扩展功能
文章信息
- 严俊杰, 刘新锐, 谢宝贵, 邓优锦
- YAN Jun-Jie, LIU Xin-Rui, XIE Bao-Gui, DENG You-Jin
- 中国野生发光真菌新记录种Neonothopanus nambi 的分离、鉴定及其形态观察
- Isolation, identification and characterization of Neonothopanus nambi (Basidiomycota, Fungi), a new record from China
- 微生物学通报, 2015, 42(9): 1703-1709
- Microbiology China, 2015, 42(9): 1703-1709
- 10.13344/j.microbiol.china.140844
-
文章历史
- 收稿日期: 2014-10-28
- 接受日期: 2015-01-19
- 优先数字出版日期(www.cnki.net): 2015-02-10
Bioluminescence is a natural phenomenon that has fascinated humans throughout history. It represents one of the oldest fields of scientific study, dating from the first written records of the ancient Greeks[1]. Recently, bioluminescence is known to exist in certain animals, plants, and fungi. Aristotle (384-322 BC) described light emissions from rotten wood and distinguished this ‘living light’ from fire for the first time in written records[2]. In 2008, Desjardin et al. updated the number of luminous fungi species from 42[3] to 64, and classified them into three distinct lineages: Omphalotus, Mycenoid and Armillaria[4]. In 2010, seven new species of luminous fungi were discovered, increasing the number of known luminescent fungi species to 71[4, 5, 6], this data had been increased to 81 until 2014[7, 8, 9]. But there are only 7 species (including Filoboletus yunnanensis, Filoboletus manipuolaris, Dictyopanus pusillus, Lampteromyces luminescens, Lampteromyces mangensis, Pleurotus prometheus and Omphalotus olearius) have been reported from China mainland[7, 10, 11, 12, 13]; although there are 15 Armillaria species was reported in China, no one of them had been reported as luminescent fungi[14].
In this paper, we report a new record of a bioluminescent fungal species, namely, Neonothopanus nambi (Speg.) R.H. Petersen & Krisai, in China, based on analysis of morphological and molecular data.
2 Materials and Methods 2.1 Microorganisms isolationFungal fruiting bodies were collected from rotting wood at the Fujian Agriculture and Forestry University (119°22′−119°24′E, 26°08′−26°09′N), Fujian, China, in July 2007 and May 2012 (identified as the same species and designated as specimens LF1 and LF3). Mycelial colonies were isolated from stipes and cultured in potato dextrose agar (PDA) slants at 25±1 °C in darkness (The strain number was designated MRC-lf3). Both two specimens were deposited in Mycological Research Center of Fujian Agriculture and Forestry University and the living culture examined was deposited in both Mycological Research Center of Fujian Agriculture and Forestry University and China Center for Type Culture Collection (CCTCCM 2013668).
2.2 Culture mediumSawdust medium: 78% sawdust, 20% bran, 1% anhydrite, 1% sugar. The ratio of material to water was 1:1.1.
2.3 Morphological analysisMorphological observations were made of specimens cultured in PDA at 25±1 °C in darkness for 4 weeks. Macro-morphology was studied at this stage, while micro-morphological examinations were made during the second week of incubation, using an Olympus BX51 microscope. Nuclear staining of mycelia and basidiospores was performed using 4 mg/L DAPI (4′,6-diamidino-2-phenylindole) and 0.001% EB (ethidium Ethidium bromide). Micro chemical reactions of the fruiting bodies to 3% KOH and 3% NH4OH were recorded from dried material revived in ethanol followed by water[15]. Basidiospores were monitored and characterized using a JSM-6380 LV scanning electron microscope (SEM), 15 KVkV. Spore statistics included: x, the arithmetic mean of spore length by spore width (± standard deviation); Q, the quotient of spore length by spore width in any one spore, indicated as a range of variation in n spores measured; and Qm, the mean length/width quotient of all spores measured (±s). The specimens were photographed using a Nikon D90 camera, and morphology was described according to fungal classification books[16-17].
