ, Angelidaki I. Influence of environmental conditions on methanogenic compositions in anaerobic biogas reactors[J]. Applied and Environmental Microbiology, 2005, 71 (1): 331–338. |
| [17] | Angel R, Claus P, Conrad R. Methanogenic archaea are globally ubiquitous in aerated soils and become active under wet anoxic conditions[J]. The ISME Journal, 2012, 6 (4): 847–862. |
| [18] | Huber R, Kurr M, Jannasch HW, et al. A novel group of abyssal methanogenic archaebacteria (Methanopyrus) growing at 110 °C[J]. Nature, 1989, 342 (6251): 833–834. |
| [19] | Offre P, Spang A, Schleper C. Archaea in biogeochemical cycles[J]. Annual Review of Microbiology, 2013, 67 (1): 437–457. |
| [20] | Demirel B, Scherer P. The roles of acetotrophic and hydrogenotrophic methanogens during anaerobic conversion of biomass to methane: a review[J]. Reviews in Environmental Science and Bio/Technology, 2008, 7 (2): 173–190. |
| [21] | Horne AJ, Lessner DJ. Assessment of the oxidant tolerance of Methanosarcina acetivorans[J]. FEMS Microbiology Letters, 2013, 343 (1): 13–19. |
| [22] | Fetzer S, Conrad R. Effect of redox potential on methanogenesis by Methanosarcina barkeri[J]. Archives of Microbiology, 1993, 160 (2): 108–113. |
| [23] | Cheeseman P, Toms-Wood A, Wolfe RS. Isolation and properties of a fluorescent compound, factor420, from Methanobacterium strain M.o.H.[J]. Journal of Bacteriology, 1972, 112 (1): 527–531. |
| [24] | Dong XZ, Chen ZJ. Psychrotolerant methanogenic archaea: diversity and cold adaptation mechanisms[J]. Science China Life Sciences, 2012, 55 (5): 415–421. |
| [25] | Zhang GS, Jiang N, Liu XL, et al. Methanogenesis from methanol at low temperatures by a novel psychrophilic methanogen, “Methanolobus psychrophilus” sp. nov., prevalent in Zoige Wetland of the Tibetan Plateau[J]. Applied and Environmental Microbiology, 2008, 74 (19): 6114–6120. |
| [26] | Franzmann PDF, Liu YT, Balkwill DL, et al. Methanogenium frigidum sp. nov., a psychrophilic, H2-using methanogen from Ace Lake, Antarctica[J]. International Journal of Systematic Bacteriology, 1997, 47 (4): 1068–1072. |
| [27] | Takai K, Nakamura K, Toki T, et al. Cell proliferation at 122 °C and isotopically heavy CH4 production by a hyperthermophilic methanogen under high-pressure cultivation[J]. Proceedings of the National Academy of Sciences of the United States of America, 2008, 105 (31): 10949–10954. |
| [28] | Jabłoński S, Rodowicz P, Łukaszewicz M. Methanogenic archaea database containing physiological and biochemical characteristics[J]. International Journal of Systematic and Evolutionary Microbiology, 2015, 65 (Pt 4): 1360–1368. |
| [29] | Bräuer SL, Cadillo-Quiroz H, Yashiro E, et al. Isolation of a novel acidiphilic methanogen from an acidic peat bog[J]. Nature, 2006, 442 (7099): 192–194. |
| [30] | Cadillo-Quiroz H, Yavitt JB, Zinder SH. Methanosphaerula palustris gen. nov., sp. nov., a hydrogenotrophic methanogen isolated from a minerotrophic fen peatland[J]. International Journal of Systematic and Evolutionary Microbiology, 2009, 59 (5): 928–935. |
| [31] | Zhilina TN, Zavarzina DG, Kevbrin VV, et al. Methanocalculus natronophilus sp. nov., a new alkaliphilic hydrogenotrophic methanogenic archaeon from a soda lake, and proposal of the new family Methanocalculaceae[J]. Microbiology, 2013, 82 (6): 698–706. |
| [32] | Sorokin DY, Abbas B, Merkel AY, et al. Methanosalsum natronophilum sp. nov., and Methanocalculus alkaliphilus sp. nov., haloalkaliphilic methanogens from hypersaline soda lakes[J]. International Journal of Systematic and Evolutionary Microbiology, 2015, 65 (10): 3739–3745. |
| [33] | Liu YC, Whitman WB. Metabolic, phylogenetic, and ecological diversity of the methanogenic archaea[J]. Annals of the New York Academy of Sciences, 2008, 1125 (1): 171–189. |
| [34] | Widdel F, Rouvière PE, Wolfe RS. Classification of secondary alcohol-utilizing methanogens including a new thermophilic isolate[J]. Archives of Microbiology, 1988, 150 (5): 477–481. |
| [35] | Imachi H, Sakai S, Nagai H, et al. Methanofollis ethanolicus sp. nov., an ethanol-utilizing methanogen isolated from a lotus field[J]. International Journal of Systematic and Evolutionary Microbiology, 2009, 59 (4): 800–805. |
