扩展功能
文章信息
- 余芸, 张永红, 王龙
- YU Yun, ZHANG Yong-Hong, WANG Long
- 日耳曼曲霉(Aspergillus germanicus) 曲霉属焦曲霉组一个我国新记录种
- Aspergillus germanicus, a new Chinese record of Aspergillus section Usti
- 微生物学通报, 2015, 42(4): 674-682
- Microbiology China, 2015, 42(4): 674-682
- 10.13344/j.microbiol.china.140769
-
文章历史
- Received: October 10, 2014
- Accepted: November 17, 2014
- Published online(www.cnki.net): November 24, 2014
2. 北京农学院动物科技学院 北京 102206;
3. 中国科学院微生物研究所真菌学国家重点实验室 北京 100101
2. College of Animal Science and Technology, Beijing University of Agriculture, Beijing 102206, China;
3. State Key Laboratory of Mycology, Institute of Microbiology, Chinese Academy of Sciences, Beijing 100101, China
Aspergillus ustus Group was formerly established as one of the 18 Groups under the genus Aspergillus according to Raper and Fennell,which accommodated five species: A. conjunctus Kwon-Chung & Fennell,A. deflectus,A. panamensis Raper & Thom,A. puniceus,and A.ustus[1]. Later,Gams et al. established section Usti Gams et al. under Subgen. Nidulantes Gams et al. to include these five species[2]. But members of the A. nidulans Group share many characters with those of theAspergillus ustus Groupin bearing small vesicles,sinuous stipes,biseriate sterigmata,and globose echinulate conidia. Thus,Kozakiewicz transferred two species,A. granulosus and A.pseudodeflectus from A. versicolor Group to A.ustus Group,because they presented some similar morphological characters of Aspergillus ustus Group. While,A.deflectus wasmoved out of Aspergillus ustus Group and a new group,namely,A.deflectus Group was established to accommodate this species,in addition,A.pulvinus Kwon & Fennell and A. silvaticus Fennell & Raper transferred from section Versicolores and section Nidulantes,respectively,were also included in this Group[3].
The study of Klich indicated that A.pseudodeflectus was closely related to A. ustus and A. nidulans (Eidam) G. Winter,but supported the placement of A. granulosus in section Versicolores rather than section Usti[4]. By contrast,the study of Peterson based on D1 and D2 regions of the nucLSU rRNA gene showed that A. granulosus clustered with A. pseudodeflectus,A. puniceus and A. ustus,and A.deflectus was not closely related to these three species. His study also showed that most species in section Nidulantes (including A.heterothallicus),section Versicolores and section Usti clustered together with 75% bootstrap support. Then Peterson eliminated section Versicolores and section Usti and transferred most members of the two sections to section Nidulantes with the exception of A.conjuctus and A. panamensis,which were moved to section Sparsi[5]. Based on morphological,chemical and molecular data,Houbraken et al.revived A. insuetus (Bainier) Thom & Church which had been regarded as a synonym of A. ustus by Raper & Fennell,and included eight species in section Usti sensu stricto[1,6]. Peterson added seven additional species,extending this section to include fifteen species[7]. Varga et al. reported another new member,A. calidoustus in this section[8]. Samson et al. proposed five new taxa including A. germanicus to this section,and again expanded it to accommodate twenty-one taxa,but excluding A. ochraceoroseus and A. versicolor[9]. Recently,Nováková et al. reported two new members[10],and Wang described one new species in this section[11]. Until now,section Usti accommodated twenty-four species.
In the monograph of Qi et al.,only three species of section Usti of Aspergillus were reported in China,i.e.,A. deflectus,A. puniceus and P.ustus[12]. Whereas,little taxonomic work had carried out on this section since then,until Wang reported one additional member of this section from China,namely,A. keveioides[11]. In the present study,two strains,AS3.15303 and AS3.15304 of Aspergilus belonging to section Usti were isolated from soil samples from Mount Tai,Shangdong Province. Based on the morphological comparisons and molecular phylogenetic analyses,they were identified as A. germanicus,which is a new record of China and thus reported here,so there are five species of section Usti discovered in China.
