扩展功能
文章信息
- 徐宁, 程海娇, 刘清岱, 刘君, 马延和
- XU Ning, CHENG Hai-Jiao, LIU Qing-Dai, LIU Jun, MA Yan-He
- 细菌Na+/H+逆向转运蛋白的研究进展
- Research progress of the Na+/H+ antiporters in bacteria
- 微生物学通报, 2015, 42(10): 2002-2011
- Microbiology China, 2015, 42(10): 2002-2011
- 10.13344/j.microbiol.china.141020
-
文章历史
- 收稿日期: 2014-12-18
- 接受日期: 2015-02-03
- 优先数字出版日期(www.cnki.net): 2015-03-16
2. 天津科技大学 食品工程与生物技术学院 天津 300457
2. College of Food Engineering and Biotechnology, Tianjin University of Science and Technology, Tianjin 300457, China
金属离子作为一种重要的无机营养元素,无论在维持普通微生物还是病原微生物生理状态和正常机能方面都发挥着不可或缺的作用。笔者前期研究曾证实,胞内铁离子和钙离子稳态是白假丝酵母等病原性微生物在宿主环境下正常存活和致病的关键因素,离子稳态的破坏会显著降低菌株的生长和致病能力[1, 2, 3, 4]。微生物与外界环境在相互作用过程中,除可能遭遇铁和钙离子等环境因素外,还经常需要应答高浓度的钠盐压力。尽管钠离子(Na+)对于微生物的生长和代谢具有重要作用,但是外界环境中Na+离子浓度过高,也会破坏细胞内的渗透压平衡,从而对细胞产生毒害作用。微生物在长期进化过程中,逐渐形成了适应高盐环境的多种生理机制:一方面,依赖于一种被称为相容性溶质机制的拒盐策略,在细胞内积累糖类、醇类、氨基酸及其衍生物等小分子极性物质,用以调节细胞内外渗透压平衡,起着渗透保护作用[5]。另一方面,利用Na+离子外排系统,将细胞内多余的Na+离子泵出至胞外,以维持胞内适宜的Na+离子,这一过程主要由位于细胞膜上的Na+/H+逆向转运蛋白(Na+/H+ antiporters)介导[6]。此外,一些耐盐微生物还具有特殊的细胞壁结构,富含酸性氨基酸,易形成负电荷区域,吸引正电荷的Na+离子,维持细胞壁的稳定性[7]。
Na+/H+逆向转运蛋白又称为Na+/H+泵,是一种膜蛋白,广泛存在于细菌、植物、动物体中,在维持细胞内pH稳态、Na+离子动态平衡和调控细胞体积等方面发挥着重要作用[6, 8, 9, 10]。同真核生物Na+/H+逆向转运蛋白功能相似,原核生物Na+/H+逆向转运蛋白也能够以跨膜的质子电化学梯度为动力,催化胞内Na+离子的泵出过程,建立跨膜的Na+离子电化学梯度,促进Na+离子梯度偶联的一系列生理生化过程,如Na+/溶质的协同转运过程和Na+离子驱动的鞭毛运动,最终实现Na+离子的循环利用[8]。本文主要关注细菌Na+/H+逆向转运蛋白,以革兰氏阴性菌大肠杆菌(Escherichia coli)、霍乱弧菌(Vibrio cholerae)和革兰氏阳性菌枯草芽孢杆菌(Bacillus subtilis)、拟坚强芽孢杆菌(Bacillus pseudofirmus)为例(图 1),从细菌Na+/H+逆向转运蛋白的种类、结构、机理和应用等方面进行综述。