| [36] | Widdel F. Growth of methanogenic bacteria in pure culture with 2-propanol and other alcohols as hydrogen donors[J]. Applied and Environmental Microbiology, 1986, 51 (5): 1056–1062. |
| [37] | Yang YL, Ladapo J, Whitman WB. Pyruvate oxidation by Methanococcus spp.[J]. Archives of Microbiology, 1992, 158 (4): 271–275. |
| [38] | Hüster R, Thauer RK. Pyruvate assimilation by Methanobacterium thermoautotrophicum[J]. FEMS Microbiology Letters, 1983, 19 (2/3): 207–209. |
| [39] | Ferry JG. CO in methanogenesis[J]. Annals of Microbiology, 2010, 60 (1): 1–12. |
| [40] | Daniels L, Fuchs G, Thauer RK, et al. Carbon monoxide oxidation by methanogenic bacteria[J]. Journal of Bacteriology, 1977, 132 (1): 118–126. |
| [41] | Oren A. The family Methanobacteriaceae[A]//Rosenberg E, DeLong EF, Lory S, et al. The Prokaryotes[M]. Berlin Heidelberg: Springer, 2014: 165-193 |
| [42] | Oren A. The family Methanosarcinaceae[A]//Rosenberg E, DeLong EF, Lory S, et al. The Prokaryotes[M]. Berlin Heidelberg: Springer, 2014: 259-281 |
| [43] | Dridi B, Fardeau ML, Ollivier B, et al. Methanomassiliicoccus luminyensis gen. nov., sp. nov., a methanogenic archaeon isolated from human faeces[J]. International Journal of Systematic and Evolutionary Microbiology, 2012, 62 (Pt 8): 1902–1907. |
| [44] | L’Haridon S, Chalopin M, Colombo D, et al. Methanococcoides vulcani sp. nov., a marine methylotrophic methanogen that uses betaine, choline and N,N-dimethylethanolamine for methanogenesis, isolated from a mud volcano, and emended description of the genus Methanococcoides[J]. International Journal of Systematic and Evolutionary Microbiology, 2014, 64 (Pt 6): 1978–1983. |
| [45] | Watkins AJ, Roussel EG, Parkes RJ, et al. Glycine betaine as a direct substrate for methanogens (Methanococcoides spp.)[J]. Applied and Environmental Microbiology, 2014, 80 (1): 289–293. |
| [46] | Watkins AJ, Roussel EG, Webster G, et al. Choline and N,N-dimethylethanolamine as direct substrates for methanogens[J]. Applied and Environmental Microbiology, 2012, 78 (23): 8298–8303. |
| [47] | Bock AK, Prieger-Kraft A, Schönheit P. Pyruvate—a novel substrate for growth and methane formation in Methanosarcina barkeri[J]. Archives of Microbiology, 1994, 161 (1): 33–46. |
| [48] | OʼBrien JM, Wolkin RH, Moench TT, et al. Association of hydrogen metabolism with unitrophic or mixotrophic growth of Methanosarcina barkeri on carbon monoxide[J]. Journal of Bacteriology, 1984, 158 (1): 373–375. |
| [49] | Miller TL, Wolin MJ. Methanosphaera stadtmaniae gen. nov., sp. nov.: a species that forms methane by reducing methanol with hydrogen[J]. Archives of Microbiology, 1985, 141 (2): 116–122. |
| [50] | Biavati B, Vasta M, Ferry JG. Isolation and characterization of “Methanosphaera cuniculi” sp. nov.[J]. Applied and Environmental Microbiology, 1988, 54 (3): 768–771. |
| [51] | Borrel G, Harris HMB, Parisot N, et al. Genome sequence of “Candidatus Methanomassiliicoccus intestinalis” issoire-Mx1, a third Thermoplasmatales-related methanogenic archaeon from human feces[J]. Genome Announcements, 2013, 1 (4): e00453–13. |
| [52] | Borrel G, OʼToole PW, Harris HMB, et al. Phylogenomic data support a seventh order of methylotrophic methanogens and provide insights into the evolution of methanogenesis[J]. Genome Biology and Evolution, 2013, 5 (10): 1769–1780. |
| [53] | Oren A. The family Methanotrichaceae[A]//Rosenberg E, DeLong EF, Lory S, et al. The Prokaryotes[M]. Berlin Heidelberg: Springer, 2014: 297-306 |
| [54] | Min H, Zinder SH. Kinetics of acetate utilization by two thermophilic acetotrophic methanogens: Methanosarcina sp. strain CALS-1 and Methanothrix sp. strain CALS-1[J]. Applied and Environmental Microbiology, 1989, 55 (2): 488–491. |
| [55] | Westermann P, Ahring BK, Mah RA. Threshold acetate concentrations for acetate catabolism by aceticlastic methanogenic bacteria[J]. Applied and Environmental Microbiology, 1989, 55 (2): 514–515. |