2 Materials and Methods 2.1 Isolation and morphologySoil samples were collected underneath the leaf litter from the foot of Mount Tai in Shandong Province located in the monsoon area of moderate-temperate zone of China (36°15′17′′N,117°06′15′′E) on 12 July,2011. The average altitude of that area is 134 m with an atmospheric pressure of 1 004.1 hPa; the annual average temperature is 12.8 ℃,with the monthly average of -1.4 ℃ in January and 26.5 ℃ in July; the period of frozen soil is from late October to late March; the frost-free period is about 200 days from late March to late September; the annual precipitation is 700 mm. (http://www.weather. com.cn). Samples were kept in sterilized plastic bags. Dilution plates were used in the isolation of the fungi[13]. Dichloran rose bengal chlortetracycline (DRBC) agar were used as the selective medium[14]. Two strains,AS3.15303 and AS3.15304 of Aspergillus were obtained and deposited at the China General Microbiological Culture Collection Center (CGMCC) in the Institute of Microbiology,Chinese Academy of Sciences,Beijing.
Cultivation was conducted using the media Czapek yeast autolysate agar (CYA),Malt Extract Agar (MEA),CYA with 20% sucrose (CY20S),yeast extract sucrose agar (YES) and 25% glycerol nitrate agar (G25N) at 25 ℃ for 7 days. The growth on CYA at 5 ℃ and 37 ℃ was also assessed[1,9,15]. Colour names followed Ridgway[16]. Wet mounts for morphological examinations were prepared using culture material from colonies grown on CYA at 25 ℃ mounted in lactophenol without dye[1]. Optical microscopic examination and photographs were performed with an Olympus BH-2 Microscope (Olympus Co. Ltd.,Japan) and a Canon Digital EOS 7D camera (Canon Co. Ltd.,Japan).
2.2 Phylogenetic analysesTotal genomic DNA extraction followed the method of Scott et al.[17]. For amplification of partial β-tubulin gene (BenA),we employed the primers bt2a and bt2b described by Glass and Donaldson[18],while amplifying the rDNA ITS1-5.8S-ITS2,the primers ITS5 and ITS4 of White et al. were used[19]. To retrieve the partial calmodulin gene (CaM) sequence,the following primers were utilized,cmdAD1: 5′-GCC GACTCTTTGACTGAAGAGC-3′,cmdAD2: 5′-GCC GATTCTTTGACCGAGGAAC-3′ and cmdAD3: 5′-G CCGATTCTTTGACCGAAGAAC-3′ (sense primers); cmdQ1: 5′-GCATCATGAGCTGGACGAACTC-3′ and cmdQ2: 5′-gcatcatgagctggacgaattc-3′ (antisense primers),in which there were six combinations,but the cmdAD1 & Q2 and cmdAD2 & Q1 were used first[20]. Polymerase chain reaction (PCR) was carried out in 20 μL reaction system: 2× EcoTaq PCR Super Mix (+dye) 8.0 μL; genomic DNA 1.0 μL; sense primer (10 μmol/L) 0.5 μL; antisense primer (10 μmol/L) 0.5 μL; ddH2O 10.0 μL (Beijing TransGen Biotech.). DNA amplification was performed in PTC-150 thermocycler (MJ Research). The thermal cycle protocol consisted of 94 ℃ for 3 min; 94 ℃ for 30 sec,50 ℃ for 30 sec,72 ℃ for 45 sec,34 cycles; 72 ℃ for 5 min; 15 ℃ for 15 min. PCR products were electrophoresed in 2.0% agarose gel with a 100 bp DNA ladder (MBI Fermentas) at 80 V for 15 min. The gel was stained in 0.5 mg/L ethidium bromide buffer for 15 min and then viewed under 254 nm UV light. The products with good results were purified and sequenced in double directions by ABI 3700 DNA analyzer (Newtsingke BioTech.). Six gene sequences were obtained and deposited at GenBank as JQ814950 to JQ814955 (Table1).
