扩展功能
文章信息
- 刘正辉, 李德豪
- LIU Zheng-Hui, LI De-Hao
- 氨氧化古菌及其对氮循环贡献的研究进展
- Ammonia-oxidizing archaea and their contribution to global nitrogen cycling: a review
- 微生物学通报, 2015, 42(4): 774-782
- Microbiology China, 2015, 42(4): 774-782
- 10.13344/j.microbiol.china.140581
-
文章历史
- 收稿日期: 2014-07-29
- 接受日期: 2014-09-10
- 优先数字出版日期(www.cnki.net): 2014-09-16
2. 广东高校石油化工污染控制与清洁生产工程技术开发中心 广东 茂名 525000
2. Technology Development Center for Petrochemical Pollution Control and Cleaner Production of Guangdong Universities,Maoming, Guangdong 525000, China
氮是生命体核酸与蛋白质必不可少的组成元素。氮素的生物地球化学循环(氮循环)对生命的存在和持续有关键作用,本质上是微生物驱动的氮素转化、利用及循环的过程,包括氮的固定、硝化作用、反硝化作用和氨化作用。其中硝化作用,即将氨氧化为亚硝态氮、进而氧化为硝态氮的过程,是氮循环的核心环节。氨氧化被认为是硝化作用的第一个限速步骤,主要是由变形菌纲中的β-变形杆菌亚门或γ-变形杆菌亚门[1, 2]所进行的专性化能自养过程,本质上由氨单加氧酶(AMO)催化完成的微生物生化反应。
近年来,随着分子生物学技术的快速发展,科学家们通过对海洋、土壤等环境的研究发现,一类古菌(即氨氧化古菌,Ammonia-oxidizing archaea,AOA)也具有编码AMO的基因[3, 4],且广泛分布于包括海洋、湖泊和土壤等在内的多种环境中[5],意味着这类古菌可能也有进行氨氧化的能力,参与环境氮循环的硝化作用环节。氨氧化古菌已经成为地理学、生物学、生态学和环境科学等交叉领域的研究热点[6, 7]。本文对氨氧化细菌和氨氧化古菌的生态分布、系统进化、存在丰度及表达活性,以及对氮循环的贡献进行综述,着重于自氨氧化古菌的发现以来近十年的研究进展。
1 氨氧化古菌的发现及检测方法19世纪末,科学家发现硝化反应由微生物参与完成[8],也通过分离培养的方法获得了自养型的原核微生物。长期以来,所有可培养的好氧自养型氨氧化菌都是β-变形杆菌亚门或γ-变形杆菌亚门[1, 2],以致人们认为所有的自养型氨氧化菌都是细菌。2004年,Venter等用鸟枪法对马尾藻海(Sargasso Sea) 的微生物宏基因组进行测序和分析后发现,奇古菌中存在氨单加氧酶的编码基因amoA序列[3];2005年,Treusch等对土壤奇古菌基因组分析时,在一个43 kb的宏基因组片段中也发现了奇古菌的氨单加氧酶编码基因amoA[4]。Könneke等在热带海洋水族馆鱼缸里成功分离到一株奇古菌Nitrosopumilus maritimus SCM1,该菌株可以以氨作为唯一能源进行自养生长[9],人类第一次获得了可培养的、具有氨氧化能力的古菌菌株。Francis设计了特异性的引物、应用PCR扩增检测的方法,发现AOA广泛存在于海洋的透光层、低氧区水柱、海岸/入海口基底部和海底沉积物等区域,提示在非极端环境的海洋中AOA可能在氮的循环中起着重要的作用。此后,古菌的氨氧化功能引起了全世界氮循环研究者的高度关注与讨论,且氨氧化古菌已经成为微生物生态学研究的一个模式微生物之一。
氨氧化古菌的检测方法主要分为两大类:一类是依赖于培养的技术,如平板分离培养、富集培养等。目前通过这些技术,已经分离到了氨氧化细菌和氨氧化古菌。然而绝大多数的环境微生物在实验室条件下是不可培养的[10],且氨氧化细菌生长缓慢,通过这种方法分离纯化相当困难,并不能获得全部的氨氧化菌株。相比而言,氨氧化细菌的研究开展由来已久,目前获得的菌株要比氨氧化古菌的要多。在氨氧化古菌的培养方面,目前分离到的纯菌株有Nitrosopumilus maritimus SCM1[9]和Nitrosocaldus yellowstonii[11];富集到的培养物有Candidatus Nitrososphaera gargensis[12]、培养物SJ[13]、Candidatus Nitrosoarchaeum limnia SFB1[14]、Candidatus Nitrosotalea devanaterra[15]、Candidatus Nitrosoarchaeum limnia BG20[16]、Candidatus Nitrosopumilus salaria BD31[17]、Candidatus Nitrosopumilus koreensis AR1和Nitrosopumilus sediminis AR2[18]。
另一类是不依赖于培养的分子微生物技术。其中基于非多聚酶链式反应(Polymerase chain reaction,PCR)的方法有荧光原位杂交(Fluorescence in situ hybridization,FISH)[19]、甘油二烷基甘油四醚脂技术(GDGT)、稳定性同位素技术(Stable isotope probing,SIP)等;而基于PCR的方法有基因克隆建库、反转录PCR (Reverse transcription PCR,RT-PCR)、定量PCR (Quantitative PCR)等。
目前,关于氨氧化细菌和古菌的生态学研究多数都通过这些基于PCR的分子生物学方法来开展,所采用的引物是16S rRNA 基因或功能基因amoA基因的两侧序列。通过对环境样品的氨单加氧酶编码基因进行PCR检测时,AOA相对于AOB容易检测到,而AOB的amoA基因可能没有检测信号。例如,Briones等在水稻根际[20]、Beman等在珊瑚[21]、Siboni等在珊瑚礁[22]、Zhang等在氨氧化菌富集培养物中[23]和Park等在表面水层样品中[24]均可扩增到古菌amoA基因,但并未扩增到细菌amoA基因。不过也有检测到AOB而检测不到AOA的例子,如Park等在废水处理厂反应器中[25]以及van der Wielen等在饮用水系统的部分样品[26]里未能成功扩增到古菌的amoA基因。扩增不成功的原因可能由于它不存在于某些样品中,或存在的数量低于PCR检测的最低限度,或者目前的引物并不适于扩增这些环境中的氨氧化酶编码基因。
最近,稳定性同位素原位示踪微生物核酸DNA/RNA技术也应用于氨氧化古菌的研究,如Wuchter等使用13C稳定性同位素培养北海水样品发现泉古菌的特异脂类同化了13C,揭示了海洋泉古菌营自养型生活[27];Jia等采用13CO2培养土壤样品时发现氨氧化细菌的amoA基因被标记了,而古菌的没有被标记,此结果表明氨氧化古菌可能没有参与土壤硝化过程,或者参与程度非常少以致未能被检测出来[28]。
此外,通过放射自显影杂交技术发现海洋泉古菌可以利用有机物如氨基酸进行异养生长[29, 30]。二次离子质谱分析技术(Nanometer-scale secondary ion mass spectrometry)可以直接观测微生物细胞对营养元素利用过程[31],极有可能应用于研究原位氨氧化古菌的代谢途径[32]。