| [56] | Jetten MSM, Stams AJM, Zehnder AJB, et al. Methanogenesis from acetate: a comparison of the acetate metabolism in Methanothrix soehngenii and Methanosarcina spp.[J]. FEMS Microbiology Letters, 1992, 88 (3/4): 181–198. |
| [57] | Jetten MSM, Stams AJM, Zehnder AJB. Acetate threshold values and acetate activating enzymes in methanogenic bacteria[J]. FEMS Microbiology Ecology, 1990, 73 (4): 339–344. |
| [58] | Rotaru AE, Shrestha PM, Liu FH, et al. A new model for electron flow during anaerobic digestion: direct interspecies electron transfer to Methanosaeta for the reduction of carbon dioxide to methane[J]. Energy & Environmental Science, 2014, 7 (1): 408–415. |
| [59] | Schink B. Syntrophic associations in methanogenic degradation[A]//Overmann J. Molecular Basis of Symbiosis: Progress in Molecular and Subcellular Biology[M]. Berlin Heidelberg: Springer, 2006, 41: 1-19 |
| [60] | López Muoz MM, Schönheit P, Metcalf WW. Genetic, genomic, and transcriptomic studies of pyruvate metabolism in Methanosarcina barkeri fusaro[J]. Journal of Bacteriology, 2015, 197 (22): 3592–3600. |
| [61] | Thauer RK. Biochemistry of methanogenesis: a tribute to Marjory Stephenson[J]. Microbiology, 1998, 144 (9): 2377–2406. |
| [62] | Schlegel K, Leone V, Faraldo-Gómez JD, et al. Promiscuous archaeal ATP synthase concurrently coupled to Na+ and H+ translocation[J]. Proceedings of the National Academy of Sciences of the United States of America, 2012, 109 (3): 947–952. |
| [63] | Welte C, Deppenmeier U. Bioenergetics and anaerobic respiratory chains of aceticlastic methanogens[J]. Biochimica et Biophysica Acta (BBA) - Bioenergetics, 2014, 1837 (7): 1130–1147. |
| [64] | Ermler U, Grabarse W, Shima S, et al. Crystal structure of methyl-coenzyme M reductase: the key enzyme of biological methane formation[J]. Science, 1997, 278 (5342): 1457–1462. |
| [65] | Lueders T, Chin KJ, Conrad R, et al. Molecular analyses of methyl-coenzyme M reductase α-subunit (mcrA) genes in rice field soil and enrichment cultures reveal the methanogenic phenotype of a novel archaeal lineage[J]. Environmental Microbiology, 2001, 3 (3): 194–204. |
| [66] | Scheller S, Goenrich M, Boecher R, et al. The key nickel enzyme of methanogenesis catalyses the anaerobic oxidation of methane[J]. Nature, 2010, 465 (7298): 606–608. |
| [67] | Evans PN, Parks DH, Chadwick GL, et al. Methane metabolism in the archaeal phylum Bathyarchaeota revealed by genome-centric metagenomics[J]. Science, 2015, 350 (6259): 434–438. |
| [68] | DiMarco AA, Bobik TA, Wolfe RS. Unusual coenzymes of methanogenesis[J]. Annual Review of Biochemistry, 1990, 59 (1): 355–394. |
| [69] | Chistoserdova L, Vorholt JA, Thauer RK, et al. C1 transfer enzymes and coenzymes linking methylotrophic bacteria and methanogenic Archaea[J]. Science, 1998, 281 (5373): 99–102. |
| [70] | Krishnakumar AM, Sliwa D, Endrizzi JA, et al. Getting a handle on the role of coenzyme M in alkene metabolism[J]. Microbiology and Molecular Biology Reviews, 2008, 72 (3): 445–456. |
| [71] | Purwantini E, Mukhopadhyay B. Conversion of NO2 to NO by reduced coenzyme F420 protects mycobacteria from nitrosative damage[J]. Proceedings of the National Academy of Sciences of the United States of America, 2009, 106 (15): 6333–6338. |
| [72] | Costa KC, Leigh JA. Metabolic versatility in methanogens[J]. Current Opinion in Biotechnology, 2014, 29 : 70–75. |
| [73] | Thauer RK, Kaster AK, Seedorf H, et al. Methanogenic archaea: ecologically relevant differences in energy conservation[J]. Nature Reviews Microbiology, 2008, 6 (8): 579–591. |
| [74] | Gottschalk G, Thauer RK. The Na+-translocating methyltransferase complex from methanogenic archaea[J]. Biochimica et Biophysica Acta (BBA)-Bioenergetics, 2001, 1505 (1): 28–36. |
| [75] | Ide T, Bäumer S, Deppenmeier U. Energy conservation by the H2: heterodisulfide oxidoreductase from Methanosarcina mazei Gö1: identification of two proton-translocating segments[J]. Journal of Bacteriology, 1999, 181 (13): 4076–4080. |
| [76] | Beifuss U, Tietze M, Bäumer S, et al. Methanophenazine: structure, total synthesis, and function of a new cofactor from methanogenic archaea[J]. Angewandte Chemie International Edition, 2000, 39 (14): 2470–2472. |