| Species 物种 | Strainsa 菌株 | GenBank accession numbers GenBank登录号 | ||
| ITS1-5.8S-ITS2 | BenA | CaM | ||
| Aspergillus amylovorus Panas. ex Samson | NRRL 5813 T | EF652503 | EF652327 | EF652415 |
| A. calidoustus Varga,Houbraken & Samson | CBS 114380 | EF591741 | EF591729 | EF591716 |
| CBS 113228 | EF591739 | EF591730 | EF591715 | |
| NRRL 26162 | EF652452 | EF652276 | EF652364 | |
| AS 3.15302 | JN982696 | JN982686 | JN982676 | |
| A. deflectus Fennell & Raper | NRRL 2206 T | EF652437 | EF652261 | EF652349 |
| A. egyptiacus Moub. & Mustafa | NRRL 5920 T | EF652504 | EF652328 | EF652416 |
| A. elongatus J. N. Rai & S. C. Agarwa | NRRL 5176 T | EF652502 | EF652326 | EF652414 |
| A. germanicus Varga,Frisvad & Samson | CBS 123887 T | FJ531146 | FJ531172 | FJ531141 |
| AS3.15303 | JQ814954 | JQ814952 | JQ814950 | |
| AS3.15304 | JQ814955 | JQ814953 | JQ814951 | |
| A. granulosus Raper & Thom | NRRL 1932 T | EF652430 | EF652254 | EF652342 |
| A. heterothallicus Kwon-Chung,Fennell & Raper | NRRL 5096 T | EF652499 | EF652323 | EF652411 |
| NRRL 5097 | EF652500 | EF652324 | EF652412 | |
| AS 3.15313 | JN982698 | JN982688 | JN982678 | |
| A. insuetus (Bainier) Thom & Church | CBS 107.25 T | EU076356 | EU076371 | EU076366 |
| CBS 119.27 | EU076355 | EU076372 | EU076367 | |
| NRRL 4876 | EF652481 | EF652305 | EF652393 | |
| NRRL 279 | EF652457 | EF652281 | EF652369 | |
| A. kassunensis Baghd. | NRRL 3752 T | EF652461 | EF652285 | EF652373 |
| A. keveii Varga,Frisvad & Samson | NRRL 1974 | EF652432 | EF652256 | EF652344 |
| CBS 561.65 | EU076352 | EU076375 | EU076364 | |
| CBS 209.92 T | EU076354 | EU076376 | EU076365 | |
| A. keveioides L. Wang | AS 3.15305 T | JN982704 | JN982694 | JN982684 |
| A. lucknowensis J.N. Rai,J.P. Tewari & S.C. Agarwal | NRRL 3491 T | EF652459 | EF652283 | EF652371 |
| A. ochraceoroseus Bartoli & Maggi | NRRL 28622 T | EF661224 | EF661113 | EF661137 |
| A. pseudodeflectus Samson & Mouch. | NRRL 6135 T | EF652507 | EF652331 | EF652419 |
| NRRL 278 | EF652456 | EF652280 | EF652368 | |
| AS3.15306 | JN982697 | JN982687 | JN982677 | |
| AS3.15307 | JN982700 | JN982690 | JN982680 | |
| AS3.15308 | JN982699 | JN982689 | JN982679 | |
| AS3.15309 | JN982701 | JN982691 | JN982681 | |
| AS3.15310 | JN982703 | JN982693 | JN982683 | |
| A. puniceus Kwon-Chung & Fennell | NRRL 5077 T | EF652498 | EF652322 | EF652410 |
| A. subsessilis Raper & Fennell | NRRL 4095 T | EF652485 | EF652309 | EF652397 |
| A. ustus (Bainier) Thom & Church | NRRL 275 T | EF652455 | EF652279 | EF652367 |
| NRRL 4991 | EF652492 | EF652316 | EF652404 | |
| AS3.15311 | JN982695 | JN982685 | JN982675 | |
| AS3.15312 | JN982702 | JN982692 | JN982682 | |
| A. versicolor (Vuill.) Tirab. | NRRL 238 T | EF652442 | EF652266 | EF652354 |
注:a:模式菌株标记有“T”.