2 氨氧化古菌的生境分布及系统发育氨氧化细菌广泛存在于陆地生态系统、水生生态系统(海洋和淡水)、工程生态系统如生物反应器等。相应地,氨氧化古菌在全球具有很广的生态位,在陆地环境中的分布非常广泛,在农业土壤[33]、温带森林土壤[34]、半干旱森林土壤[35]、稻田土壤[36]、牧草土壤、淡水植物Littorella uniflora根系土壤[37]以及高原永久冻结带[38]等陆地生态系统中都检测到AOA。在水生生态统中的各个区域都有AOA的分布,包括海洋透光层[39, 40, 41]和深海缺氧层[42, 43, 44, 45]、深海沉积物[24, 43]、珊瑚和暗礁[21]、江河口[46, 47]、地下河入海口[48]、淡水及沉积物[47, 49, 50],以及南北两极的海水[51]。在工程系统中也发现有AOA的存在,如Park等工程系统废水生物反应器[25]、Zhang等在污水处理厂的活性污泥里[52]、Wells等在活性污泥生物反应器15%的样品中[53]、Gao等在污水处理厂中都检测到AOA[54, 55]。
在系统发育与进化方面,通过16S rRNA基因的系统发育,Brochier-Armanet等将中温泉古菌(主要包括AOA)从泉古菌中独立开来作为一个新的门Thaumarchaeota[56],国内张丽梅等建议意译称为奇古菌[57]。现在古菌主要有3个门,分别是:广古菌门Euryarchaeota、泉古菌门Crenarchaeota和奇古菌门Thaumarchaeota。进一步的研究揭示了AOA主要有两个聚类:奇古菌Group I.1a分支和奇古菌Group I.1b分支[58]。其中,奇古菌Group I.1a分支包括了海洋、大多数水体以及沉积物来源的AOA;奇古菌Group I.1b分支涵盖了土壤及其他陆地生态系统来源的AOA (图 1)[59]。而Weidler等通过amoA基因的系统发育表明,来源于热泉的AOA独立成一个分支,不同于来源海洋和土壤的AOA分支[60]。Cao等收集了来源于全球海洋、海洋底泥、淡水、土壤等生境的6 200条古菌amoA基因的序列进行系统发育分析,发现来源于海洋的分支明显与其他来源的区分开来[61]。
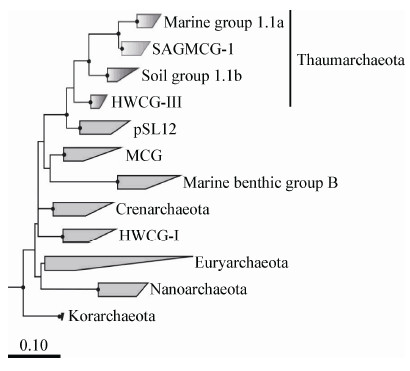
|
|
图 1
基于奇古菌16S rRNA基因的系统发育
Figure 1
Phylogeny of Thaumarchaeota based on archaeal 16S rRNA gene sequences[59]
注:系统发育树上所有节点中黑点表示Bootstrap values大于80%,标尺表示每个核苷酸位点的替换率为0.10.SAGMCG-1:南非金矿泉古泉类群1;pSL12:热泉古菌;HWCG-I:热泉古菌类群I;HWCG-III:热泉古菌类群III;MCG:Miscellaneous crenarchaeotic group;Thaumarchaeota:奇古菌门;Crenarchaeota:泉古菌门;Euryarchaeota:广古菌门;Korarchaeota:初古菌门;Nanoarchaeota:微古菌门. Note: Nodes on phylogenetic tree supported by bootstrap values greater than 80% are indicated with block dot; The scale bar represents 0.10 changes per sequence position. SAGMCG-1: South African gold mine crenarchaeotic group 1; pSL12: Hot spring crenarchaea; HWCG-I: Hot water crenarchaeotic group I; HWCG-III: Hot water crenarchaeotic group III; MCG: Miscellaneous crenarchaeotic group. |
基于微生物分子生态学的研究发现,地球环境中存在大量泉古菌(Crenarchaeota)[62],在海洋中约占微型浮游生物数量的20%,在土壤中约占原核生物量的5%[63]。自然环境条件下每个AOA细胞持有一个拷贝amoA基因[40],而每个AOB持有约2个
拷贝amoA基因[64],通过关键功能基因amoA的定量PCR分析可以推测特定生态环境中氨氧化微生物的分布丰度信息。
在水生生态系统中古菌amoA的基因数量比细菌的要高。Wuchter等在北海及北大西洋的研究显示古菌amoA的拷贝数量是细菌的10-1 000倍[39];Mincer等在Monterey海湾和北太平洋发现古菌amoA的拷贝数量是细菌的几个数量级[40];Caffrey等发现在多数河口地方古菌amoA拷贝数要比细菌的高约80倍[47];研究者也发现在淡水河湖等生境中AOA数量远高于AOB[49, 65, 66]。但也存在情形相反的例子,如Caffrey等在Weeks海湾河口[47]、Santoro等在地下河入海口[48]、Jiang等在中国青海湖的湖底沉积物中[67]、Mosier等在圣弗兰西克海 湾[68]发现细菌amoA拷贝数要略高于古菌的相应拷贝数。总体而言,在水生生态系统中AOA具有类群数量优势。
土壤环境中AOA与AOB的比例也相当高,总体高出2-3个数量级。Leininger等对12个来源不同的土壤的研究发现,古菌amoA基因的拷贝数与细菌amoA的比例高达3 000倍[33];Herrmann等对淡水植物Littorella uniflora根系土壤的研究发现古菌与细菌的amoA拷贝数比例在500-8 000倍之 间[37];Isobe等在中国华南3个森林土壤样品中只检测到AOA而没有AOB[69];Alves等在北极土壤中11个样品中5个样品只能检测氨氧化古菌,另外4个样品中古菌amoA基因数比细菌的高[70];de Gannes等在热带土壤中检测和定量了AOB相对于AOA显得很低,或者未能检测出AOB[71]。但是也有研究表明AOB的存在丰度高于AOA的例子,如Zhang等在珠穆朗玛峰永久冻结带土壤中发现细菌的amoA拷贝数是古菌的1-2个数量级[38]。
4 氨氧化古菌对氮循环的贡献(生态学意义) 4.1 菌株培养及功能验证古菌具有氨氧化功能的直接证据也来源于实验室条件下环境微生物的成功培养。2005年,Könneke等从海洋水族馆中分离到一株中温泉古菌 Nitrosopumilus maritimus SCM1,属于化能自养型,可以以NH4+作为唯一能源,通过将NH4+氧化成为NO2−而获得生长[9];这是人类在实验室条件下第一次获得了氨氧化古菌纯培养菌株,该发现在“Nature”杂志上进行了报道。2008年de la Torre在美国黄石国家森林公园的热泉中(74 °C)富集到的喜温泉古菌Nitrosocaldus yellowstoni[11],也是通过氨氧化作用获取能源的自养菌,证实了生长温度高至74 °C的条件下依然存在微生物的硝化作用。此外,Hatzenpichler等对Siberian Garga热泉进行了长达6年的富集培养工作,终于获得了氨氧化古菌Group I.1b类群中的Candidatus Nitrososphaera gargensis培养物[12];Park等在东海北极海洋沉积物里富集了Group I.1a的培养物[13]。后来研究者在其他地方也成功富集了氨氮化古菌。至今为止,尚未有从淡水水体生态系统中分离到氨氧化古菌的报道。
Martens-Habbena等研究了泉古菌 Nitrosopumilus maritimus SCM1的氨氧化动力学,发现AOA的半饱和速率常数Km与海洋原位氨氧化作用的Km非常接近[72],且AOA对底物铵态氮的亲和力相当高,进行氨氧化作用的最低铵态氮浓度比AOB最低生长浓度还要低100倍以上[73]。因此AOA在与其他微生物竞争底物铵态氮时占有绝对的优势,不难理解在氨氮浓度极低的贫营养水体中AOA丰度远高于AOB。最近,Könneke等[74]发现可以通过羟基丙酸/羟基丁酸循环更高效地利用能源进行CO2的固定,将无机碳转化为有机碳营自养生活,从而从生物化学的角度解释了AOA能够在寡营养条件下生存和繁荣的机制。