| [77] | Heiden S, Hedderich R, Setzke E, et al. Purification of a two-subunit cytochrome-b-containing heterodisulfide reductase from methanol-grown Methanosarcina barkeri[J]. European Journal of Biochemistry, 1994, 221 (2): 855–861. |
| [78] | Buan NR, Metcalf WW. Methanogenesis by Methanosarcina acetivorans involves two structurally and functionally distinct classes of heterodisulfide reductase[J]. Molecular Microbiology, 2010, 75 (4): 843–853. |
| [79] | Murakami E, Deppenmeier U, Ragsdale SW. Characterization of the intramolecular electron transfer pathway from 2-hydroxyphenazine to the heterodisulfide reductase from Methanosarcina thermophila[J]. The Journal of Biological Chemistry, 2001, 276 (4): 2432–2439. |
| [80] | Meuer J, Kuettner HC, Zhang JK, et al. Genetic analysis of the archaeon Methanosarcina barkeri Fusaro reveals a central role for Ech hydrogenase and ferredoxin in methanogenesis and carbon fixation[J]. Proceedings of the National Academy of Sciences of the United States of America, 2002, 99 (8): 5632–5637. |
| [81] | Kaster AK, Moll J, Parey K, et al. Coupling of ferredoxin and heterodisulfide reduction via electron bifurcation in hydrogenotrophic methanogenic archaea[J]. Proceedings of the National Academy of Sciences of the United States of America, 2011, 108 (7): 2981–2986. |
| [82] | Lie TJ, Costa KC, Lupa B, et al. Essential anaplerotic role for the energy-converting hydrogenase Eha in hydrogenotrophic methanogenesis[J]. Proceedings of the National Academy of Sciences of the United States of America, 2012, 109 (38): 15473–15478. |
| [83] | Tersteegen A, Hedderich R. Methanobacterium thermoautotrophicum encodes two multisubunit membrane-bound [NiFe] hydrogenases[J]. European Journal of Biochemistry, 1999, 264 (3): 930–943. |
| [84] | Costa KC, Lie TJ, Jacobs MA, et al. H2-independent growth of the hydrogenotrophic methanogen Methanococcus maripaludis[J]. mBio, 2013, 4 (2): e00062–13. |
| [85] | Thauer RK. The Wolfe cycle comes full circle[J]. Proceedings of the National Academy of Sciences of the United States of America, 2012, 109 (38): 15084–15085. |
| [86] | Hedderich R, Whitman WB. Physiology and biochemistry of the methane-producing archaea[A]//Rosenberg E, DeLong EF, Lory S, et al. The Prokaryotes[M]. Berlin Heidelberg: Springer, 2013: 635-662 |
| [87] | Ferry JG. Enzymology of one-carbon metabolism in methanogenic pathways[J]. FEMS Microbiology Reviews, 1999, 23 (1): 13–38. |
| [88] | Ferguson Jr DJ, Gorlatova N, Grahame DA, et al. Reconstitution of dimethylamine: coenzyme M methyl transfer with a discrete corrinoid protein and two methyltransferases purified from Methanosarcina barkeri[J]. The Journal of Biological Chemistry, 2000, 275 (37): 29053–29060. |
| [89] | Burke SA, Krzycki JA. Reconstitution of monomethylamine: coenzyme M methyl transfer with a corrinoid protein and two methyltransferases purified from Methanosarcina barkeri[J]. The Journal of Biological Chemistry, 1997, 272 (26): 16570–16577. |
| [90] | Sauer K, Harms U, Thauer RK. Methanol: coenzyme M methyltransferase from Methanosarcina barkeri[J]. European Journal of Biochemistry, 1997, 243 (3): 670–677. |
| [91] | Burke SA, Lo SL, Krzycki JA. Clustered genes encoding the methyltransferases of methanogenesis from monomethylamine[J]. Journal of Bacteriology, 1998, 180 (13): 3432–3440. |
| [92] | Hagemeier CH, Krüer M, Thauer RK, et al. Insight into the mechanism of biological methanol activation based on the crystal structure of the methanol-cobalamin methyltransferase complex[J]. Proceedings of the National Academy of Sciences of the United States of America, 2006, 103 (50): 18917–18922. |
| [93] | Maeder DL, Anderson I, Brettin TS, et al. The Methanosarcina barkeri genome: comparative analysis with Methanosarcina acetivorans and Methanosarcina mazei reveals extensive rearrangement within methanosarcinal genomes[J]. Journal of Bacteriology, 2006, 188 (22): 7922–7931. |
| [94] | Deppenmeier U, Johann A, Hartsch T, et al. The genome of Methanosarcina mazei: evidence for lateral gene transfer between bacteria and archaea[J]. Journal of Molecular Microbiology and Biotechnology, 2002, 4 (4): 453–461. |