Raw sequences were proof-read and edited manually with BioEdit 7.0.9[21]. Edited sequences were aligned using MUSCLE implemented in MEGA version 5[22]. Thirty-nine strains of eighteen species in section Usti (Table1) were analyzed with A. versicolor as the outgroup using the Maximum-Likelihood (ML) and Neighbor-Joining (NJ) methods,respectively,with Kimura-2 model and subjected to 1 000 bootstrap replicates.
3 Results 3.1 Description of Aspergillus germanicus (Figure1)On CYA at 25 ℃ after 7 days: Colonies attaining 30-32 mm in diam.,low,plane,umbonate in central areas; velutinous; conidiogenesis sparse,distributed in central areas,coloured Smoke Gray to Drab Gray (R. Pl. XLVI); mycelia white; no exudates; soluble pigment moderate,Pale Green-Yellow (R. Pl. V),reverse Green-Yellow.
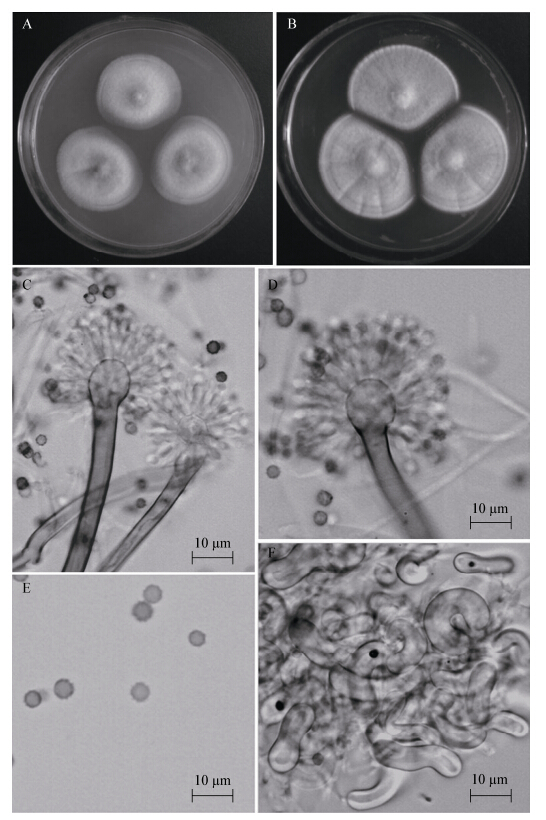
|
|
Fig.1
Morphology of A. germanicus AS3.15303
图1
日耳曼曲霉AS3.15303的形态
Note: A-B: Colonies on CYA and YES at 25 ℃ after 7 days; C-D: Conidiophores on CYA; E: Conidia on CYA; F: Hülle cells on MEA. 注:A-B:在培养基CYA和YES上于25 ℃培养7 d的菌落;C-D:在CYA上的分生孢子梗;E:在CYA上的分生孢子;F:在MEA上的壳细胞. |
On Malt Extract Agar (MEA) at 25 ℃ after 7 days: Colonies 36-38 mm in diam.,floccose; conidiogenesis sparse,distributed in centra areas,near Smoke Gray to Drab Gray (R. Pl. XLVI); mycelia white,hülle cells sparse,aggregated into conspicuous,small white masses; no exudate and soluble pigment; reverse Baryta Yellow (R. Pl. IV).
On yeast extract sucrose agar (YES) at 25 ℃ after 7 days: Colonies 38-40 mm in diam.,low,radially and irregularly sulcate; velutinous with slightly floccose in centers; conidiogenesis moderate,Smoke Gray to Drab Gray (R. Pl. XLVI); mycelia white with Pale Viridine Yellow tint (R. Pl. V); no exudates; soluble pigment light,Pale Green-Yellow (R. Pl. V); reverse Green-Yellow.
Colonies on CYA with 20% sucrose (CY20S) at 25 ℃ 7 days: Colonies reaching 33-35 mm in diam.,low,plane,velvety with floccose in centers; condiogenesis sparse,Smoke Grey to Drab Gray (R. Pl. XLVI); mycelia white; exudate and soluble pigment absent; reverse Green-Yellow.
