扩展功能
文章信息
- 魏子艳, 金德才, 邓晔
- WEI Zi-Yan, JIN De-Cai, DENG Ye
- 环境微生物宏基因组学研究中的生物信息学方法
- Bioinformatics tools and applications in the study of environmental microbial metagenomics
- 微生物学通报, 2015, 42(5): 890-901
- Microbiology China, 2015, 42(5): 890-901
- 10.13344/j.microbiol.china.140992
-
文章历史
- 收稿日期: 2014-12-10
- 接受日期: 2015-02-04
- 优先数字出版日期(www.cnki.net): 2015-03-03
2. 中国科学院大学 北京 100049
2. University of Chinese Academy of Sciences, Beijing 100049, China
目前,人类社会已进入全新的信息时代,崭新的生物科技时代正在逐步到来。以基因工程技术为代表的新学科、新应用技术的兴起和迅猛发展是这个时期的重要标志之一。生物信息学是20世纪80年代末随着人类基因组计划的启动而兴起的一门新的交叉学科[1],也是两个时代交叉点的必然产物。它致力于运用信息学技术和手段来解决现代生物学中的信息问题,是所有生物数据收集和存储、信息分析和挖掘的总称[2, 3, 4, 5]。近两年来一个新的信息学名词“大数据”越来越多地被提及,它被用于描述在信息爆炸时代所产生的海量数据,并以此统称与之相关的存储、查询、分析等各种信息技术手段[6-8]。与此同时,在生物技术方面,以高通量测序技术为核心的新一代生物大分子检测技术也正迅速改变着生物学研究的面貌。大量生物大分子序列、结构数据的产生必然需要高效的数据处理分析手段[9],而生物信息学技术恰好能够满足这一需求[10, 11, 12],因此,生物信息学在生命科学各个领域的应用已成为不可逆转的必然趋势[2, 13, 14, 15, 16]。本文将重点介绍生物信息学在环境生物技术领域主要是宏基因组学研究领域中的运用及当前存在的技术问题。
1 宏基因组学 1.1 组学技术的概念和起源环境中微生物群落组成、结构、动态和功能的深入研究,使生物组学技术在环境微生物研究领域得到了迅速发展[16, 17, 18, 19, 20, 21, 22]。生物组学是指以基因组、转录组、蛋白组、代谢组等生物大分子群体作为研究对象的学科,它们不同于以往仅研究少数几个基因、蛋白质或生化通路的分子生物学方法,而是注重研究环境生物系统组成之间的相互关系、系统结构和功能的关联、生物群落各物种间的关系、以及群落结构与生态系统的关联等整体上的科学问 题[5, 20, 23]。其中,以测序技术和基因芯片技术为基础的宏基因组学是目前最为关键和成熟的组学方法,它也为其他组学的研究提供了基础。目前关于宏转录组学、宏蛋白组学和宏代谢组学的研究仍处于起步阶段,但它们却显示出巨大的发展前景(图 1)。

|
| 图 1 环境微生物组学技术与环境地化参数结合以揭示自然状态下微生物群落的组成和功能 Figure 1 Microbial community composition and function in natural condition were revealed by environmental microbial omics technologies and environmental geochemistry measurement |
随着宏基因组学研究的深入,研究者们逐渐意识到生活在土壤、淡水、海水、空气、甚至人体等环境中的微生物,其系统发育的多样性和复杂度远远超过我们以往的认识[24, 25]。高通量组学技术的出现掀起了一场环境微生物领域的革命,同时能够帮助我们进一步了解微生物群落的遗传潜力和功能活动规律。而高度复杂的微生物群落组成和庞大的数据,使信息分析从组学技术诞生的那一刻起就成为了它在应用上的瓶颈[7,17]。
1.2 宏基因组学宏基因组学(也称元基因组学),是环境样品中所有微生物基因组集合的研究技术和方法[26]。根据分析对象和实验目的,环境微生物的宏基因组研究基本上可以分为核糖体rDNA (细菌和古细菌16S rDNA或真菌18S、28S rDNA和ITS)的分类和鉴定,功能基因(比如固氮还原酶nifH基因和氨基氧化酶amoA基因等)的多样性和分类分析,以及全部宏基因组DNA的整体测序和分析等。而从实验手段来看,目前环境微生物的宏基因组研究主要以高通量检测技术为主,以基因芯片技术和高通量测序技术为代表。这两种技术各有优势和缺陷[14, 27]。基因芯片技术是基于已有的DNA序列设计芯片探针,所以它能从样品中筛选出已知物种或有明确功能的基因信息,经过系统分析得到这些已知物种或功能的生态分布或变化趋势,但是它无法检测到未知物种或功能基因,所以很难用于估算生境中的物种和个体总量,因此也被称为封闭体系[28]。相对而言,高通量测序技术是一种开放体系,对其合理运用可以获取某一特定基因的大多数操作分类单元(OTU)及其个体数量、或者宏基因组中的大片段DNA的信息,从而能够准确的反映生境中微生物群落的组成、结构以及遗传进化关系等。
目前宏基因组学在环境微生物研究中已经占据了主导地位[12, 29, 30],测序通量的增加和成本的降低将进一步扩大这一技术的应用范围[17]。毫无疑问,在过去的20年中,DNA测序技术是驱动环境微生物学领域发展的关键技术之一。相对基因芯片技术,高通量测序技术的应用更为普遍[31-32],因此本综述侧重高通量测序技术背景下微生物宏基因组分析中生物信息学的应用。
2 微生物宏基因组学大数据的处理过程生物信息分析在微生物宏基因组学和其他组学分析中占据十分重要的地位[12, 33, 34, 35, 36, 37, 38, 39, 40, 41]。可以说,面对海量的大数据资料,没有生物信息学技术与方法的应用,研究工作将寸步难行。而随着大规模测序技术的发展以及数据积累程度的增加,其难度和重要性也将逐渐增大[33, 42]。
宏基因组学大数据分析的各个环节都需要运用信息学和生物信息学技术(图 2)。首先是大数据的存储,包括环境样品的采集(采集地点、样本类型、地理环境、气候季节等)和处理信息(实验条件、处理时间等),样品的地球物理化学参数,测序信息(测序反应条件、测序仪器、测序深度等)和大量的序列数据;经过分类和整理之后的数据,需要进入标准化的数据库进行保存,以备后续分析使用。其次是大数据的前处理,即海量序列的基础分析,包括序列质量控制、序列拼接、序列的物种分类学分析、序列功能的预测和相对定量分析。大数据的前处理是宏基因组学研究的基础,其速度和准确性都将对实验进度和最终结论产生很大影响。最后,经过基础分析的数据需要进一步进行信息分析、比对与提炼,进而分析微生物的群落组成与多样性、群落功能与遗传变异、群落结构与物种间的相互关联、群落与环境的相互作用,最终为环境变化的预测和治理提供理论依据(图 2)。