| [95] | Galagan JE, Nusbaum C, Roy A, et al. The genome of M. acetivorans reveals extensive metabolic and physiological diversity[J]. Genome Research, 2002, 12 (4): 532–542. |
| [96] | Krätzer C, Carini P, Hovey R, et al. Transcriptional profiling of methyltransferase genes during growth of Methanosarcina mazei on trimethylamine[J]. Journal of Bacteriology, 2009, 191 (16): 5108–5115. |
| [97] | Hao B, Gong WM, Ferguson TK, et al. A new UAG-encoded residue in the structure of a methanogen methyltransferase[J]. Science, 2002, 296 (5572): 1462–1466. |
| [98] | Paul L, Ferguson Jr DJ, Krzycki JA. The trimethylamine methyltransferase gene and multiple dimethylamine methyltransferase genes of Methanosarcina barkeri contain in-frame and read-through amber codons[J]. Journal of Bacteriology, 2000, 182 (9): 2520–2529. |
| [99] | Sprenger WW, Hackstein JHP, Keltjens JT. The energy metabolism of Methanomicrococcus blatticola: physiological and biochemical aspects[J]. Antonie van Leeuwenhoek, 2005, 87 (4): 289–299. |
| [100] | Fricke WF, Seedorf H, Henne A, et al. The genome sequence of Methanosphaera stadtmanae reveals why this human intestinal archaeon is restricted to methanol and H2 for methane formation and ATP synthesis[J]. Journal of Bacteriology, 2006, 188 (2): 642–658. |
| [101] | Deppenmeier U, Lienard T, Gottschalk G. Novel reactions involved in energy conservation by methanogenic archaea[J]. FEBS Letters, 1999, 457 (3): 291–297. |
| [102] | Bäumer S, Ide T, Jacobi C, et al. The F420H2 dehydrogenase from Methanosarcina mazei is a redox-driven proton pump closely related to NADH dehydrogenases[J]. The Journal of Biological Chemistry, 2000, 275 (24): 17968–17973. |
| [103] | Kulkarni G, Kridelbaugh DM, Guss AM, et al. Hydrogen is a preferred intermediate in the energy-conserving electron transport chain of Methanosarcina barkeri[J]. Proceedings of the National Academy of Sciences of the United States of America, 2009, 106 (37): 15915–15920. |
| [104] | Sapra R, Bagramyan K, Adams MWW. A simple energy-conserving system: proton reduction coupled to proton translocation[J]. Proceedings of the National Academy of Sciences of the United States of America, 2003, 100 (13): 7545–7550. |
| [105] | Lang K, Schuldes J, Klingl A, et al. New mode of energy metabolism in the seventh order of methanogens as revealed by comparative genome analysis of “Candidatus Methanoplasma termitum”[J]. Applied and Environmental Microbiology, 2015, 81 (4): 1338–1352. |
| [106] | Kröninger L, Berger S, Welte C, et al. Evidence for the involvement of two heterodisulfide reductases in the energy- conserving system of Methanomassiliicoccus luminyensis[J]. The FEBS Journal, 2015 : . |
| [107] | Smith KS, Ingram-Smith C. Methanosaeta, the forgotten methanogen?[J]. Trends in Microbiology, 2007, 15 (4): 150–155. |
| [108] | Grahame DA. Catalysis of acetyl-CoA cleavage and tetrahydrosarcinapterin methylation by a carbon monoxide dehydrogenase-corrinoid enzyme complex[J]. The Journal of Biological Chemistry, 1991, 266 (33): 22227–22233. |
| [109] | Grahame DA, DeMoll E. Partial reactions catalyzed by protein components of the acetyl-CoA decarbonylase synthase enzyme complex from Methanosarcina barkeri[J]. The Journal of Biological Chemistry, 1996, 271 (14): 8352–8358. |
| [110] | Fischer R, Thauer RK. Methyltetrahydromethanopterin as an intermediate in methanogenesis from acetate in Methanosarcina barkeri[J]. Archives of Microbiology, 1989, 151 (5): 459–465. |
| [111] | Alber BE, Ferry JG. A carbonic anhydrase from the archaeon Methanosarcina thermophila[J]. Proceedings of the National Academy of Sciences of the United States of America, 1994, 91 (15): 6909–6913. |
| [112] | Schlegel K, Müller V. Evolution of Na+ and H+ bioenergetics in methanogenic archaea[J]. Biochemical Society Transactions, 2013, 41 (1): 421–426. |
| [113] | Schlegel K, Welte C, Deppenmeier U, et al. Electron transport during aceticlastic methanogenesis by Methanosarcina acetivorans involves a sodium-translocating Rnf complex[J]. The FEBS Journal, 2012, 279 (24): 4444–4452. |