On 25% glycerol nitrate agar (G25N) at 25 ℃ in 7 days: Colonies 21-25 mm,low,plane; velutinous; conidiogenesis sparse,Pale Vinaceous-Fawn (R. Pl. XL); no exudate and pigment; reverse light yellow.
On CYA at 37 ℃ in 7 days: Colonies 5-7 mm.
On CYA at 5 ℃ in 7 days: No growth.
Conidial heads globose,(80-)120-150 μm; conidiophores arising from substratum and surface hyphae,stipes brown-coloured,heavy-walled,180-240(-360)×(5-8) μm; vesicles ellipsoidal to spathulate,light-brown,thin-walled,(13-16)×(9-11) μm,fertile over the most parts of vesicles; biseriate,metulae (3.5-5.0)×(2-3) μm; phialides ampuliform,5.5-7.0 μm,with short collula; conidia globose 3-4 μm,echinulate,brown-coloured in mass; hülle cells irregularly elongate,thick-walled,commonly (10-)20-30(-40)×(5-8) μm.
Isolates examined: AS 3.15303 and AS 3.15304.
Sustratum: Soil. China,Shandong Province,foot of Mount Tai,from soil,12 July 2011,collected by Yong-Hong Zhang.
3.2 PCR amplicons and Phylogenies (Figure 2-3)The PCR amplification of CaM,BenA and ITS1-5.8S-ITS2 regions yielded ca. 700,400 and 600 bp replicons,respectively. The primers for CaM generated nearly the full length ofCaM gene sequence,namely,from the 2nd nucleotide of the codon for the 9th amino acid Gln (Q) to the 3rd nucleotide of the codon for the 140th amino acid Asn (N),and the trimmed alignments of the three gene sequences were respectively 501,462 and 558 characters with gaps. Figures 2-3 are the ML phylograms based on partial CaM andBenA,respectively. The NJ trees resulted from the three genes are not shown.

|
|
Fig.2
The ML phylogram yielded from partial CaM sequence data set
图2
基于钙调蛋白基因部分序列用最大似然法推导出的系统发育树
Note: The bootstrap percentages over 70% derived from 1 000 replicates are indicated at the nodes. Bar=0.05 substitutions per nucleotide position. Ex-type strains are marked with “T”. GenBank accession numbers are in parentheses. Aspergillus germanicus is in bold-face type. 注:用自展法进行1 000次重复取样得到的各分支自展支持率大于等于70%的标注在分支节点处;标尺=0.05个替代每核苷酸位点;模式菌株标记有“T”;GenBank登录号在圆括号中;日耳曼曲霉用粗体标出. |
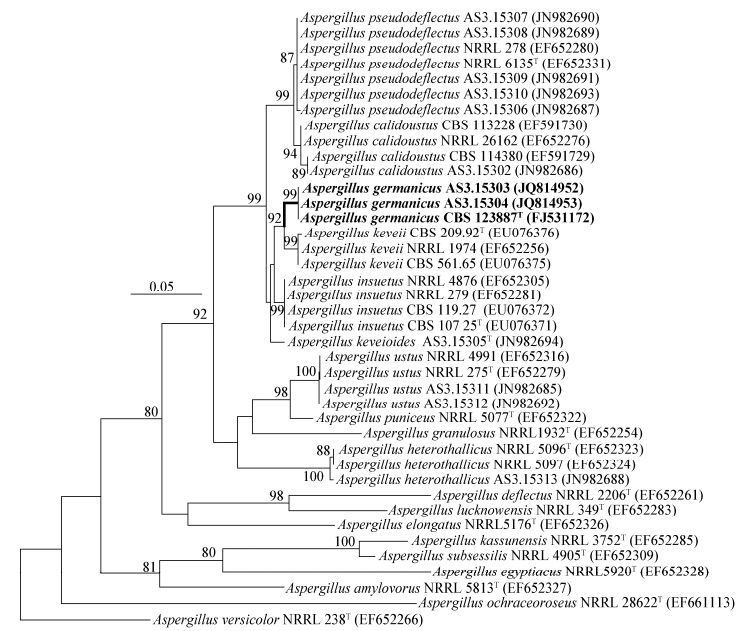
|
|
Fig.3
The ML phylogram yielded from partial BenA sequence data set
图3
基于β-微管蛋白基因部分序列用最大似然法推导出的系统发育树
Note: The bootstrap percentages over 70% derived from 1 000 replicates are indicated at the nodes. Bar=0.05 substitutions per nucleotide position. Ex-type strains are marked with “T”. GenBank accession numbers are in parentheses. 注:用自展法进行1 000次重复取样得到的各分支自展支持率大于等于70%的标注在分支节点处;标尺=0.05个替代每核苷酸位点;模式菌株标记有“T”;GenBank登录号在圆括号中. |