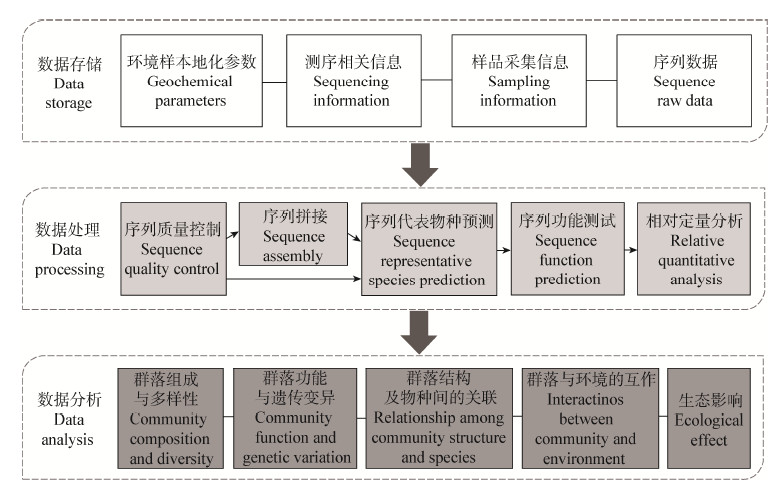
|
| 图 2 宏基因组研究中信息处理的运用流程 Figure 2 Data process in the study of metagenomics |
近年来,宏基因组学作为环境微生物学的前沿工具,被广泛应用于土壤、海洋、河湖水、肠道,以及极端生境如沙漠、苔原、深海底床、酸矿、生物反应器等一系列环境样品的微生物群落组成的分析研究[4,43-46]。其中,以对16S rRNA基因扩增进行检测的测序技术最为常用[47, 48, 49, 50, 51, 52, 53]。通过对16S rRNA基因的测序和分析可以获得环境中各个细菌种类的相对丰度和多样性水平,从而了解环境中微生物群落的组成和结构。除16S rRNA基因测序之外,也可以针对性地选择微生物群落功能基因进行扩增测序,比如固氮还原酶nifH基因和氨基氧化酶amoA基因等[54, 55, 56, 57]。这类研究揭示了各个功能菌群的构成和多样性,同时也能够在更高的分类尺度上(比如种的水平或者菌株的水平)对微生物的群落展开更细致的研究。而更准确含义上的宏基因组学特指针对整个环境样本DNA进行所有基因组的分 析[31, 58],这方面的研究是当前环境生物学的热点,大量的研究结果层出不穷[59, 60, 61, 62, 63],系统的综述也不胜枚举[31, 58, 64, 65, 66, 67]。
目前针对大规模测序技术,主要是扩增子测序和宏基因组全测序,宏基因组学研究中常用的生物信息学平台如表 1所示[17, 19, 20, 21, 29, 30, 31, 46, 68]。
| 平台及入口 Pipeline and its entrance | 适用范围 Range of application | 重要工具及其功能 Important tools and its functions |
| ARB[69] http://www.arb-home.de/ | 扩增子测序 (rDNA amplicon sequence) | Sequencher:基因组拼装 PT Server/SINA:序列比对 MARK:构建系统发育树 BLAST:识别相关序列 |
| Genboree Microbiome Toolset[70] http://genboree.org/theCommons/projects/ pub-gen-microbiome | RDP Classifier:物种聚类 cd-hit, mothur, and uclust:创建OTU表 Chimera Slayer:嵌合体检测 UniFrac:进化分析 | |
| Mothur[71] http://www.mothur.org/ | RDP:质量控制 NAST, SINA, and RDP aligners:序列比对 DOTUR, CD-HIT and SONS:序列分配,估算丰富性和多样性 ∫-LIBSHUFF/TreeClimber/UniFrac:群落结构检测 | |
| Orione[72] http://orione.crs4.it/ | FastX and FASTQC:质量控制 de Bruijn graph, ABySS and SPAdes/SSAKE, Edena:基因组拼装 Glimmer and tRNAscan-SE:基因组注释 | |
| PHYLOSHOP[73] http://omics.informatics.indiana.edu/mg/phyloshop/ | HMMER search:基因组预测 ChimericSlayer:嵌合体检测 RDP, NCBI or Hugenholtz:物种分类 Fast UniFrac:细菌群落的组成和结构比较 | |
| Visualization and Analysis of Microbial Population Structures (VAMPS)[74] http://vamps.mbl.edu/ | BioPerl scripts:质量控制和物种分类 UCLUST, oligotyping, SLP and CROP:OTU分类 Taxonomy Tables/Heatmap Comparison:群落可视化工具 | |
| Quantitative Insights Into Microbial Ecology (QIIME)[75, 76] http://qiime.org/index.html | Denoiser/AmpliconNoise:质量控制 PyNAST/Infernal:序列比对 ChimeraSlayer:嵌合体检测 RDP Classifier/RTAX and USEARCH:物种分类 FastTree/RAxML and pplacer:构建系统发育树 Emperor:比较分析 | |
| Ribosomal Database Project (RDP)[77, 78] http://rdp.cme.msu.edu/ | RDP Aligner:序列比对 RDP Classifier:物种分类 Tree Builder:构建系统发育树 Defined Community Analysis and Chimera Check:群落分析和嵌合体检测 | |
| BioBakery https://bitbucket.org/biobakery/biobakery/ wiki/biobakery_wiki | 宏基因组全测序 (shotgun metagenome sequence) | MetaPhlAn:群落组成分析 PICRUSt:基因组功能预测 PhyloPhlAn:构建系统发育树 GraPhlAn:可视化分类和系统发育信息 |
| Cloud Virtual Resource (CLoVR)[79] http://clovr.org/ | Celera assembler/Velvet:基因组拼装 Glimmer3:基因组预测 BLASTN against RefSeq:物种分类 BLASTX against COG:功能分类 BLASTX against UniREF100 and COG, HMMER search against Pfam and TIGRfam:功能注释 Metastats:比较分析 | |
| Community Cyberinfrastructure for Advanced Microbial Ecology Research and Analysis (CAMERA)[80] https://portal.camera.calit2. net/gridsphere/ gridsphere?cid=microgenometab | GIS Query:数据查询 QC Filter and 454 Duplicate Clustering:质量控制 454 Read Assembly:基因组拼装 Metagenomic Data Annotation and Clustering workflow:功能注释和聚类 | |