2.4 DNA extraction, amplification and sequencingGenomic DNA was extracted, using an improved Hexadecyl trimethyl ammonium Bromide (CTAB) method[18], from mycelia cultured in liquid PDA and fruiting body, respectively. ITS sequences containing the 18S rRNA partial sequence, the internal transcribed spacer regions 1 and 2, the 5.8S rRNA complete sequence and the 28S rDNA rRNA partial sequence were amplified by polymerase chain reaction (PCR) using universal primers ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′)[19, 20, 21]. The PCR reaction mixture contained 5 μl 10×buffer (with Mg2+), 4 μl (10 μmol/L) dNTPs, 2 μl (5 μmol/L) each primer, 1 μl (5 U) Taq DNA polymerase, 34 μl H2O, and 2 μl genomic DNA in a total volume of 50 μl. Samples were incubated in a thermal cycler at 95°C for 3 min, followed by 35 cycles of 94°C for 1 min, 55°C for 30 s, 72 °C for 1 min; followed by a final extension at 72°C for 10 min.
Amplifications that resulted in a single product were purified with Universal DNA Purification Kit (Tiangen, Beijing, China). Purified fragments were then cloned into a pMD18-T vector (Takara, Dalian, China), propagated in E. coli DH5α, and sequenced using an ABI-PRISM3730 sequencer at Sangon Biotech Co., Ltd (Shanghai, China).
2.5 Phylogenetic analysesAll 24 sequences used in this study were downloaded from GenBank, the sequences accession numbers and information on their origins were obtained from the NCBI database, and bioluminescence information was obtained from Desjardin et al[4, 5]. Sequence information is listed in Table 1.
| Strain | Sequences’ origin | GenBank number | Bioluminescence |
| Neonothopanus gardneri | Brazil | JF344713 | + |
| Neonothopanus nambi | Vietnam | JN571729 | + |
| Neonothopanus nambi | Vietnam | JN571726 | + |
| Neonothopanus nambi | Vietnam | JN571728 | + |
| Neonothopanus nambi | PuertoRico | AF525074 | + |
| Neonothopanus nambi | Malaysia | DQ444306 | + |
| Neonothopanus nambi | Malaysia | DQ444307 | + |
| MRC-lf3 | China | KC514805 | + |
| Omphalotus iμudens | USA | AY313271 | + |
| Omphalotus olivascens | USA | AY313281 | + |
| Omphalotus nidiformis | * | EU424307 | + |
| Omphalotus illudens | * | JN571727 | + |
| Omphalotus illudens | USA | AY313273 | + |
| Marasmieμus opacus | USA | AY256703 | - |
| Marasmiellus opacus | USA | DQ450005 | - |
| Marasmiellus ramealis | Sweden | DQ450030 | - |
| Marasmiellus stenophy llus | USA | DQ450032 | - |
| Marasmielus aff. pluvius | USA | DQ450029 | - |
| Marasmius brunneoaurantiacus | * | FJ917620 | - |
| Marasmius occultatus | * | FJ917622 | - |
| Marasmius sp.Z37 | * | JN198388 | - |
| Marasmius sp.RR-2010a | * | FJ917629 | - |
| Pleurotus citrinopileatus | * | EU424285 | - |
| Pleurotus salmoneostramineus | * | EU424302 | - |
The obtained sequences and other sequences downloaded from the GenBank database were aligned with MEGA 5.10 using ClustalW, followed by manual adjustment[22]. Phylogenetic analysis was performed with MEGA 5.10 using Maximum Composite Likelihood with a transition to transversion ratio. Phylogenetic trees were constructed using the Maximum Likelihood method. Bootstrap tests were performed using 1 000 replicates[22].
3 Results 3.1 Morphological observationsGrowth habit: Fungal specimens were discovered growing at the base of rotting stumps and roots of hardwood trees (Figure1A).