| [114] | Wang MY, Tomb JF, Ferry JG. Electron transport in acetate-grown Methanosarcina acetivorans[J]. BMC Microbiology, 2011, 11 (1): 165. |
| [115] | Welte C, Deppenmeier U. Membrane-bound electron transport in Methanosaeta thermophila[J]. Journal of Bacteriology, 2011, 193 (11): 2868–2870. |
| [116] | Cheng SA, Xing DF, Call DF, et al. Direct biological conversion of electrical current into methane by electromethanogenesis[J]. Environmental Science & Technology, 2009, 43 (10): 3953–3958. |
| [117] | Rotaru AE, Shrestha PM, Liu FH, et al. Direct interspecies electron transfer between Geobacter metallireducens and Methanosarcina barkeri[J]. Applied and Environmental Microbiology, 2014, 80 (15): 4599–4605. |
| [118] | Rother M, Metcalf WW. Anaerobic growth of Methanosarcina acetivorans C2A on carbon monoxide: an unusual way of life for a methanogenic archaeon[J]. Proceedings of the National Academy of Sciences of the United States of America, 2004, 101 (48): 16929–16934. |
| [119] | Lessner DJ, Li LY, Li QB, et al. An unconventional pathway for reduction of CO2 to methane in CO-grown Methanosarcina acetivorans revealed by proteomics[J]. Proceedings of the National Academy of Sciences of the United States of America, 2006, 103 (47): 17921–17926. |
| [120] | Barker HA. Studies upon the methane-producing bacteria[J]. Archiv für Mikrobiologie, 1936, 7 (1/5): 420–438. |
| [121] | Wolfe RS. Methanogens: a surprising microbial group[J]. Antonie van Leeuwenhoek, 1979, 45 (3): 353–364. |
| [122] | Schnellen CGTP. Onderzoekingen over de methaangisting[D]. Delft: Doctoral Dissertation of Delft University of Technology, 1947 |
| [123] | Hungate RE. Chapter IV a roll tube method for cultivation of strict anaerobes[J]. Methods in Microbiology, 1969, 3B : 117–132. |
| [124] | Woese CR, Fox GE. Phylogenetic structure of the prokaryotic domain: the primary kingdoms[J]. Proceedings of the National Academy of Sciences of the United States of America, 1977, 74 (11): 5088–5090. |
| [125] | Iino T, Tamaki H, Tamazawa S, et al. Candidatus Methanogranum caenicola: a novel methanogen from the anaerobic digested sludge, and proposal of Methanomassiliicoccaceae fam. nov. and Methanomassiliicoccales ord. nov., for a methanogenic lineage of the class thermoplasmata[J]. Microbes and Environments, 2013, 28 (2): 244–250. |
| [126] | Liu Y. Methanobacteriales[A]//Timmis KN. Handbook of Hydrocarbon and Lipid Microbiology[M]. Berlin Heidelberg: Springer, 2010: 559-571 |
| [127] | Borrel G, Joblin K, Guedon A, et al. Methanobacterium lacus sp. nov., isolated from the profundal sediment of a freshwater meromictic lake[J]. International Journal of Systematic and Evolutionary Microbiology, 2012, 62 (Pt 7): 1625–1629. |
| [128] | Krivushin KV, Shcherbakova VA, Petrovskaya LE, et al. Methanobacterium veterum sp. nov., from ancient Siberian permafrost[J]. International Journal of Systematic and Evolutionary Microbiology, 2010, 60 (2): 455–459. |
| [129] | Lauerer G, Kristjansson JK, Langworthy TA, et al. Methanothermus sociabilis sp. nov., a second species within the Methanothermaceae growing at 97 °C[J]. Systematic and Applied Microbiology, 1986, 8 (1/2): 100–105. |
| [130] | Liu Y. Methanococcales[A]//Timmis KN. Handbook of Hydrocarbon and Lipid Microbiology[M]. Berlin Heidelberg: Springer, 2010: 573-581 |
| [131] | Oren A. The family Methanococcaceae[A]//Rosenberg E, DeLong EF, Lory S, et al. The Prokaryotes[M]. Berlin Heidelberg: Springer, 2014: 215-224 |
| [132] | Oren A. The family Methanocaldococcaceae[A]//Rosenberg E, DeLong EF, Lory S, et al. The Prokaryotes[M]. Berlin Heidelberg: Springer, 2014: 201-208 |
| [133] | Stewart LC, Jung JH, Kim YT, et al. Methanocaldococcus bathoardescens sp. nov., a hyperthermophilic methanogen isolated from a volcanically active deep-sea hydrothermal vent[J]. International Journal of Systematic and Evolutionary Microbiology, 2015, 65 (Pt 4): 1280–1283. |
| [134] | Liu Y. Methanomicrobiales[A]//Timmis KN. Handbook of Hydrocarbon and Lipid Microbiology[M]. Berlin Heidelberg: Springer, 2010: 583-593 |