| EBI Metagenomics[81] https://www.ebi.ac.uk/metagenomics/ | BiopythonSeqIO package:质量控制 InterProScan 5:功能预测 RDP classifier and Greengenes:分类分析 | |
| Galaxy[82, 83] http://galaxyproject.org/ | UCSC:基因组注释 Fetch Alignments/Multiple Alignments:序列比对 Plotting tool:数据绘图 Phylogenetic Tree:构建系统发育树 | |
| Integrated Microbial Genomes System for Metagenomes (IMG/M)[84] https://img.jgi.doe.gov/cgi-bin/m/main.cgi | Lucy and DUST:质量控制 CRT and PILER-CR:基因组预测 Pfams, COGs and hmmsearch:功能注释 SNP VISTA:SNP可视化 Abundance Comparison tool:丰富度比较 | |
| JCVI Metagenomics Reports (METAREP)[76] http://www.jcvi.org/metarep/ | SOAP de novo assembler:基因组拼装 JPMAP/HUMAnN:基因组注释 NCBI taxonomy (family level)/KEGG pathways (pathway level):聚类分析 METASTATS:统计学检验 Compare Page: 不同功能和分类水平的多重比较 | |
| MEtaGenome ANalyzer (MEGAN)[87, 88] http://ab.inf.uni-tuebingen.de/software/megan5/ | BLAST:序列比较 NCBI taxonomy:物种分类 SEED/KEGG/COG/EGGNOG:功能分析 PCoA:分类和功能分析 | |
| MetaGenomics Rapid Annotation using Subsystem (MG-RAST)[89] http://metagenomics.anl.gov/ | SolexaQA/DRISEE/Bowtie:质量控制 FragGeneScan:基因组预测 NCBI taxonomy:物种分类 SEED FIFfams:功能分类 LCA:分类注释 SEED:基因组注释 Analysis page:功能分析 | |
| MetAMOS[90] http://marbl.github.io/metAMOS/ | FastQC and Bambus 2:质量控制 HMP:基因组拼装 FCP and Bowtie:基因组注释 BLAST:功能注释 Ruffus:后置处理 | |
| MOCAT[91] http://vm-lux.embl.de/~kultima/MOCAT/ | FastX and SolexaQA:质量控制 SOAPaligner/USEARCH:序列比对 SOAPdenovo and BWA:基因组拼装 Prodigal/MetaGeneMark:基因组预测 mOTU:物种分类 | |
| Parallel-META[92, 93] http://www.computationalbioenergy.org/parallel-meta.html | POSLX thread, OpenMP, and CUDA:基因组预测和注释 Here Velvet:基因组拼装 GO-term annotation and SEED annotation:功能分析 Krona:分类结构可视化 SVG:功能结构可视化 | |
| Rapid Analysis of Multiple Metagenomes with a Clustering and Annotation Pipeline (RAMMCAP)[94] http://weizhong-lab.ucsd.edu/rammcap/cgi-bin/rammcap.cgi | CD-HIT-454:质量控制 CD-HIT-EST:序列聚类 CD-HIT:ORFs聚类 HMMER/RPS-BLAST:ORFs注释 | |
| Short Oligonucleotide Analysis Package (SOAP)[95, 96] http://soap.genomics.org.cn/ | SOAPaligner/soap2/SOAP3/GPU:序列比对 SOAPsv:扫描结构变异 SOAPdenovo:基因组拼装 | |
| Simple Metagenomics Analysis Shell formicrobial communities (Smash Community)[97] http://www.bork.embl.de/software/smash/ | Lucy:质量控制 Arachne and Celere:基因组拼装 GeneMark and MetaGene:基因组预测 | |
| WebCARMA[98, 99] http://wwww.cebitec.uni-bielefeld.de/webcarma. cebitec.uni-bielefeld.de/ | BLAST and HMMER:物种分类 HMMER variant:功能分类 | |
| WebMGA[100] http://weizhongli-lab.org/metagenomic-analysis/ | QC-filter and SolexaQA:质量控制 CD-HIT-EST, CD-HIT, H-CD-HIT and CD-HIT-454:序列聚类 HMMER3 and RPS-BLAST:功能注释 FNA-stat and FAA-stat:序列统计 |
从宏基因组技术诞生的那一刻起,信息分析一直是其研究的主要瓶颈[36, 39, 40, 42, 101]。随着测序成本的下降和测序深度的增加,其分析难度将会越来越大,制约效应也将会越来越明显[7]。如图 3A所示,从2010年到2019年,预计的单位测序成本将会以指数关系下降,但其中计算成本下降的幅度会远慢于测序成本本身。针对某单一环境的宏基因组研究而言,实验的设计和完成会耗费更多的时间和成本,但测序本身的消耗将会大大下降,而用于数据分析的时间和精力会随着测序通量的增加而迅速增加(图 3B)。

|
|
图 3
2010−2019年10年内宏基因组研究中测序经费和时间成本估算
Figure 3
Estimation of sequencing of financial and time cost in metagenomic study in next ten years (2010−2019)
注:A:平均每一条原始序列(reads)所需测序和计算的平均成本(美元)估算;B:未来10年内,实验、测序和数据分析的平均时间长度的估算.