1 Na+/H+逆向转运蛋白的类型和特征Na+/H+逆向转运蛋白作为细菌主要的Na+外排系统,也是细菌中对Na+胁迫的主要适应性应答系统。在长期的进化过程中,微生物形成了多种类型的Na+/H+逆向转运蛋白,依据其蛋白质结构特点,主要分为单亚基型和多亚基型两大类,其中绝大多数属于单亚基型。目前,已经报道多个Na+/H+逆向转运蛋白成员,包括Cpa1家族、Cpa2家族、Cpa3家族、Mfs家族、NhaA家族和NhaB、NhaC、NhaD、CaCA等[8, 11, 12]。

|
|
图 1
4种模式菌株Na+/H+逆向转运蛋白家族示意图
Figure 1
Schematic diagrams of the Na+/H+ antiporter family in four physiologically distinct bacteria
注:A:革兰氏阴性嗜中性肠道菌大肠杆菌;B:革兰氏阴性嗜中性致病菌霍乱弧菌;C:革兰氏阳性嗜中性菌枯草芽孢杆菌;D:革兰氏阳性嗜碱菌拟坚强芽孢杆菌. Note:A: E. coli, a Gram-negative neutrophilic enteric bacilli; B: V. cholera, a Gram-negative neutrophilic pathogen; C: B. subtilis, a Gram-positive neutrophilic organism; D: B. pseudofirmus, a Gram-positive alkaliphilic bacterium. |
阳离子:质子逆向转运蛋白-1家族(The cation:proton antiporter-1 family,Cpa1 family)广泛分布于革兰氏阳性菌、革兰氏阴性菌、真菌、植物和动物等膜系统上,具有Na+/H+逆向转运蛋白活性,主要参与调控胞内pH稳态[13]。在动物和高等植物中,Cpa1家族成员得到了较为详细的研究,但是对细菌Cpa1家族成员的认识相对较少。YjcE和CvrA (YcgO)是大肠杆菌中鉴定出的两个Cpa1家族成员[14]。研究发现,YjcE和CvrA在呼吸作用驱动的Na+离子输出过程中作用较小,基因的缺失对菌株在50−500 mmol/L Na+离子浓度下的生长速率并没有显著影响。CvrA蛋白能够参与调控细胞体积的变化,对低渗条件下菌株生长是必需的。有趣的是,在枯草芽孢杆菌ATCC 9372中发现了Cpa1家族新成员NhaG,该蛋白具有Na+/H+逆向转运活性,但是未能在枯草芽孢杆菌168中鉴定出其同源物,表明不同来源的同一菌株可能存在进化差异[15]。NhaK (YvgP)是枯草芽孢杆菌168中唯一鉴定出的Cpa1家族成员,该蛋白同时具有Na+ (K+,Li+,Rb+)/H+逆向转运活性,Mg2+、Ca2+或Mn2+离子存在会抑制其活性[16]。NhaP是铜绿假单胞菌(Pseudomonas aeruginosa)中发现的一种Cpa1家族Na+/H+逆向转运蛋白,是菌株Na+离子输出系统的主要组分[17]。霍乱弧菌中也鉴定出两种NhaP型Na+/H+逆向转运蛋白,其中NhaP1蛋白在较低的环境pH条件下对于维持菌株的生长和胞内pH稳态是必需的,而NhaP2蛋白对K+离子具有更高的亲和性[18]。通过全基因组序列分析,我们发现拟坚强芽孢杆菌中也含有两个潜在的NhaP型Na+/H+逆向转运蛋白(NhaP和NhaP1),推测可能参与离子转运和细胞体积调控过程,其生物学功能尚有待进一步阐释[19]。