4.2 功能基因数量与氨氧化作用的耦合相关性氨氧化微生物的功能基因amoA拷贝数的定量,揭示了这些微生物可能在氨氧化过程中发挥着作用,参与氮的生物地球化学循环[33, 39, 40]。从线性耦合相关性来看,在一些低氨氮浓度水体和极端环境中古菌amoA基因拷贝数与氨氧化速率存在很好的相关性,许多研究者推测AOA对硝化作用的贡献可能大于AOB。例如在加利福尼亚海湾[41]和荷兰西斯海尔德河口[75]等水体中,亚硝酸盐浓度和氨氧化速率均与泉古菌amoA基因数量呈显著正相关;弗罗里达州9个不同河口底层水体和沉积物中,硝化速率与泉古菌amoA基因数量也呈正相关,但与AOB的amoA数量没有显著的相关性[47];Wuchter等发现北海泉古菌amoA基因数量与海水中铵态氮浓度呈显著负相关[39]。然而有些生境中也有不存在相关性的情形,如Bernhard等[76]在麻萨诸塞州(Massachusetts)东北部的Plum Island Sound河口/海湾发AOA与潜在的硝化速率不存在相关性;Di等[77]在铵态氮浓度较高的草原土壤中发现,铵态氮浓度升高对AOA的amoA基因拷贝数影响不大,而促使AOB的amoA基因拷贝数呈正相方向增加。因此,古菌amoA基因的大量存在或相当高的丰度并不意味着该基因一定在环境中发挥作用,我们对基因丰度数据的解读必须慎重对待[78]。
4.3 表达活性古菌和细菌的amoA基因可能只在特殊的环境条件下才可以表达,且基因产物执行其在氨氧化过程中相应的生态功能,因此对amoA基因的表达活性进行研究的意义相当突出。在不少水体和土壤生境中,AOA的表达活性要高于AOB的表达活性,如Jiang等[67]在中国青海湖样品的反转录定量PCR结果表明在有氧的水体中AOA的表达丰度比AOB的略高一些,Church等[79]在太平洋中上层海域中发现绝大多数奇古菌的amoA基因都具有表达活性,推测AOA是太平洋氮循环的积极贡献者。Lam 等[42]利用反转录定量PCR比较了黑海泉古菌和γ-AOB的amoA基因表达差异,通过统计分析发现泉古菌amoA的表达可以解释74.5%的NO2−(氨氧化作用的产物)变化。Treusch等[4]对土壤进行了反转录PCR检测到古菌amoA的mRNA。Leininger等[33]对西欧和北欧的土壤进行反转录定量PCR分析表明古菌amoA的表达丰度高于细菌相应基因的表达。Chen等[36]发现在淹水稻田土壤中,AOA的amoA基因数量以及转录水平均显著高于AOB。Tourna等[80]研究发现土壤中AOA的amoA基因数量及其转录活性都高于AOB的相应部分。此外,Jung等[81]采用稳定性同位素技术证实了土壤硝化作用和反硝化过程中产生的N2O部分来源于AOA,即AOA参与氮循环。但是也有相反的例子,如在中国青海湖的湖底沉积物中AOB的表达丰度比AOA的稍高一点[67];在高氮草原土壤中,是AOB的而不是AOA的数量和活性随土壤铵态氮浓度的升高而增大,与土壤硝化活性呈正相关关系[77, 82]。
有研究者为了确认氨氧化过程中是古菌还是细菌在起作用,将硝化抑制剂如乙炔等也用于土壤氨氧化途径的阻断研究。Jia等[28]利用高浓度氨和乙炔抑制氨氧化途径进行实验时,发现古菌amoA基因拷贝数虽然占优势,但在功能上氨氧化细菌是最主要的。Offre等[83]利用不同浓度的乙炔来抑制农业土壤微生物的氨氧化活性时,AOA的生长受到抑制,群落生长与硝化速率的相关性最好,表明AOA而非AOB是氨氧化过程的参与者。Di等[77]发现土壤氨氧化古菌尽管在数量上与氨氧化细菌相当或占优势,但其丰度和活性并不随氨态氮源增加而变化,其对硝化抑制剂的反应也不敏感;相反,随氨态氮源增加,氨氧化细菌数量增加3-10倍,硝化活性增加177倍,且对硝化抑制剂反应敏感。在土壤中AOB在高氨氮的生境中发挥作用,而AOA则喜好低氨氮的土壤生境[82]。Meyer等[84]发现土壤中的硝化作用是由氨氧化细菌而不是氨氧化古菌驱动的。
近年来Liu等[49]对中国东江流域展开了氨氧化微生物的研究,发现东江淡水水体也存在AOA。东江AOA的丰度比AOB的要高2-4个数量级;通过主成分分析(PCA)表明AOA的amoA基因的丰度与水体氨氮浓度存在一定的负相关性。AOA/AOB的比率在氨氮浓度较低的水体环境中较高,表明AOA比AOB更偏好氨氮浓度较低的生境。淡水AOA的发现是对揭开氨氧化古菌全球地理分布生境的补充和贡献,同时也提示了AOA在东江水体氨循环中可能起着重要的作用。然而,我们对AOA在淡水水体氮循环中的贡献是建立在分子生态学及环境水质分析的层面上的。在AOA和AOB的活性证明上尚未有更有力的证据,如amoA基因转录水平的变化,水样硝化速率和反硝化速率与AOA和AOB表达活性的关系如何等等。我们将进一步深入研究AOA在淡水水体氮循环中的贡献,阐明AOA在淡水生态系统中的生态学意义。
5 展望目前对于AOA在氮的生物地球化学循环中贡献的了解依然相当有限,目前主要是通过分子生物学方法发现AOA中存在着与AOB参与硝化作用的关键酶氨单加氧酶编码基因amoA,而对于AOA氨氧化的独特生物化学过程还知之甚少。虽然已经在海洋(海水及沉积物)和热泉中都分离或富集到了AOA,但如何保持纯培养物在分离和富集过程中的连续生长依然相当困难,目前从陆地生态系统分离或富集培养获得的AOA还很少。在氨氧化的生态功能方面,有学者提出观点认为奇古菌amoA基因的高丰度与氨氧化过程并不相关,而可能与混合营养类型相关,这种观点可以多大程度地解释现有的氨氧化微生物分布丰度差异还需深入探讨。由于古菌在细胞大小上要比细菌小得多,且细胞结构和代谢生理也明显区别于细菌,会不会致使古菌在细胞水平上的表达活性与细菌的相比也有着显著的差异,这有待进一步的研究。今后多种研究方法如纯菌培养、分子生物学、宏基因组学以及生物地球化学等的结合,对于阐明AOA在氮的生物地球化学循环中的贡献将会起着重要的作用。
| [1] | Head IM, Hiorns WD, Embley TM, et al. The phylogeny of autotrophic ammonia-oxidizing bacteria as determined by analysis of 16S ribosomal-RNA gene sequences[J]. Journal of General Microbiology, 1993, 139(Pt6): 1147-1153 |
| [2] | Kowalchuk GA, Stephen JR. Ammonia-oxidizing bacteria: a model for molecular microbial ecology[J]. Annual Review of Microbiology, 2001, 55: 485-529 |
| [3] | Venter JC, Remington K, Heidelberg JF, et al. Environmental genome shotgun sequencing of the Sargasso Sea[J]. Science, 2004, 304(5667): 66-74 |