Note: A: Estimate of the average sequencing and calculation cost of each raw sequence (read); B: Estimate of the average time cost of experiment, sequencing and data analysis in the next decade. |
在数据存储和数据处理的层面上,rDNA和扩增序列的分析难度较小,基本可以在个人电脑或者小型服务器上完成,但宏基因组全测序的分析却主要受限于计算技术的发展。宏基因组全序列的分析难度包括:(1) 数据存储的容量。目前Illumina公司HiSeq 2000测序仪一次运行将产生6×109左右序列(100 bp×2端),而一般的分析将产生10倍以上的数据量。因此,一次测序将增加(10−20)×1012的数据量。当样本数量十分庞大的时候,往往还需要结合多次的测序结果进行研究,如此巨大的数据量将会对数据存储设备提出严峻的挑战。(2) 序列的拼接。目前比较成熟的序列拼接算法都是基于一个或少数几个基因组的数据(如Genovo[102],MetaVelvet[103],MAP[104]等),而对HiSeq 2000所产生的宏基因组数据无能为力,其最主要的原因是所有的拼接算法都需要庞大的内存资源[105],而这一需求远大于市面上最大的单服务器所支持的内存数量(4 Tb左右)。因此,大部分的宏基因组序列拼接工作必须以牺牲时间的单样本序列拼接串联方式进行。此外,每个计算核心和其匹配内存的效率也决定了整个拼接的效率。(3) 序列拼接、基因和基因功能预测的准确性同速度之间的矛盾。拼接和基因预测算法往往需要耗费大量的运算资源,而很多近似或高速算法往往以牺牲准确性作为代价。因此,如何在保证准确性的前提下提高速度是决定宏基因组分析质量的关键。
在宏基因组数据挖掘的层面上,目前的难点主要体现在:(1) 物种多样性(taxonomy diversity)、功能多样性(functional diversity)和遗传多样性(genetic diversity)的估算。生物多样性(biodiversity)一直都是生态学研究的重点,而其所属的各类多样性的描述或计算都有相应的方法[106]。然而,在分子生态学领域内,特别是宏基因组出现之后,相应的定义和算法却很不完善。例如微生物物种数量的估算,因为稀有物种的大量检出,经典的估算方法如Chao等[107]都会产生严重的偏差[108]。所以如何有效地利用微生物组学数据估计和描述环境中的微生物群落多样性是所有研究者共同面临的难题。(2) 宏观生态理论在分子生态中的运用。现代生态学经过20世纪的发展已经积累了大量成熟的理论和模型,近年来随着计算能力的增强,新的生态理论和模型更是层出不穷。然而大部分的生态群落理论还是建立在宏观生态的基础之上,用以揭示动物、植物和人类社会现象所体现出来的自然规律。这些理论是否也适用于微观领域,现在还没有明确的结果支持。(3) 微生物物种间关联的不确定性。生物群落的结构不仅包括多样性和物种数量上的分布,而且应该包含物种间的相互作用关系,而这些关系在物质、能量、信息循环中起到了至关重要的作用[109]。然而,由于微生物群落庞杂、细小和难以培养的属性,微生物物种间的相互作用往往无法像宏观生态中予以观察和定性,因此也给相关的研究工作提出了挑战。
5 宏基因组技术在我国近期的发展方向作为环境微生物研究的重要组成部分,微生物宏基因组学中的生物信息分析在我国的开展和研究仍需要大力加强。而这部分所涉及的领域十分广泛,不仅仅包括环境科学、生态学和环境微生物学,而且需要用到大量的生物信息学、统计学、超级计算机技术和比较基因组学,其中很多学科也才刚刚兴起并在迅速发展之中。所以,随着微生物组学技术的普及,在未来数十年之内数据分析的基础平台建设将对我国环境微生物学的发展提供保障,而分析技术本身的研究和发展也大有可为。
首先,近期内应该着重加强基础性分析和存储平台的建设。对于大部分研究者而言,宏基因组的信息分析并不是其研究领域,如何能够快速有效地获得测序后的分析结果,使得分析平台的建设十分必要。而随着宏基因组技术的发展,新的算法和计算平台也在不断出现。有效的整合通用的算法和分析手段,比较和平衡不同算法之间的准确性和速度的矛盾,也需要基础分析平台的建设。此外,数据的整合和保存需要一个统一的存储空间。为了规范环境样本信息,有效存取海量数据信息,提供更多公用的数据源,我们需要建立规范的宏基因组存储平台。分析和存储平台的建设应该结合计算机技术的最新发展趋势,有效利用超级计算技术、云存储技术等新的信息技术,从而为宏基因组技术的广泛应用提供坚实的基础。
其次,针对基础的生物信息学算法研究,应该抓住分析的主要瓶颈步骤,重点突破。例如,复杂生物背景下,超大规模序列的拼接是目前无法逾越的障碍。如何高效地结合实验技术和超算技术的发展,准确、快速地对微生物群落的宏基因组进行有效的拼接和重组,依然有大量的研究工作可以挖掘。此外,新的计算机技术,如图形处理器(GPU)和超算技术的发展也为宏基因组分析提供了更多更快速的解决方案。如何有效的利用这些新的技术和资源,为大型生物信息运算提供通用的算法和接口,也值得进一步发展和研究。
最终,微生物群落宏基因组学的信息分析目标还是要阐明微生物群落组成、结构、功能、以及群落与环境的相互作用,所以如何有效的利用和挖掘微生物宏基因组学的数据来建立分子生态的理论,是微生物生态信息分析的重点任务。在这个方向上,可以借鉴宏观生态学建立起来的生态理论和模型,将其用于宏基因组鉴定出来的微生物群落,并通过改进这些理论和模型来理解和改造微生物群落,从而为预测环境变化提供依据,为我国环境的修复和治理提供理论基础。
目前,微生物群落宏基因组学的研究仍处于初级起步阶段,但是随着实验技术成本的下降和生物信息学技术的日趋成熟,宏基因组学的应用将会更加广泛。此外,宏基因组学、宏转录组学、宏蛋白组学及宏代谢组学的并行应用使我们可以在不同层面上研究微生物的群落结构。这些组学方法在微生物研究中将会有广阔的应用前景,包括整体微生物多样性及其活动规律的揭示,以及对特殊生境下可能发挥重要功能的未知微生物的探索。
| [1] | Wu M.The development of bioinformatics[J]. Bulletin of Chinese Academy of Sciences, 1998(3):183-186(in Chinese) 吴旻.生物信息学的发展[J]. 中国科学院院刊, 1998(3): 183-186 |
| [2] | Orozco A, Morera J, Jimenez S, et al.A review of bioinformatics training applied to research in molecular medicine, agriculture and biodiversity in Costa Rica and Central America[J]. Briefings in Bioinformatics, 2013, 14(5):661-670 |
| [3] | Li H, Wang CJ.Application of bioinformatics in the research of toxicogenomics[J]. China Journal of Bioinformatics, 2010, 8(4): 330-333(in Chinese) 李宏, 王崇均.生物信息学在毒理基因组学研究中的应用[J]. 生物信息学, 2010, 8(4):330-333 |
| [4] | Petrosino JF, Highlander S, Luna RA, et al.Metagenomic pyrosequencing and microbial identification[J]. Clinical Chemistry, 2009, 55(5):856-866 |
| [5] | Espindola FS, Calabria LK, Alves de Rezende AA, et al. Bioinformatic resources applied on the omic sciences as genomic, transcriptomic, proteomic, interatomic and metabolomic[J]. Bioscience Journal, 2010, 26(3):463-477 |
| [6] | Microbiota meet big data[J]. Nature Chemical Biology, 2014, 10(8):605 |
| [7] | Hunter CI, Mitchell A, Jones P, et al.Metagenomic analysis:the challenge of the data bonanza[J]. Briefings in Bioinformatics, 2012, 13(6):743-746 |
| [8] | Schneider MV, Watson J, Attwood T, et al.Bioinformatics training:a review of challenges, actions and support requirements[J]. Briefings in Bioinformatics, 2010, 11(6): 544-551 |
| [9] | Bai Y, Iwasaki Y, Kanaya S, et al.A novel bioinformatics method for efficient knowledge discovery by BLSOM from big genomic sequence data[J]. BioMed Research International, 2014(2014): e765648 |