| [135] | Oren A. The family Methanocalculaceae[A]//Rosenberg E, DeLong EF, Lory S, et al. The Prokaryotes[M]. Berlin Heidelberg: Springer, 2014: 195-199 |
| [136] | Oren A. The family Methanocorpusculaceae[A]//Rosenberg E, DeLong EF, Lory S, et al. The Prokaryotes[M]. Berlin Heidelberg: Springer, 2014: 225-230 |
| [137] | Oren A. The family Methanomicrobiaceae[A]//Rosenberg E, DeLong EF, Lory S, et al. The Prokaryotes[M]. Berlin Heidelberg: Springer, 2014: 231-246 |
| [138] | Weng CY, Chen SC, Lai MC, et al. Methanoculleus taiwanensis sp. nov., a methanogen isolated from deep marine sediment at the deformation front area near Taiwan[J]. International Journal of Systematic and Evolutionary Microbiology, 2015, 65 (Pt 3): 1044–1049. |
| [139] | Chen SC, Chen MF, Lai MC, et al. Methanoculleus sediminis sp. nov., a methanogen from sediments near a submarine mud volcano[J]. International Journal of Systematic and Evolutionary Microbiology, 2015, 65 (7): 2141–2147. |
| [140] | Oren A. The family Methanoregulaceae[A]//Rosenberg E, DeLong EF, Lory S, et al. The Prokaryotes[M]. Berlin Heidelberg: Springer, 2014: 253-258 |
| [141] | Bräuer SL, Cadillo-Quiroz H, Ward RJ, et al. Methanoregula boonei gen. nov., sp. nov., an acidiphilic methanogen isolated from an acidic peat bog[J]. International Journal of Systematic and Evolutionary Microbiology, 2011, 61 (1): 45–52. |
| [142] | Oren A. The family Methanospirillaceae[A]//Rosenberg E, DeLong EF, Lory S, et al. The Prokaryotes[M]. Berlin Heidelberg: Springer, 2014: 283-290 |
| [143] | Zhou LG, Liu XL, Dong XZ. Methanospirillum psychrodurum sp. nov., isolated from wetland soil[J]. International Journal of Systematic and Evolutionary Microbiology, 2014, 64 (Pt 2): 638–641. |
| [144] | Parshina SN, Ermakova AV, Bomberg M, et al. Methanospirillum stamsii sp. nov., a psychrotolerant, hydrogenotrophic, methanogenic archaeon isolated from an anaerobic expanded granular sludge bed bioreactor operated at low temperature[J]. International Journal of Systematic and Evolutionary Microbiology, 2014, 64 (1): 180–186. |
| [145] | Kurr M, Huber R, König H, et al. Methanopyrus kandleri, gen. and sp. nov. represents a novel group of hyperthermophilic methanogens, growing at 110 °C[J]. Archives of Microbiology, 1991, 156 (4): 239–247. |
| [146] | Slesarev AI, Mezhevaya KV, Makarova KS, et al. The complete genome of hyperthermophile Methanopyrus kandleri AV19 and monophyly of archaeal methanogens[J]. Proceedings of the National Academy of Sciences of the United States of America, 2002, 99 (7): 4644–4649. |
| [147] | Liu Y. Methanosarcinales[A]//Timmis KN. Handbook of Hydrocarbon and Lipid Microbiology[M]. Berlin Heidelberg: Springer, 2010: 595-604 |
| [148] | Garrity GM, Labeda DP, Oren A. Judicial commission of the international committee on systematics of prokaryotes XIIth international (IUMS) congress of bacteriology and applied microbiology[J]. International Journal of Systematic and Evolutionary Microbiology, 2011, 61 (11): 2775–2780. |
| [149] | Zhang GS, Zhang F, Ding G, et al. Acyl homoserine lactone-based quorum sensing in a methanogenic archaeon[J]. The ISME Journal, 2012, 6 (7): 1336–1344. |
| [150] | Li LY, Zheng MY, Ma HL, et al. Significant performance enhancement of a UASB reactor by using acyl homoserine lactones to facilitate the long filaments of Methanosaeta harundinacea 6Ac[J]. Applied Microbiology and Biotechnology, 2015, 99 (15): 6471–6480. |
| [151] | Cheng L, Qiu TL, Yin XB, et al. Methermicoccus shengliensis gen. nov., sp. nov., a thermophilic, methylotrophic methanogen isolated from oil-production water, and proposal of Methermicoccaceae fam. nov.[J]. International Journal of Systematic and Evolutionary Microbiology, 2007, 57 (12): 2964–2969. |
| [152] | Wagner D, Schirmack J, Ganzert L, et al. Methanosarcina soligelidi sp. nov., a desiccation- and freeze-thaw-resistant methanogenic archaeon from a Siberian permafrost-affected soil[J]. International Journal of Systematic and Evolutionary Microbiology, 2013, 63 (Pt 8): 2986–2991. |