In the phylogenetic trees inferred from ITS1-5.8S-ITS2 data,A. germanicus and A. insuetus and A. keveioides were not discriminated using both ML and NJ methods (not shown). Whereas,the trees yielded from partial CaM and BenA data indicated that the two Chinese isolates with the ex-type of A. germanicus CBS 123887 formed one single clade with 99%-100% bootstrap support according to both ML and NJ methods. Moreover,the ML and NJ phylogenetic trees resulted from CaM sequences also showed that A. germanicus was related to A. insuetus,A. keveii,A.keveioides (e. g.,Figure2 of the ML tree),but the ML and NJ trees generated by BenA sequences both indicated that A. germanicus was most related to A. keveii (e. g.,Figure3 of the ML tree).
4 DiscussionWe constructed phylograms respectively based on the individual gene instead of using the combined sequences of the three genes,because according to the concept of Genealogical Concordance Phylogenetic Species (GCPSR),when different gene trees are concordant in topology,the concordant branches represent species[23]. In this study,the two phylograms inferred from CaM and BenA both indicated that our two isolates together with the ex-type of A.germanicus formed one single clade with 100% and 99% bootstrap supports,respectively,which confirmed the correct identification of our two isolates (Figures 1-2). Although the phylogram resulted from ITS1-5.8S-ITS2 could not distinguish A. germanicus from A. insuetus and A.keveioides,yet the branch containing the above three species and A.keveii had no bootstrap support,which meant that this branch could be polyphyletic. Samson et al. argued the less variation of ITS1-5.8S-ITS2 in the phylogenetic analyses of aspergilli,they also discussed CaM,BenA and RPB2 genes,and recommended CaM as the supplementary genetic marker in Aspergillus phylogenetics[24].
Samson et al. proposed the species A. germanicus based on only one isolate,namely,the ex-type CBS 123887[9]. The poor sporulation,yellow reverse,spathulate vesicles,smooth-walled conidiophores,and globose echinulate conidia are the striking similar characters as those of our two isolates,though there are some subtle differences between the ex-type and our isolates. For example,the ex-type grows more slowly on CYA (about 22-26 mm in diam. after 7 days at 25 ℃) than ours (about 30-32 mm in diam. after 7 days at 25 ℃),and the vesicles of the ex-type are smaller which are about 14-22 mm in diam. than those of our isolates which are about 30-32 mm in diam. But these differences only indicate the variation among different strains,the above morphological identity and the molecular evidence (Figure 2-3) both verified the identification of our two isolates as A. germanicus.