| [10] | Davenport CF, Tummler B.Advances in computational analysis of metagenome sequences[J]. Environmental Microbiology, 2013, 15(1):1-5 |
| [11] | Hong H, Zhang W, Shen J, et al.Critical role of bioinformatics in translating huge amounts of next-generation sequencing data into personalized medicine[J]. Science China Life Sciences, 2013, 56(2):110-118 |
| [12] | Kunin V, Copeland A, Lapidus A, et al.A bioinformatician's guide to metagenomics[J]. Microbiology and Molecular Biology Reviews, 2008, 72(4):557-578 |
| [13] | Bellazzi R, Diomidous M, Sarkar IN, et al.Data analysis and data mining:current issues in biomedical informatics[J]. Methods of Information in Medicine, 2011, 50(6):536-544 |
| [14] | Yang MQ, Athey BD, Arabnia HR, et al.High-throughput next-generation sequencing technologies foster new cutting-edge computing techniques in bioinformatics[J]. BMC Genomics, 2009, 10(Suppl 1):I1 |
| [15] | Yang JY, Yang MQ, Zhu MM, et al.Promoting synergistic research and education in genomics and bioinformatics[J]. BMC Genomics, 2008, 9(Suppl 1):I1 |
| [16] | Sharon I, Banfield JF.Genomes from metagenomics[J]. Science, 2013, 342(6162):1057-1058 |
| [17] | Kim M, Lee KH, Yoon SW, et al.Analytical tools and databases for metagenomics in the next-generation sequencing era[J]. Genomics&Informatics, 2013, 11(3):102-113 |
| [18] | Chistoserdova L.Is metagenomics resolving identification of functions in microbial communities?[J]. Microbial Biotechnology, 2014, 7(1):1-4 |
| [19] | Logares R, Haverkamp TH, Kumar S, et al.Environmental microbiology through the lens of high-throughput DNA sequencing:synopsis of current platforms and bioinformatics approaches[J]. Journal of Microbiological Methods, 2012, 91(1): 106-113 |
| [20] | Segata N, Boernigen D, Tickle TL, et al.Computational meta'omics for microbial community studies[J]. Molecular Systems Biology, 2013(9):666 |
| [21] | Seifert J, Herbst FA, Halkjaer Nielsen P, et al.Bioinformatic progress and applications in metaproteogenomics for bridging the gap between genomic sequences and metabolic functions in microbial communities[J]. Proteomics, 2013, 13(18/19): 2786-2804 |
| [22] | Kumar R, Eipers P, Little RB, et al.Getting started with microbiome analysis:sample acquisition to bioinformatics[J]. Current Protocols in Human Genetics, 2014.DOI: 10.1002/0471142905.hg1808s82 |
| [23] | Ulrich-Merzenich G, Panek D, Zeitler H, et al.New perspectives for synergy research with the"omic"-technologies[J]. Phytomedicine:International Journal of Phytotherapy and Phytopharmacology, 2009, 16(6/7):495-508 |
| [24] | Curtis TP, Sloan WT.Exploring microbial diversity-a vast below[J]. Science, 2005, 309(5739):1331-1333 |
| [25] | Hugenholtz P, Goebel BM, Pace NR.Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity[J]. Journal of Bacteriology, 1998, 180(18): 4765-4774 |
| [26] | Handelsman J, Rondon MR, Brady SF, et al.Molecular biological access to the chemistry of unknown soil microbes:a new frontier for natural products[J]. Chemistry&Biology, 1998, 5(10):R245-R249 |
| [27] | Van Vliet AH.Next generation sequencing of microbial transcriptomes:challenges and opportunities[J]. FEMS Microbiology Letters, 2010, 302(1):1-7 |
| [28] | Zhou JZ, He ZL, Yang YF, et al.Highthroughput metagenomic technologies for complex microbial community analysis:open and closed formats[J]. mBio, 2015, 6(1):02288-14 |
| [29] | De Filippo C, Ramazzotti M, Fontana P, et al.Bioinformatic approaches for functional annotation and pathway inference in metagenomics data[J]. Briefings in Bioinformatics, 2012, 13(6): 696-710 |
| [30] | Teeling H, Glockner FO.Current opportunities and challenges in microbial metagenome analysis-a bioinformatic perspective[J]. Briefings in Bioinformatics, 2012, 13(6):728-742 |
| [31] | Di Bella JM, Bao Y, Gloor GB, et al.High throughput sequencing methods and analysis for microbiome research[J]. Journal of Microbiological Methods, 2013, 95(3):401-414 |
| [32] | Liu LY, Cui HF, Tian G.Application of high throughput sequencing in metagenomics[J]. Chinese Medicinal Biotechnology, 2013, 8(3):196-200(in Chinese) 刘莉扬, 崔鸿飞, 田埂.高通量测序技术在宏基因组学中的应用[J]. 中国医药生物技术, 2013, 8(3):196-200 |
| [33] | Blow N.Metagenomics:exploring unseen communities[J]. Nature, 2008, 453(7195):687-690 |
| [34] | Daniel R.The metagenomics of soil[J]. Nature Reviews Microbiology, 2005, 3(6):470-478 |