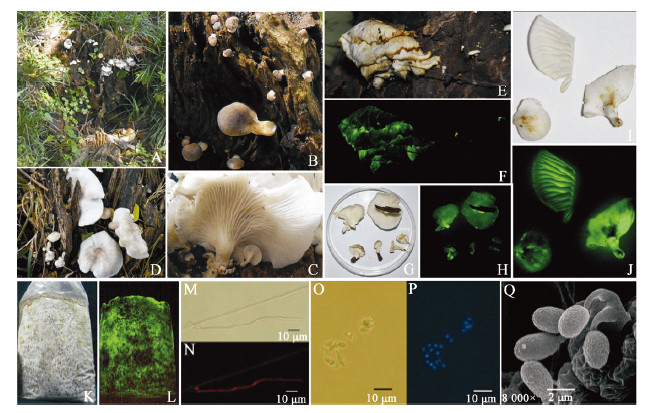
|
| Figure 1 The luminous fungus Neonothopanus nambi (LF3) 图 1 发光真菌Neonothopanus nambi (LF3) Note: A: The habitat of LF3; B and C: Primordial and first developmental stages of the fruiting bodies on sunny and shady slopes, respectively; D: Middle developmental stage of the fruiting bodies (sunny slope); E and F: Late developmental stage of fruiting with daylight exposure and 10 min dark exposure, respectively; G and H: Mature fruiting bodies grown on sunny slopes with daylight exposure or 10 min dark exposure, respectively; I and J: Mature fruiting bodies grown on shady slopes with daylight exposure or 10 min dark exposure, respectively; K and L: Colonies grown on sawdust media with daylight exposure or 10 min dark exposure, respectively; M and N: Microstructure of mycelia, unstained or stained by EB, respectively; O-Q: Microstructure of basidiospores, unstained, stained by DAPI, or observed by SEM, respectively. |
Fruiting bodies were solitary,clumped,and comprised overlaid layers. They could be divided into four different developmental stages: primordia,first developmental stage,middle developmental stage,and late development stage. The primordial stage was characterized by the presence of white or brown dots (Figure1B). The first developmental stage presented the following: a semicircular pileus with a spatulate,rotund and approximately circular shape; stipe was central and eccentrically to centrally lateral; both the pileus and stipe were white or light tan in color on shady slopes and brown on sunny slopes; lamellae were decurrent,white,and somewhat dense (Figure1B,1C). The middle development stage was characterized by the following: pileus exhibited rotundity and an irregular round or reniform shape with involute edges; 10-50 mm in width,0.3−0.7 mm thick; presence of radial stripes and a taupe coloration on sunny slopes,and white with some tan and light yellow patches on shady slopes; lamellae were decurrent,white,divergent and 0.1−0.5 mm broad (Figure1D,1G,1I). The pileus was smooth,glabrous,lacked a veil or chap,and presented a hyaline or pale yellowish appearance in 3% KOH but greenish in 3% NH4OH; stipes were clavate and solid with tough flesh,and were fairly short and thin (3-7 mm wide and 1-10 mm long). No annulus or volva was present. The context was fleshy,thin,white,odorless; the white part of the fruiting body was strongly luminescent (bright yellowish green; Figure1F,1H,1J); in all development stages.
Basidiospores presented the following characteristics: spore prints were white; spores were hyaline, inamyloid, ellipsoid, and ruffled at the surface. Spore dimensions were, (3.4−5.5) μm×(2.1−3.4) μm, [x=(4.8±1.0) μm×(2.7±0.5) μm; Q=1.4−2.1; Qm=1.75±0.18; n=20]. Four basidiospores were observed in each Basidia and the Pleurocystidia was absent; DAPI staining revealed that the spores were uninucleate (Figure 1O, 1P, 1Q).
The mycelium exhibited the following characteristics: white in color, vigorous growth, and bright yellowish green luminescence. A dark brown liquid was produced after one month when grown in a PDA culture medium (Figure 1K, 1L). Microscopic observations revealed that the mycelium had branch, septal and clamp connection, 2.5−6.2 μm in diameter. EB staining showed that cells were binucleate (Figure 1M, 1N).
N. nambi is distinguished by the ability to form luminescent and pleurotoid-like basidiomes with decurrent and distant lamellae, eccentric and solid stipes, inamyloid and ellipsoid basidiospores (4.0−6.5) μm×(2.8−4.0) μm, non-gelatinized, inamyloid hyphae with clamp connections, and white to grayish-tan fruiting bodies accommodate this isolate in N. nambi (Petersen & Krisai-Greilhuber 1999)[23].