| [1] | Raper KB,Fennell DI.The Genus Aspergillus[M].Baltimore:Williams&Wilkins,1965:1-686 |
| [2] | Gams W,Christensen M,Onions AH,et al.Infrageneric taxa of Aspergillus[A]//Samson RA,Pitt JI.Advances in Penicillium and Aspergillus Sytematics[M].New York:Plenum Press,1985:55-61 |
| [3] | Kozakiewicz Z.Aspergillus species in stored products[J].Mycological Papers,1989,161:1-188 |
| [4] | Klich MA.Morphological studies of Aspergillus section Versicolores and related species[J].Mycologia,1993,85(1):100-107 |
| [5] | Peterson SW.Phylogenetic relationships in Aspergillus based on rDNA sequence analysis[A]//Samson RA,Pitt JI.Integration of Modern Taxonomic Methods for Penicillium and Aspergillus Classification[M].Amsterdam:Harwood Academic Publishers,2000:323-355 |
| [6] | Houbraken J,Due M,Warga J,et al.Polyphasic taxonomy of Aspergillus section Usti[J].Studies in Mycology,2007,59:107-128 |
| [7] | Peterson SW.Phylogenetic analysis of Aspergillus species using DNA sequences from four loci[J].Mycologia,2008,100(2):205-226 |
| [8] | Varga J,Houbraken J,van der Lee HAL,et al.Aspergillus calidoustus sp.nov.,causative agent of human infections previously assigned to Aspergillus ustus[J].Eukaryotic Cell,2008,7(4):630-638 |
| [9] | Samson RA,Varga J,Meijer M,et al.New taxa in Aspergillus section Usti[J].Studies in Mycology,2011,69:81-97 |
| [10] | Nováková A,Hubka V,Saiz-Jimenez C,et al.Aspergillus baeticus sp.nov.and Aspergillus thesauricus sp.nov.,two species in section Usti from Spanish caves[J].International Journal of Systematic and Evolutionary Microbiology,2012,62(Pt 11):2778-2785 |
| [11] | Wang L.Aspergillus keveioides,a new species of Aspergillus sect.Usti from Shandong Province,China[J].Mycosystema,2013,32(Suppl.):136-144 |
| [12] | Qi ZT,Kong HZ,Sun ZM.Flora Fungorum Sinicorum Vol.5:Aspergillus et Teleomorphi Cognati[M].Beijing:Science Press,1997:55-59(in Chinese) |
| [13] | Malloch D.Moulds Their Isolation,Cultivation and Identification[M].Toronto:University of Toronto Press,1981:1-97 |
| [14] | King DA,Hocking AD,Pitt JI.Dichloran-rose bengal medium for enumeration and isolation of molds from foods[J]Journal of Applied and Environmental Microbiology,1997,37(5):959-964 |
| [15] | Pitt JI,Hocking AD.Fungi and Food Spoilage[M].New York:Springer Science+Business Media,2009:41-44 |
| [16] | Ridgway R.Color Standards and Color Nomenclature[M].Washington DC:Published by the Author,1912:1-53 |
| [17] | Scott J,Malloch D,Wong B,et al.DNA heteroduplex fingerprinting in Penicillium[A]//Samson RA,Pitt JI.Integration of Modern Taxonomic Methods for Penicillium and Aspergillus Classification[M].Amsterdam:Harwood Academic Publishers,2000:225-236 |
| [18] | Glass NL,Donaldson GC.Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous Ascomycetes[J].Applied and Environmental Microbiology,1995,61(4):1323-1330 |
| [19] | White TJ,Bruns T,Lee S,et al.Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics[A]//Innis MA,Gelfand GH.PCR Protocols:a Guide to Methods and Applications[M].San Diego:Academic Press,1990:315-322 |
| [20] | Wang B,Wang L.Penicillium kongii,a new terverticillate species isolated from plant leaves in China[J].Mycologia,2013,105(6):1547-1554 |
| [21] | Hall TA.Bioedit:a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT[J].Nucleic Acids Symposium Series,1999,41:95-98 |
| [22] | Tamura K,Peterson D,Peterson N,et al.MEGA5:molecular evolutionary genetics analysis using maximum likelihood,evolutionary distance,and maximum parsimony methods[J].Molecular Biology and Evolution,2011,28(10):2731-2739 |
| [23] | Taylor JW,Jacobson DJ,Kroken S,et al.Phylogenetic species recognition and species concepts in fungi[J].Fungal Genetics and Biology,2000,31:21-32 |
| [24] | Samson RA,Visagie CM,Houbraken J,et al.Phylogeny,identification and nomenclature of the genus Aspergillus[J].Studies in Mycology,2014,78:141-173 |
 2015, Vol. 42
2015, Vol. 42