| [35] | Kunin V, Copeland A, Lapidus A, et al.A bioinformatician's guide to metagenomics[J]. Microbiology and Molecular Biology Reviews, 2008, 72(4):557-578 |
| [36] | Logares R, Haverkamp TH, Kumar S, et al.Environmental microbiology through the lens of high-throughput DNA sequencing:synopsis of current platforms and bioinformatics approaches[J]. Journal of Microbiological Methods, 2012, 91(1): 106-113 |
| [37] | Oremland RS, Capone DG, Stolz JF, et al.Whither or wither geomicrobiology in the era of'community metagenomics'[J]. Nature Reviews Microbiology, 2005, 3(7):572-578 |
| [38] | Simon C, Daniel R.Achievements and new knowledge unraveled by metagenomic approaches[J]. Applied Microbiology and Biotechnology, 2009, 85(2):265-276 |
| [39] | Simon C, Daniel R.Metagenomic analyses:past and future trends[J]. Applied and Environmental Microbiology, 2011, 77(4): 1153-1161 |
| [40] | Kuczynski J, Lauber CL, Walters WA, et al.Experimental and analytical tools for studying the human microbiome[J]. Nature Reviews Genetics, 2012, 13(1):47-58 |
| [41] | Luo C, Rodriguez-R LM, Konstantinidis KT.A user's guide to quantitative and comparative analysis of metagenomic datasets[J]. Metagenomics, Metatranscriptomics, and Metaproteomics, 2013, 531:525-547 |
| [42] | Teeling H, Glockner FO.Current opportunities and challenges in microbial metagenome analysis-a bioinformatic perspective[J]. Briefings in Bioinformatics, 2012, 13(6):728-742 |
| [43] | Fierer N, Leff JW, Adams BJ, et al.Cross-biome metagenomic analyses of soil microbial communities and their functional attributes[J]. Proceedings of the National Academy of Sciences of the United States of America, 2012, 109(52):21390-21395 |
| [44] | Xu Z, Hansen MA, Hansen LH, et al.Bioinformatic approaches reveal metagenomic characterization of soil microbial community[J]. PLoS One, 2014, 9(4):e93445 |
| [45] | Fang H, Cai L, Yang Y, et al.Metagenomic analysis reveals potential biodegradation pathways of persistent pesticides in freshwater and marine sediments[J]. The Science of the Total Environment, 2014, 470/471:983-992 |
| [46] | Morgan XC, Huttenhower C.Meta'omic analytic techniques for studying the intestinal microbiome[J]. Gastroenterology, 2014, 146(6):1437-1448 |
| [47] | Rousk J, Bååth E, Brookes PC, et al.Soil bacterial and fungal communities across a pH gradient in an arable soil[J]. The ISME Journal, 2010, 4(10):1340-1351 |
| [48] | Roesch LFW, Fulthorpe RR, Riva A, et al.Pyrosequencing enumerates and contrasts soil microbial diversity[J]. The ISME Journal, 2007, 1(4):283-290 |
| [49] | Nacke H, Thürmer A, Wollherr A, et al.Pyrosequencing-based assessment of bacterial community structure along different management types in German forest and grassland soils[J]. PLoS One, 2011, 6(2):e17000 |
| [50] | Sogin ML, Morrison HG, Huber JA, et al.Microbial diversity in the deep sea and the underexplored "rare biosphere"[J]. Proceedings of the National Academy of Sciences of the United States of America, 2006, 103(32):12115-12120 |
| [51] | Breitbart M, Hoare A, Nitti A, et al.Metagenomic and stable isotopic analyses of modern freshwater microbialites in Cuatro Cienegas, Mexico[J]. Environmental Microbiology, 2008, 11(1): 16-34 |
| [52] | Serino M, Luche E, Gres S, et al.Metabolic adaptation to a high-fat diet is associated with a change in the gut microbiota[J]. Gut, 2012, 61(4):543-553 |
| [53] | Murphy E, Cotter P, Healy S, et al.Composition and energy harvesting capacity of the gut microbiota:relationship to diet, obesity and time in mouse models[J]. Gut, 2010, 59(12): 1635-1642 |
| [54] | Leininger S, Urich T, Schloter M, et al.Archaea predominate among ammonia-oxidizing prokaryotes in soils[J]. Nature, 2006, 442(7104):806-809 |
| [55] | Mou X, Sun S, Edwards RA, et al.Bacterial carbon processing by generalist species in the coastal ocean[J]. Nature, 2008, 451(7179):708-711 |
| [56] | Pegard A, Miquel C, Valentini A, et al.Universal DNA-based methods for assessing the diet of grazing livestock and wildlife from feces[J]. Journal of Agricultural and Food Chemistry, 2009, 57(13):5700-5706 |
| [57] | Kowalczyk R, Taberlet P, Coissac E, et al.Influence of management practices on large herbivore diet-case of European bison in Białowieża Primeval Forest (Poland)[J]. Forest Ecology and Management, 2011, 261(4):821-828 |