| [153] | Ganzert L, Schirmack J, Alawi M, et al. Methanosarcina spelaei sp. nov., a methanogenic archaeon isolated from a floating biofilm of a subsurface sulphurous lake[J]. International Journal of Systematic and Evolutionary Microbiology, 2014, 64 (Pt 10): 3478–3484. |
| [154] | Singh N, Kendall MM, Liu YT, et al. Isolation and characterization of methylotrophic methanogens from anoxic marine sediments in Skan Bay, Alaska: description of Methanococcoides alaskense sp. nov., and emended description of Methanosarcina baltica[J]. International Journal of Systematic and Evolutionary Microbiology, 2005, 55 (6): 2531–2538. |
| [155] | Cha IT, Min UG, Kim SJ, et al. Methanomethylovorans uponensis sp. nov., a methylotrophic methanogen isolated from wetland sediment[J]. Antonie van Leeuwenhoek, 2013, 104 (6): 1005–1012. |
| [156] | Lomans BP, Maas R, Luderer R, et al. Isolation and characterization of Methanomethylovorans hollandica gen. nov., sp. nov., isolated from freshwater sediment, a methylotrophic methanogen able to grow on dimethyl sulfide and methanethiol[J]. Applied and Environmental Microbiology, 1999, 65 (8): 3641–3650. |
| [157] | Wu SY, Lai MC. Methanogenic archaea isolated from Taiwan’s Chelungpu fault[J]. Applied and Environmental Microbiology, 2011, 77 (3): 830–838. |
| [158] | Sakai S, Conrad R, Imachi H. The family Methanocellaceae[A]//Rosenberg E, DeLong EF, Lory S, et al. The Prokaryotes[M]. Berlin Heidelberg: Springer, 2014: 209-214 |
| [159] | Schütz H, Seiler W, Conrad R. Influence of soil temperature on methane emission from rice paddy fields[J]. Biogeochemistry, 1990, 11 (2): 77–95. |
| [160] | Sakai S, Imachi H, Sekiguchi Y, et al. Isolation of key methanogens for global methane emission from rice paddy fields: a novel isolate affiliated with the clone cluster Rice Cluster I[J]. Applied and Environmental Microbiology, 2007, 73 (13): 4326–4331. |
| [161] | Erkel C, Kube M, Reinhardt R, et al. Genome of Rice Cluster I archaea—the key methane producers in the rice rhizosphere[J]. Science, 2006, 313 (5785): 370–372. |
| [162] | Lü Z, Lu YH. Complete genome sequence of a thermophilic methanogen, Methanocella conradii HZ254, isolated from Chinese rice field soil[J]. Journal of Bacteriology, 2012, 194 (9): 2398–2399. |
| [163] | McCalley CK, Woodcroft BJ, Hodgkins SB, et al. Methane dynamics regulated by microbial community response to permafrost thaw[J]. Nature, 2014, 514 (7523): 478–481. |
| [164] | Mondav R, Woodcroft BJ, Kim EH, et al. Discovery of a novel methanogen prevalent in thawing permafrost[J]. Nature Communications, 2014, 5 : 3212. |
| [165] | Borrel G, Harris HMB, Tottey W, et al. Genome sequence of “Candidatus Methanomethylophilus alvus” Mx1201, a methanogenic archaeon from the human gut belonging to a seventh order of methanogens[J]. Journal of Bacteriology, 2012, 194 (24): 6944–6945. |
| [166] | Borrel G, Parisot N, Harris HMB, et al. Comparative genomics highlights the unique biology of Methanomassiliicoccales, a Thermoplasmatales-related seventh order of methanogenic archaea that encodes pyrrolysine[J]. BMC Genomics, 2014, 15 (1): 679. |
| [167] | Schink B. Energetics of syntrophic cooperation in methanogenic degradation[J]. Microbiology and Molecular Biology Reviews, 1997, 61 (2): 262–280. |
| [168] | Stams AJM, Plugge CM. Electron transfer in syntrophic communities of anaerobic bacteria and archaea[J]. Nature Reviews Microbiology, 2009, 7 (8): 568–577. |
| [169] | Sieber JR, Le HM, McInerney MJ. The importance of hydrogen and formate transfer for syntrophic fatty, aromatic and alicyclic metabolism[J]. Environmental Microbiology, 2014, 16 (1): 177–188. |
| [170] | Tarnocai C, Canadell JG, Schuur EAG, et al. Soil organic carbon pools in the northern circumpolar permafrost region[J]. Global Biogeochemical Cycles, 2009, 23 (2): . |
| [171] | Li K, Liu RH, Sun C. A review of methane production from agricultural residues in China[J]. Renewable and Sustainable Energy Reviews, 2016, 54 : 857–865. |
| [172] | Chen Y, Cheng JJ, Creamer KS. Inhibition of anaerobic digestion process: a review[J]. Bioresource Technology, 2008, 99 (10): 4044–4064. |
 2016, Vol. 43
2016, Vol. 43