| [4] | Treusch AH, Leininger S, Kletzin A, et al. Novel genes for nitrite reductase and Amo-related proteins indicate a role of uncultivated mesophilic crenarchaeota in nitrogen cycling[J]. Environmental Microbiology, 2005, 7(12): 1985-1995 |
| [5] | Francis CA, Roberts KJ, Beman JM, et al. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean[J]. Proceedings of the National Academy of Sciences of the United States of America, 2005, 102(41): 14683-14688 |
| [6] | Hu AY, Jiao NZ. Ammonia-oxidizing archaea—a hotspot of environmental microbial ecology[J]. Progress in Natural Science, 2009, 19(4): 370-379 (in Chinese) 胡安谊, 焦念志. 氨氧化古菌——环境微生物生态学研究的 一个前沿热点[J]. 自然科学进展, 2009, 19(4): 370-379 |
| [7] | He JZ, Zhang LM. Advances in ammonia-oxidizing microorganisms and global nitrogen cycle[J]. Acta Ecologica Sinica, 2009, 29(1): 406-415 (in Chinese) 贺纪正, 张丽梅. 氨氧化微生物生态学与氮循环研究进展[J]. 生态学报, 2009, 29(1): 406-415 |
| [8] | Winogradsky S. Recherches Sur Les Organismes de laNitrification[M]. Java: Impr. Charaire, 1890, 110: 1013-1016 |
| [9] | Könneke M, Bernhard AE, de la Torre JR, et al. Isolation of an autotrophic ammonia-oxidizing marine archaeon[J]. Nature, 2005, 437(7058): 543-546 |
| [10] | Sharma R, Ranjan R, Kapardar RK, et al. “Unculturable” bacterial diversity: an untapped resource[J]. Current Science, 2005, 89(1): 72-77 |
| [11] | de la Torre JR, Walker CB, Ingalls AE, et al. Cultivation of a thermophilic ammonia oxidizing archaeon synthesizing crenarchaeol[J]. Environmental Microbiology, 2008, 10(3): 810-818 |
| [12] | Hatzenpichler R, Lebecleva EV, Spieck E, et al. A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring[J]. Proceedings of the National Academy of Sciences of the United States of America, 2008, 105(6): 2134-2139 |
| [13] | Park BJ, Park SJ, Yoon DN, et al. Cultivation of autotrophic ammonia-oxidizing archaea from marine sediments in co-culture with sulfur-oxidizing bacteria[J]. Applied and Environmental Microbiology, 2010, 76(22): 7575-7587 |
| [14] | Blainey PC, Mosier AC, Potanina A, et al. Genome of a low-salinity ammonia-oxidizing archaeon determined by single-cell and metagenomic analysis[J]. PLoS One, 2011, 6(2): e16626 |
| [15] | Lehtovirta-Morley LE, Stoecker K, Vilcinskas A, et al. Cultivation of an obligate acidophilic ammonia oxidizer from a nitrifying acid soil[J]. Proceedings of the National Academy of Sciences of the United States of America, 2011, 108(38): 15892-15897 |
| [16] | Mosier AC, Allen EE, Kim M, et al. Genome sequence of “Candidatus Nitrosoarchaeum limnia” BG20, a low-salinity ammonia-oxidizing archaeon from the San Francisco Bay estuary[J]. Journal of Bacteriology, 2012, 194(8): 2119-2120 |
| [17] | Mosier AC, Allen EE, Kim M, et al. Genome sequence of “Candidatus Nitrosopumilus salaria” BD31, an ammonia-oxidizing archaeon from the San Francisco Bay estuary[J]. Journal of Bacteriology, 2012, 194(8): 2121-2122 |
| [18] | Park SJ, Ghai R, Martin-Cuadrado AB, et al. Genomes of two new ammonia-oxidizing archaea enriched from deep marine sediments[J]. PLoS One, 2014, 9(5): e96449 |
| [19] | Jurgens G, Glockner FO, Amann R, et al. Identification of novel Archaea in bacterioplankton of a boreal forest lake by phylogenetic analysis and fluorescent in situ hybridization[J]. FEMS Microbiology Ecology, 2000, 34(1): 45-56 |