| [58] | Gilbert JA, Dupont CL.Microbial metagenomics:beyond the genome[J]. Annual Review of Marine Science, 2011(3):347-371 |
| [59] | Frias-Lopez J, Shi Y, Tyson GW, et al.Microbial community gene expression in ocean surface waters[J]. Proceedings of the National Academy of Sciences of the United States of America, 2008, 105(10):3805-3810 |
| [60] | Wu GD, Chen J, Hoffmann C, et al.Linking long-term dietary patterns with gut microbial enterotypes[J]. Science, 2011, 334(6052):105-108 |
| [61] | Turnbaugh PJ, Hamady M, Yatsunenko T, et al.A core gut microbiome in obese and lean twins[J]. Nature, 2009, 457(7228): 480-484 |
| [62] | Arumugam M, Raes J, Pelletier E, et al.Enterotypes of the human gut microbiome[J]. Nature, 2011, 473(7346):174-180 |
| [63] | Seth S, Valimaki N, Kaski S, et al.Exploration and retrieval of whole-metagenome sequencing sample[J]. Bioinformatics, 2014, 30(17):2471-2479 |
| [64] | Chistoserdova L.Is metagenomics resolving identification of functions in microbial communities?[J]. Microbial Biotechnology, 2014, 7(1):1-4 |
| [65] | Marx CJ.Can you sequence ecology?Metagenomics of adaptive diversification[J]. PLoS Biology, 2013, 11(2):e1001487 |
| [66] | Riesenfeld CS, Schloss PD, Handelsman J.Metagenomics: genomic analysis of microbial communities[J]. Annual Review of Genetics, 2004(38):525-552 |
| [67] | Sharon I, Banfield JF.Genomes from metagenomics[J]. Science, 2013, 342(6162):1057-1058 |
| [68] | Ye DD, Fan MM, Guang Q, et al.A review on the bioinformatics pipelines for metagenomic research[J]. Zoological Research, 2012, 33(6):574-585(in Chinese) 叶丹丹, 樊萌萌, 关琼, 等.宏基因组研究的生物信息学平台 现状[J]. 动物学研究, 2012, 33(6):574-585 |
| [69] | Ludwig W, Strunk O, Westram R, et al.ARB:a software environment for sequence data[J]. Nucleic Acids Research, 2004, 32(4):1363-1371 |
| [70] | Riehle K, Coarfa C, Jackson A, et al.The Genboree Microbiome Toolset and the analysis of 16S rRNA microbial sequences[J]. BMC Bioinformatics, 2012, 13(Suppl 13):S11 |
| [71] | Schloss PD, Westcott SL, Ryabin T, et al.Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities[J]. Applied and Environmental Microbiology, 2009, 75(23): 7537-7541 |
| [72] | Cuccuru G, Orsini M, Pinna A, et al.Orione, a web-based framework for NGS analysis in microbiology[J]. Bioinformatics, 2014, 30(13):1928-1929 |
| [73] | Shah N, Tang HX, Doak TG, et al.Comparing bacterial communities inferred from 16S rRNA gene sequencing and shotgun metagenomics[J]. Pacific Symposium on Biocomputing, 2011:165-176 |
| [74] | Huse SM, Welch DB, Voorhis A, et al.VAMPS:a website for visualization and analysis of microbial population structures[J]. Bioinformatics, 2014(15):e41 |
| [75] | Caporaso JG, Kuczynski J, Stombaugh J, et al.QⅡME allows analysis of high-throughput community sequencing data[J]. Nature Method, 2010, 7(5):335-336 |
| [76] | Kuczynski J, Stombaugh J, Walters WA, et al.Using QⅡME to analyze 16S rRNA gene sequences from microbial communities[J]. Current Protocols in Bioinformatics, 2011.DOI: 10.1002/0471250953.bi1007s36 |
| [77] | Cole JR, Wang Q, Cardenas E, et al.The ribosomal database project:improved alignments and new tools for rRNA analysis[J]. Nucleic Acids Research, 2009, 37:D141-D145 |
| [78] | Cole JR, Wang Q, Fish JA, et al.Ribosomal database project: data and tools for high throughput rRNA analysis[J]. Nucleic Acids Research, 2014, 42:D633-D642 |
| [79] | Angiuoli SV, Matalka M, Gussman A, et al.CloVR:a virtual machine for automated and portable sequence analysis from the desktop using cloud computing[J]. BMC Bioinformatics, 2011, 12:e356 |
| [80] | Sun S, Chen J, Li W, et al.Community cyberinfrastructure for advanced microbial ecology research and analysis:the CAMERA resource[J]. Nucleic Acids Research, 2011, 39:546-551 |
| [81] | Hunter S, Corbett M, Denise H, et al.EBI metagenomics-a new resource for the analysis and archiving of metagenomic data[J]. Nucleic Acids Research, 2014, 42:600-606 |
| [82] | Blankenberg D, Von Kuster G, Coraor N, et al.Galaxy:a web-based genome analysis tool for experimentalists[J]. Current Protocols in Molecular Biology, 2010.DOI: 10.1002/0471142727.mb1910s89 |