3.2 ITS sequence and phylog enetic analysisThe ITS sequences of fruiting body (LF3) and mycelia culture (MRC-lf3) were exactly the same,and the length was 726 bp (GenBank number: KC514805). The final ITS dataset comprised 24 sequences and 706 characters including gaps,after excluding regions deemed too ambiguous for alignment. Phylogenetic analysis of the aligned sequences was performed using the Maximum Likelihood method (Figure2). MRC-lf3 and six N. nambi ITS sequences were identified in the same clade (Bootstrap value=99%),and clustered with Neonothopanus gardneri in the Neonothopanus genus (Bootstrap value=89%). The Neonothopanus clade is strongly supported (Bootstrap value=88%) as the sister clade to Omphalotus. Both clades were identified to be Omphalotus lineages,belonging to Omphalotaceae. Four Marasmiaceae species and two Pleurotaceae species were used as out-groups; they were in the different clades to Omphalotaceae.
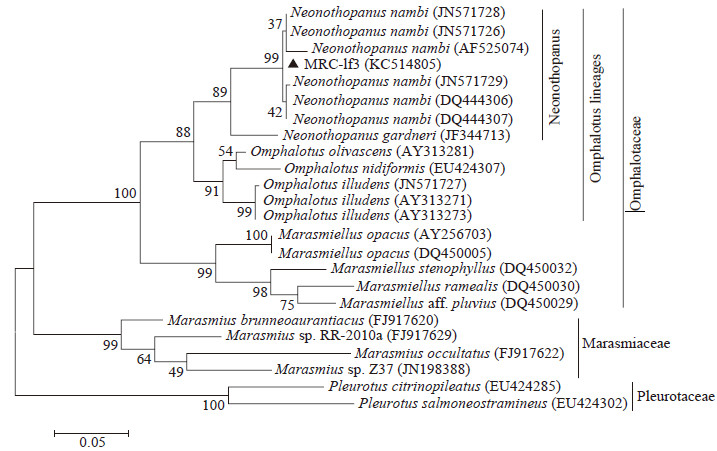
|
| Figure 2 Maximum Likelihood tree obtained from heuristic research based on the ITS sequences 图 2 基于ITS序列构建的系统进化树 Note:All of sequences were downloaded from NCBI database (http://www.ncbi.nlm.nih.gov/pubmed/).Taxon name was composed of "Organism (NCBI ID)" .(Scale bar indicates evolutionary distance.Values above branches indicate the degree of bootstrap support from a 1000 replicate analysis. |
The ITS sequences of 24 strains built by the Maximum Likelihood tree showed our isolate is N. nambi, a species not previously recorded from China; this result is supported by previous studies[24].
4 DiscussionNeonothopanus was established based on N. nambi, which was formally recognized as Agaricus nambi[23]. According to Species Fungorum 2014 (http://www.indexfungorum.org/Names/Names.asp), there are three species, namely, N. nambi, N. gardneri and N. hygrophanus belonging to the Neonothopanus genus. Both N. gardneri and N. hygrophanus were transferred to the Neonothopanus genus in 2011[25, 26]. And several main differences could be found among these three species. For instance, N. nambi and N. hygrophanus forms white to pale grayish tan (brown) basidiomes and ellipsoid basidiospores, but N. gardneri forms yellow basidiomes and globose basidiospores; the fruiting body of N. nambi and N. gardneri could luminescent, but N. hygrophanus has not been explicitly reported as bioluminescent[9, 25, 26].
N. nambi has been reported in Vietnam, South America, Central America and the Caribbean region, Australasia including Papua New Guinea and New Caledonia, South Asia and Southeastern Asia, but it is a new species in China[4, 9, 24, 27]. Compared with the 19 specimens which had been described by Audrey et al[9], the specimens we collected have some special and new characteristics, such as: both the pileus and stipe were brown on sunny slopes when young, and weakly, sometimes no luminescent; and the basidiospores were ruffled at the surface according to scanning electron microscope results.