| [20] | Briones AM, Okabe S, Umemiya Y, et al. Influence of different cultivars on populations of ammonia-oxidizing bacteria in the root environment of rice[J]. Applied and Environmental Microbiology, 2002, 68(6): 3067-3075 |
| [21] | Beman JM, Roberts KJ, Wegley L, et al. Distribution and diversity of archaeal ammonia monooxygenase genes associated with corals[J]. Applied and Environmental Microbiology, 2007, 73(17): 5642-5647 |
| [22] | Siboni N, Ben-Dov E, Sivan A, et al. Global distribution and diversity of coral-associated Archaea and their possible role in the coral holobiont nitrogen cycle[J]. Environmental Microbiology, 2008, 10(11): 2979-2990 |
| [23] | Zhang Y, Ruan XH, den Camp HJMO, et al. Diversity and abundance of aerobic and anaerobic ammonium-oxidizing bacteria in freshwater sediments of the Xinyi River (China)[J]. Environmental Microbiology, 2007, 9(9): 2375-2382 |
| [24] | Park SJ, Park BJ, Rhee SK. Comparative analysis of archaeal 16S rRNA and amoA genes to estimate the abundance and diversity of ammonia-oxidizing archaea in marine sediments[J]. Extremophiles, 2008, 12(4): 605-615 |
| [25] | Park HD, Wells GF, Bae H, et al. Occurrence of ammonia-oxidizing archaea in wastewater treatment plant bioreactors[J]. Applied and Environmental Microbiology, 2006, 72(8): 5643-5647 |
| [26] | van der Wielen P, Voost S, van der Kooij D. Ammonia-oxidizing bacteria and archaea in groundwater treatment and drinking water distribution systems[J]. Applied and Environmental Microbiology, 2009, 75(14): 4687-4695 |
| [27] | Wuchter C, Schouten S, Boschker HTS, et al. Bicarbonate uptake by marine Crenarchaeota[J]. FEMS Microbiology Letters, 2003, 219(2): 203-207 |
| [28] | Jia Z, Conrad R. Bacteria rather than Archaea dominate microbial ammonia oxidation in an agricultural soil[J]. Environmental Microbiology, 2009, 11(7): 1658-1671 |
| [29] | Ouverney CC, Fuhrman JA. Marine planktonic Archaea take up amino acids[J]. Applied and Environmental Microbiology, 2000, 66(11): 4829-4833 |
| [30] | Agogue H, Brink M, Dinasquet J, et al. Major gradients in putatively nitrifying and non-nitrifying Archaea in the deep North Atlantic[J]. Nature, 2008, 456(7223): 788-791 |
| [31] | Li T, Wu TD, Mazeas L, et al. Simultaneous analysis of microbial identity and function using NanoSIMS[J]. Environmental Microbiology, 2008, 10(3): 580-588 |
| [32] | Jia ZJ, Weng JH, Lin XG, et al. Microbial ecology of archaeal ammonia oxidation—a review[J]. Acta Microbiologica Sinica, 2010, 50(4): 431-437 (in Chinese) 贾仲君, 翁佳华, 林先贵, 等. 氨氧化古菌的生态学研究进展[J]. 微生物学报, 2010, 50(4): 431-437 |
| [33] | Leininger S, Urich T, Schloter M, et al. Archaea predominate among ammonia-oxidizing prokaryotes in soils[J]. Nature, 2006, 442(7104): 806-809 |
| [34] | Onodera Y, Nakagawa T, Takahashi R, et al. Seasonal change in vertical distribution of ammonia-oxidizing archaea and bacteria and their nitrification in temperate forest soil[J]. Microbes and Environments, 2010, 25(1): 28-35 |
| [35] | Adair KL, Schwartz E. Evidence that ammonia-oxidizing archaea are more abundant than ammonia-oxidizing bacteria in semiarid soils of northern Arizona, USA[J]. Microbial Ecology, 2008, 56(3): 420-426 |
| [36] | Chen XP, Zhu YG, Xia Y, et al. Ammonia-oxidizing archaea: important players in paddy rhizosphere soil?[J]. Environmental Microbiology, 2008, 10(8): 1978-1987 |