1.2 Cpa2 家族阳离子:质子逆向转运蛋白-2家族(The cation: proton antiporter-2 family,Cpa2 family)主要存在于细菌和古细菌等原核生物中,较少出现在真核生物膜系统中[13]。KefB和KefC是大肠杆菌中研究较为详细的Cpa2家族成员,是一种谷胱甘肽门控的K+离子输出蛋白,在胞质酸化和保护细胞抵御活性亲电物质方面具有重要作用[20]。NapA是海氏肠杆菌(Enterococcushirae)中鉴定出的一种主要的Cpa2家族逆向转运蛋白,该蛋白的缺失使菌株丧失Na+/H+逆向转运活性,无法在高盐环境下生长[21]。GerN是蜡样芽胞杆菌(Bacillus cereus)中发现的NapA型同源物,同时具有Na+/H+和Na+/H+-K+逆向转运活性[22]。全基因组序列分析揭示,拟坚强芽孢杆菌中同样含有潜在的NapA型Na+/H+逆向转运蛋白,但是其功能尚不清楚[19]。
1.3 Cpa3家族阳离子:质子逆向转运蛋白-3家族(The cation:proton antiporter-3 family,Cpa3 family),广泛存在于具有不同生理学功能的原核生物中[8, 11]。Mrp系统(Multiple resistance and pH-related antiporter)作为一种单价阳离子:质子逆向转运蛋白,是跨膜转运蛋白Cpa-3家族的最重要成员[23]。该系统广泛存在于细菌和古细菌等原核生物中,早期由于命名系统的紊乱,又被赋予了多种别称,如Mnh、Pha、Sha和Sno等。Mrp家族能够依靠呼吸作用或者ATP酶水解形成的跨膜质子电化学梯度为动力,催化胞质内单价阳离子(如Na+、K+、Li+等)输出,同时偶联细胞膜外H+质子输入过程。
典型的Mrp系统是一种多亚基型转运蛋白复合物,由同一操纵子的6或7个高度保守基因(mrpA−mrpG)编码,每个亚基对激活复合体的完整功能都是必需的[23, 24]。根据mrp操纵子中mrpA和mrpB基因的排列方式,可分为两类(Group I和Group II)。第一类中,mrp操纵子由7个基因构成,含有单独mrpA和mrpB基因,并且mrpA基因中含有mrpB结构域,在革兰氏阳性菌、革兰氏阴性菌及古细菌中均有发现。第二类中,mrp操纵子由6个基因构成,不含有单独的mrpB基因,但是mrpA基因中含有两个mrpB结构域,主要存在于革兰氏阳性菌中。Mrp逆向转运蛋白首次发现于嗜碱菌Bacillus halodurans C-125,由7个亚基组成,具有Na+/H+逆向转运活性,在维持胞内pH稳态方面发挥着至关重要的作用[25]。在嗜中性枯草芽孢杆菌中,Mrp蛋白具有很高的Na+-Li+/H+逆向转运活性,在维持细胞对Na+离子和胆酸盐抗性方面发挥着核心作用,但是在Na+和K+依赖性pH稳态调控过程中作用较小[26]。然而,嗜碱拟坚强芽孢杆菌Mrp蛋白无论在Na+离子胁迫应答还是胞内pH稳态调控方面都扮演着极其重要的角色[8, 27]。
大多数微生物中只含有一种Mrp系统,但是一些菌株中含有多个Mrp操作子。例如,中华根瘤菌中含有两套Mrp系统(Pha1和Pha2),Pha1蛋白的缺失会导致菌株固氮能力缺陷和K+离子敏感性,而Pha2蛋白缺失导致菌株Na+离子敏感性[28]。基因组序列分析揭示,人类病原菌金黄色葡萄球菌(Staphylococcus aureus)也含有两套Mrp系统(Mnh1和Mnh2),均由7个亚基构成,推测可能与其耐受高盐环境有关[29]。