As a species of Neonothopanus, N. nambi has been extensively studied. In 2009, a luminous fungus isolated from dead wood in rainforests in southern Vietnam was successfully cultivated on a large scale and then identified as N. nambi based on a combination of morphological and molecular data[24, 28]. This made N. nambi an important species in research on bioluminescent systems, bioactive compounds, and related applications[29, 30, 31]. However, few photographs of N. nambi in the wild have been reported[9], especially with regards to the entire developmental process. Our research addressed this deficiency by photographing the entire process from sporulation and mycelial production to fruiting body development. A notable finding was that illumination intensity changed the color of the pileus. The pileus became light tan in color and exhibited a partial or complete inability to emit light when grown under high illumination conditions. This phenomenon can provide certain reference values in the application of N. nambi in bioluminescence research.
| [1] | Lee J. Bioluminescence: the first 3000 years (review)[J]. Journal of Siberian Federal University Biology, 2008(3): 194-205 |
| [2] | Harvey EN. A History of Luminescence from the Earliest Times Until 1900[M]. Philadelphia: American Philosophical Society, 1957: 1-692 |
| [3] | Wassink EC. Luminescence in Fungi[M]//Herring PJ. Bioluminescence in Action. London: Academic Press, 1978: 171-197 |
| [4] | Desjardin DE, Oliveira AG, et al. Fungi Bioluminescence Revisited[J]. Photochemical & Photobiological Sciences, 2008, 7(2): 170-182 |
| [5] | Desjardin DE, Perry BA, Lodge DJ, et al. Luminescent Mycena: new and noteworthy species[J]. Mycological Society of America, 2010, 102(2): 459-477 |
| [6] | Stevani CV, Oliveira AG, Mendes LF, et al. Current status of research on fungal bioluminescence: biochemistry and prospects for ecotoxicological application[J]. Photochemistry and Photobiology, 2013, 89(6): 1318-1326 |
| [7] | Shih YS, Chen CY, Lin WW, et al. Mycena kentingensis, a new species of luminous mushroom in Taiwan, with reference to its culture method[J]. Mycological Progress, 2014, 13(2): 429-435 |
| [8] | Chew AL, Tan YS, Desjardin DE, et al. Four new bioluminescent taxa of Mycena sect. Calodontes from Peninsular Malaysia[J]. Mycological Society of America, 2014, 106(5): 976-988 |
| [9] | Chew AL, Desjardin DE, Tan YS, et al. Bioluminescent fungi from Peninsular Malaysia—a taxonomic and phylogenetic overview[J]. Fungal Diversity, 2014: 1-39 |
| [10] | Zang M. Some new species of higher fungi from Xizang (Tibet) of China[J]. Acta Botanica Yunnanica, 1979, 1(2): 101-105 (in Chinese) 臧穆. 我国西藏高等真菌数新种[J]. 云南植物研究, 1979, 1(2): 101-105 |
| [11] | Liu JZ, Hu XW. A new species of Lampteromyces from Hunan[J]. Acta Scientiarum Naturalium Universitatis Normalis Hunanensis, 1993, 16(2): 188-189 (in Chinese) 李建宗, 胡新文. 湖南亮菌属一新种[J]. 湖南师范大学自然 科学学报, 1993, 16(2): 188-189 |
| [12] | Liu PG, Yang ZL. Studies of classification and geographic distribution on Lashia-complex from the Southern and Southeastern Yunnan, China[J]. Acta Botanica Yunnanica, 1994, 16(1): 47-52 (in Chinese) 刘培贵, 杨祝良. 滇南及滇东南胶孔菌复合群的分类地理研 究[J]. 云南植物研究, 1994, 16(1): 47-52 |