| [37] | Herrmann M, Saunders AM, Schramm A. Archaea dominate the ammonia-oxidizing community in the rhizosphere of the freshwater macrophyte Littorella uniflora[J]. Applied and Environmental Microbiology, 2008, 74(10): 3279-3283 |
| [38] | Zhang LM, Wang M, Prosser JI, et al. Altitude ammonia-oxidizing bacteria and archaea in soils of Mount Everest[J]. FEMS Microbiology Ecology, 2009, 70(2): 208-217 |
| [39] | Wuchter C, Abbas B, Coolen MJL, et al. Archaeal nitrification in the ocean[J]. Proceedings of the National Academy of Sciences of the United States of America, 2006, 103(33): 12317-12322 |
| [40] | Mincer TJ, Church MJ, Taylor LT, et al. Quantitative distribution of presumptive archaeal and bacterial nitrifiers in Monterey Bayand the North Pacific Subtropical Gyre[J]. Environmental Microbiology, 2007, 9(5): 1162-1175 |
| [41] | Beman JM, Popp BN, Francis CA. Molecular and biogeochemical evidence for ammonia oxidation by marine Crenarchaeota in the Gulf of California[J]. ISME Journal, 2008, 2(4): 429-441 |
| [42] | Lam P, Jensen MM, Lavik G, et al. Linking crenarchaeal and bacterial nitrification to anammox in the Black Sea[J]. Proceedings of the National Academy of Sciences of the United States of America, 2007, 104(17): 7104-7109 |
| [43] | Nakagawa T, Mori K, Kato C, et al. Distribution of cold-adapted ammonia-oxidizing microorganisms in the deep-ocean of the northeastern Japan Sea[J]. Microbes and Environments, 2007, 22(4): 365-372 |
| [44] | Coolen MJL, Abbas B, van Bleijswijk J, et al. Putative ammonia-oxidizing Crenarchaeota in suboxic waters of the Black Sea: a basin-wide ecological study using 16S ribosomal and functional genes and membrane lipids[J]. Environmental Microbiology, 2007, 9(4): 1001-1016 |
| [45] | Cao H, Hong Y, Li M, et al. Phylogenetic diversity and ecological pattern of ammonia-oxidizing archaea in the surface sediments of the Western Pacific[J]. Microbial Ecology, 2011, 62(4): 813-823 |
| [46] | Beman JM, Francis CA. Diversity of ammonia-oxidizing archaea and bacteria in the sediments of a hypernutrified subtropical estuary: Bahia del Tobari, Mexico[J]. Applied and Environmental Microbiology, 2006, 72(12): 7767-7777 |
| [47] | Caffrey JM, Bano N, Kalanetra K, et al. Ammonia oxidation and ammonia-oxidizing bacteria and archaea from estuaries with differing histories of hypoxia[J]. ISME Journal, 2007, 1(7): 660-662 |
| [48] | Santoro AE, Francis CA, de Sieyes NR, et al. Shifts in the relative abundance of ammonia-oxidizing bacteria and archaea across physicochemical gradients in a subterranean estuary[J]. Environmental Microbiology, 2008, 10(4): 1068-1079 |
| [49] | Liu Z, Huang S, Sun G, et al. Diversity and abundance of ammonia-oxidizing archaea in the Dongjiang River, China[J]. Microbiological Research, 2011, 166(5): 337-345 |
| [50] | French E, Kozlowski JA, Mukherjee M, et al. Ecophysiological characterization of ammonia-oxidizing archaea and bacteria from freshwater[J]. Applied and Environmental Microbiology, 2012, 78(16): 5773-5780 |
| [51] | Kalanetra KM, Bano N, Hollibaugh JT. Ammonia-oxidizing archaea in the Arctic Ocean and Antarctic coastal waters[J]. Environmental Microbiology, 2009, 11(9): 2434-2445 |
| [52] | Zhang T, Jin T, Yan Q, et al. Occurrence of ammonia-oxidizing archaea in activated sludges of a laboratory scale reactor and two wastewater treatment plants[J]. Journal of Applied Microbiology, 2009, 107(3): 970-977 |