| [83] | Goecks J, Nekrutenko A, Taylor J, et al.Galaxy:a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences[J]. Genome Biology, 2010, 11(8):R86 |
| [84] | Markowitz VM, Ivanova NN, Szeto E, et al.IMG/M:a data management and analysis system for metagenomes[J]. Nucleic Acids Research, 2008, 36:534-538 |
| [85] | Goll J, Thiagarajan M, Abubucket S, et al.A case study for large-scale human microbiome analysis using JCVI's metagenomics reports (METAREP)[J]. PLoS One, 2012, 7(6): e29044 |
| [86] | Goll J, Rusch DB, Tanenbaum DM, et al.METAREP:JCVI metagenomics reports-an open source tool for high-performance comparative metagenomics[J]. Bioinformatics, 2010, 26(20):2631-2632 |
| [87] | Huson DH, Auch AF, Qi J, et al.MEGAN analysis of metagenomic data[J]. Genome Research, 2007, 17(3):377-386 |
| [88] | Huson DH, Weber N.Microbial community analysis using MEGAN[J]. Microbial Metagenomics, Metatranscriptomics, and Metaproteomics, 2013, 531:465-485 |
| [89] | Meyer F, Paarmann D, D Souza M, et al.The metagenomics RAST server-a public resource for the automatic phylogenetic and functional analysis of metagenomes[J]. BMC Bioinformatics, 2008(9):e386 |
| [90] | Treangen TJ, Koren S, Sommer DD, et al.MetAMOS:a modular and open source metagenomic assembly and analysis pipeline[J]. Genome Biology, 2013, 14(1):R2 |
| [91] | Kultima JK, Sunagawa S, Li J, et al.MOCAT:A metagenomics assembly and gene prediction toolkit[J]. PLoS One, 2012, 7(10): e47656 |
| [92] | Su X, Xu J, Ning K.Parallel-META:efficient metagenomic data analysis based on high-performance computation[J]. BMC Systems Biology, 2012, 6(Suppl 1):S16 |
| [93] | Su XQ, Pan WH, Song BX, et al.Parallel-META 2.0:enhanced metagenomic data analysis with functional annotation, high performance computing and advanced visualization[J]. PLoS One, 2014, 9(3):e89323 |
| [94] | Li W.Analysis and comparison of very large metagenomes with fast clustering and functional annotation[J]. BMC Bioinformatics, 2009(10):e359 |
| [95] | Li R, Li Y, Kristiansen K, et al.SOAP:short oligonucleotide alignment program[J]. Bioinformatics, 2008, 24(5):713-714 |
| [96] | Li R, Yu C, Li Y, et al.SOAP2:an improved ultrafast tool for short read alignment[J]. Bioinformatics, 2009, 25(15): 1966-1967 |
| [97] | Arumugam M, Harrington ED, Foerstner KU, et al.Smash Community:a metagenomic annotation and analysis tool[J]. Bioinformatics, 2010, 26(23):2977-2978 |
| [98] | Gerlach W, Junemann S, Tille F, et al.WebCARMA:a web application for the functional and taxonomic classification of unassembled metagenomic reads[J]. BMC Bioinformatics, 2009, 10:e430 |
| [99] | Gerlach W, Stoye J.Taxonomic classification of metagenomic shotgun sequences with CARMA3[J]. Nucleic Acids Research, 2011, 39(14):e91 |
| [100] | Wu S, Zhu Z, Fu L, et al.WebMGA:a customizable web server for fast metagenomic sequence analysis[J]. BMC Genomics, 2011, 12:e444 |
| [101] | Scholz MB, Lo CC, Chain PS.Next generation sequencing and bioinformatic bottlenecks:the current state of metagenomic data analysis[J]. Current Opinion in Biotechnology, 2012, 23(1):9-15 |
| [102] | Laserson J, Jojic V, Koller D.Genovo:de novo assembly for metagenomes[J]. Journal of Computational Biology, 2011, 18(3): 429-443 |
| [103] | Namiki T, Hachiya T, Tanaka H, et al.MetaVelvet:an extension of Velvet assembler to de novo metagenome assembly from short sequence reads[J]. Nucleic Acids Research, 2012, 40(2):e155 |
| [104] | Lai B, Ding R, Li Y, et al.A de novo metagenomic assembly program for shotgun DNA reads[J]. Bioinformatics, 2012, 28(11): 1455-1462 |
| [105] | Santamaria M, Fosso B, Consiglio A, et al.Reference databases for taxonomic assignment in metagenomics[J]. Briefings in Bioinformatics, 2012, 13(6):682-695 |
| [106] | Hawksworth DL.Biodiversity:Measurement and Estimation[M]. Verlag:Springer, 1995:5-12 |
| [107] | Chao A.Nonparametric estimation of the number of classes in a population[J]. Scandinavian Journal of Statistics, 1984, 11(4): 265-270 |
| [108] | Haegeman B, Hamelin J, Moriaty J, et al.Robust estimation of microbial diversity in theory and in practice[J]. The ISME Journal, 2013, 7(6):1092-1101 |
| [109] | Zhou J, Deng Y, Luo F, et al.Phylogenetic molecular ecological network of soil microbial communities in response to elevated CO2[J]. mBio, 2011, 2(4):e00122-11 |
 2015, Vol. 42
2015, Vol. 42