| [13] | Liu PG. Luminous fungi[J]. Chinese Biodiversity, 1995, 3(2):109-112 (in Chinese) 刘培贵. 发光真菌[J]. 生物多样性, 1995, 3(2): 109-112 |
| [14] | Zhao J, Dai YC, Qin GF, et al. New biological species of Armillaria from China[J]. Mycosystema, 2008, 27(2): 156-170 (in Chinese) 赵俊, 戴玉成, 秦国夫, 等. 新的蜜环菌生物种[J]. 菌物学报, 2008, 27(2): 156-170 |
| [15] | Zhao R, Desjardin DE, Soytong K, et al. A monograph of Micropsalliota in Northern Thailand based on morphological and molecular data[J]. Fungal Diversity, 2010, 45(1): 33-79 |
| [16] | Deng SQ. Chinese Fungi[M]. Beijing: Science Press, 1964: 1-808 (in Chinese) 邓叔群. 中国的真菌[M]. 北京: 科学出版社, 1964: 1-808 |
| [17] | Wei JC. Fungi Appraisal Manual[M]. Shanghai: Shanghai Scientific & Technical Publishers, 1979: 780 (in Chinese) 魏景超. 真菌鉴定手册[M]. 上海: 上海科学技术出版社, 1979: 1-780 |
| [18] | Porebski S, Bailey LG, Baum BR. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components[J]. Plant Molecular Biology Reporter, 1997, 15(1): 8-15 |
| [19] | Gardes M, Bruns TD. ITS primers with enhanced specificity for basidiomycetes―application to the identification of mycorrhizae and rusts[J]. Molecular Ecology, 1993, 2(2): 113-118 |
| [20] | Lin X, Huang Y, Fang M, et al. Cytotoxic and antimicrobial metabolites from marine lignicolous fungi, Diaporthe sp.[J]. FEMS Microbiology Letters, 2005, 251(1): 53-58 |
| [21] | White TJ, Bruns T, Lee S, et al. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics[J]. PCR Protocols: a Guide to Methods and Applications, 1990, 18: 315-322 |
| [22] | Tamura K, Peterson D, Peterson N, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods[J]. Molecular Biology and Evolution, 2011, 28(10): 2731-2739 |
| [23] | Petersen RH, Krisai-Greilhuber I. Type specimen studies in Pleurotus[J]. Persoonia, 1999, 17(2): 201-219 |
| [24] | Vydryakova GA, Van DT, Shoukouhi P, et al. Intergenomic and intragenomic ITS sequence heterogeneity in Neonothopanus nambi (Agaricales) from Vietnam[J]. Mycology: An International Journal on Fungal Biology, 2012, 3(2): 89-99 |
| [25] | Capelari M, Desjardin DE, Perry BA, et al. Neonothopanus gardneri: a new combination for a bioluminescent agaric from Brazil[J]. Mycological Society of America, 2011, 103(6): 1433-1440 |
| [26] | Eyi Ndong HE, Degreef J, De Kesel A. Champignons comestibles des forêts denses d’Afrique Centrale[J]. Taxonomie et Identification. ABC Taxa, 2011, 10: 168-169 |
| [27] | Yang ZL, Feng B. The genus Omphalotus (Omphalotaceae) in China[J]. Mycosystema, 2013, 32(3): 545-556 |
| [28] | Van DT. Physiological and cultural properties of the luminous fungus Omphalotus aff. illudent[J]. Journal of Siberian Federal University, 2009, 2: 157-171 |
| [29] | Bua-art S, Saksirirat W, Hiransalee A, et al. Effect of bioactive compound from luminescent mushroom (Neonothopanus nambi Speg.) on root-knot nematode (Meloidogyne incognita Chitwood) and non-target organisms[J]. KKU Research Journal, 2011, 16(4): 331-341 |
| [30] | Bondar VS, Puzyr AP, Purtov KV, et al. The luminescent system of the luminous fungus Neonothopanus nambi[J]. Doklady. Biochemistry and Biophysics, 2011, 438: 138-140 |
| [31] | Kanokmedhakul S, Lekphrom R, Kanokmedhakul K, et al. Cytotoxic sesquiterpenes from luminescent mushroom Neonothopanus nambi[J]. Tetrahedron, 2012, 68(39): 8261-8266 |
 2015, Vol. 42
2015, Vol. 42