| [53] | Wells GF, Park HD, Yeung CH, et al. Ammonia-oxidizing communities in a highly aerated full-scale activated sludge bioreactor: betaproteobacterial dynamics and low relative abundance of Crenarchaea[J]. Environmental Microbiology, 2009, 11(9): 2310-2328 |
| [54] | Gao J, Luo X, Wu G, et al. Abundance and diversity based on amoA genes of ammonia-oxidizing archaea and bacteria in ten wastewater treatment systems[J]. Applied Microbiology and Biotechnology, 2014, 98(7): 3339-3354 |
| [55] | Gao JF, Luo X, Wu GX, et al. Quantitative analyses of the composition and abundance of ammonia-oxidizing archaea and ammonia-oxidizing bacteria in eight full-scale biological wastewater treatment plants[J]. Bioresource Technology, 2013, 138: 285-296 |
| [56] | Brochier-Armanet C, Boussau B, Gribaldo S, et al. Mesophilic crenarchaeota: proposal for a third archaeal phylum, the Thaumarchaeota[J]. Nature Reviews Microbiology, 2008, 6(3): 245-252 |
| [57] | Zhang LM, He JZ. A novel archaeal phylum: Thaumarchaeota— a review[J]. Acta Microbiologica Sinica, 2012, 52(4): 411-421 (in Chinese) 张丽梅, 贺纪正. 一个新的古菌类群——奇古菌门 (Thaumarchaeota)[J]. 微生物学报, 2012, 52(4): 411-421 |
| [58] | Nicol GW, Schleper C. Ammonia-oxidising Crenarchaeota: important players in the nitrogen cycle?[J]. Trends in Microbiology, 2006, 14(5): 207-212 |
| [59] | Stahl DA, de la Torre JR. Physiology and diversity of ammonia-oxidizing archaea[J]. Annual Review of Microbiology, 2012, 66: 83-101 |
| [60] | Weidler GW, Gerbl FW, Stan-Lotter H. Crenarchaeota and their role in the nitrogen cycle in a subsurface radioactive thermal spring in the Austrian central Alps[J]. Applied and Environmental Microbiology, 2008, 74(19): 5934-5942 |
| [61] | Cao H, Auguet JC, Gu JD. Global ecological pattern of ammonia-oxidizing archaea[J]. PLoS One, 2013, 8(2): e52853 |
| [62] | Fuhrman JA, Mccallum K, Davis AA. Novel major archaebacterial group from marine plankton[J]. Nature, 1992, 356(6365): 148-149 |
| [63] | Cavicchioli R, DeMaere MZ, Thomas T. Metagenomic studies reveal the critical and wide-ranging ecological importance of uncultivated archaea: the role of ammonia oxidizers[J]. Bioessays, 2007, 29(1): 11-14 |
| [64] | Chain P, Lamerdin J, Larimer F, et al. Complete genome sequence of the ammonia-oxidizing bacterium and obligate chemolithoautotroph Nitrosomonas europaea[J]. Journal of Bacteriology, 2003, 185(9): 2759-2773 |
| [65] | Auguet JC, Triado-Margarit X, Nomokonova N, et al. Vertical segregation and phylogenetic characterization of ammonia-oxidizing Archaea in a deep oligotrophic lake[J]. ISME Journal, 2012, 6(9): 1786-1797 |
| [66] | Bollmann A, Bullerjahn GS, McKay RM. Abundance and diversity of ammonia-oxidizing archaea and bacteria in sediments of trophic end members of the Laurentian Great Lakes, Erie and Superior[J]. PLoS One, 2014, 9(5): e97068 |
| [67] | Jiang HC, Dong HL, Yu BS, et al. Diversity and abundance of ammonia-oxidizing archaea and bacteria in Qinghai Lake, northwestern China[J]. Geomicrobiology Journal, 2009, 26(3): 199-211 |
| [68] | Mosier AC, Francis CA. Relative abundance and diversity of ammonia-oxidizing archaea and bacteria in the San Francisco Bay estuary[J]. Environmental Microbiology, 2008, 10(11): 3002-3016 |
| [69] | Isobe K, Koba K, Suwa Y, et al. High abundance of ammonia-oxidizing archaea in acidified subtropical forest soils in southern China after long-term N deposition[J]. FEMS Microbiology Ecology, 2012, 80(1): 193-203 |
| [70] | Alves RJE, Wanek W, Zappe A, et al. Nitrification rates in Arcticsoils are associated with functionally distinct populations of ammonia-oxidizing archaea[J]. ISME Journal, 2013, 7(8): 1620-1631 |
| [71] | de Gannes V, Eudoxie G, Hickey WJ. Impacts of edaphic factors on communities of ammonia-oxidizing archaea, ammonia-oxidizing bacteria and nitrification in tropical soils[J]. PLoS One, 2014, 9(2): e89568 |
| [72] | Martens-Habbena W, Berube PM, Urakawa H, et al. Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria[J]. Nature, 2009, 461(7266): 976-979 |
| [73] | Bollmann A, Bar-Gilissen MJ, Laanbroek HJ. Growth at low ammonium concentrations and starvation response as potential factors involved in niche differentiation among ammonia-oxidizing bacteria[J]. Applied and Environmental Microbiology, 2002, 68(10): 4751-4757 |
| [74] | Könneke M, Schubert DM, Brown PC, et al. Ammonia-oxidizing archaea use the most energy-efficient aerobic pathway for CO2 fixation[J]. Proceedings of the National Academy of Sciences of the United States of America, 2014, 111(22): 8239-8244 |
| [75] | Sahan E, Muyzer G. Diversity and spatio-temporal distribution of ammonia-oxidizing archaea and bacteria in sediments of the Westerschelde estuary[J]. FEMS Microbiology Ecology, 2008, 64(2): 175-186 |
| [76] | Bernhard AE, Landry ZC, Blevins A, et al. Abundance of ammonia-oxidizing archaea and bacteria along an estuarine salinity gradient in relation to potential nitrification rates[J]. Applied and Environmental Microbiology, 2010, 76(4): 1285-1289 |
| [77] | Di HJ, Cameron KC, Shen JP, et al. Nitrification driven by bacteria and not archaea in nitrogen-rich grassland soils[J]. Nature Geoscience, 2009, 2(9): 621-624 |
| [78] | Prosser JI, Nicol GW. Relative contributions of archaea and bacteria to aerobic ammonia oxidation in the environment[J]. Environmental Microbiology, 2008, 10(11): 2931-2941 |
| [79] | Church MJ, Wai B, Karl DM, et al. Abundances of crenarchaeal amoA genes and transcripts in the Pacific Ocean[J]. Environmental Microbiology, 2010, 12(3): 679-688 |
| [80] | Tourna M, Freitag TE, Nicol GW, et al. Growth, activity and temperature responses of ammonia-oxidizing archaea and bacteria in soil microcosms[J]. Environmental Microbiology, 2008, 10(5): 1357-1364 |
| [81] | Jung MY, Well R, Min D, et al. Isotopic signatures of N2O produced by ammonia-oxidizing archaea from soils[J]. ISME Journal, 2014, 8(5): 1115-1125 |
| [82] | Di HJ, Cameron KC, Shen JP, et al. Ammonia-oxidizing bacteria and archaea grow under contrasting soil nitrogen conditions[J]. FEMS Microbiology Ecology, 2010, 72(3): 386-394 |
| [83] | Offre P, Prosser JI, Nicol GW. Growth of ammonia-oxidizing archaea in soil microcosms is inhibited by acetylene[J]. FEMS Microbiology Ecology, 2009, 70(1): 99-108 |
| [84] | Meyer A, Focks A, Radl V, et al. Influence of land use intensity on the diversity of ammonia oxidizing bacteria and archaea in soils from grassland ecosystems[J]. Microbial Ecology, 2014, 67(1): 161-166 |
 2015, Vol. 42
2015, Vol. 42