1.4 Mfs 家族细菌能够通过耐药泵将药物主动泵出胞外,可以有效地抵御抗生素及其他药物的毒性作用。主要协助蛋白转运超家族(Major facilitator superfamily,Mfs family)是由12或14个跨膜螺旋区组成的转运蛋白,通过质子交换提供向细胞外泵出药物所需的能量,作用底物包括氨基糖苷类、四环素类和氯霉素类等药物。MdfA是大肠杆菌中鉴定出的一种Drug-Na+-K+/H+逆向转运蛋白,能够将胞内药物泵出,在菌株抵御药物压力方面具有重要作用[30]。枯草芽孢杆菌中TetB是一种Tetracycline-Na+-K+/H+逆向转运蛋白,能够以跨膜的质子电化学梯度为动力,催化胞内四环素、Na+离子和K+离子输出过程,在Na+和K+依赖性pH稳态调控过程中发挥着核心作用[8]。
1.5 NhaA 家族大肠杆菌NhaA是微生物界发现的第一种Na+/H+逆向转运蛋白,也是目前研究最为详细的Na+/H+逆向转运蛋白[31, 32, 33, 34]。NhaA是大肠杆菌和许多肠道细菌的最主要Na+/H+逆向转运蛋白,在维持胞内pH稳态方面发挥着至关重要的作用,该蛋白缺失会导致菌株在高盐环境压力下的生长缺陷[34, 35]。NhaA蛋白活性具有pH依赖性,能够感应并传导外界环境pH信号,只有当胞内pH值达到6.5时,才会激活NhaA蛋白活性。研究发现,nhaA基因含有P1和P2两个启动子,LysR调控因子NhaR能够以Na+离子依赖性方式直接结合P1启动子,从而参与激活nhaA基因的表达,而P2启动子在对数期有较低组成性表达,不受Na+离子和NhaR影响[35, 36]。此外,NhaA家族Na+/H+逆向转运蛋白在许多致病微生物中也有发现,如霍乱弧菌、破伤风杆菌、幽门螺杆菌、伤寒沙门氏菌等[12, 37, 38]。
1.6 其他家族NhaB是大肠杆菌中发现的第2种Na+/H+逆向转运蛋白,由504个氨基酸残基构成,也能形成12个跨膜区域[39, 40]。NhaB主要在较低Na+离子浓度和较低pH值环境下发挥作用,能够弥补NhaA在此条件下活性较低的不足,与NhaA蛋白协同参与环境压力应答过程。ChaA是大肠杆菌中鉴定的第3种Na+/H+逆向转运蛋白。ChaA是Ca2+/H+逆向转运蛋白家族(the calcium cation antiporter family,CaCA family)的重要成员,主要在碱性条件下参与离子运输,既能转运Ca2+离子也能转运Na+离子[41]。研究发现,当NhaA、NhaB和ChaA蛋白同时缺失时,大肠杆菌在0.2 mol/L NaCl条件下基本丧失存活能力[42]。因此,常以大肠杆菌Na+/H+逆向转运蛋白基因缺失菌株E. coli KNabc为异源宿主系统,通过功能互补策略,筛选和鉴定新型的Na+/H+逆向转运蛋白[43, 44]。
枯草芽孢杆菌含有2个NhaC型Na+/H+逆向转运蛋白,分别命名为NhaC (YheL)和MleN (YqkI)[45, 46]。枯草芽孢杆菌中NhaC在维持Na+离子依赖性的pH稳态方面作用有限,并且不参与高盐耐受性应答。MleN蛋白不仅具有Na+/H+逆向转运活性,还能偶联苹果酸-乳酸逆向转运能力,增强菌株在较低质子动力势时转运苹果酸的能力。拟坚强芽孢杆菌是一种极端兼性嗜碱菌,具有广泛的pH适应性,能够在胞外pH 7.5−11.4的环境条件下生长[19]。拟坚强芽孢杆菌NhaC5是从嗜碱菌中鉴定出的第一个Na+/H+逆向转运蛋白,由462个氨基酸残基组成[47]。该蛋白对Na+离子具有高亲和性,主要参与低pH条件下Na+离子的输出,对于菌株的嗜碱特性并不是必需的。此外,基因组序列分析表明,拟坚强芽孢杆菌还含有另外5种NhaC家族成员(NhaC1−4和NhaC6),对其功能的研究将会进一步揭示拟坚强芽孢杆菌的嗜碱特性[19]。
NhaD型Na+/H+逆向转运蛋白广泛地存在于细菌中,但是对其详细的生理学功能尚不清楚。科学家最早在弧菌属中鉴定出NhaD转运蛋白,发现其能够补救E. coli KNabc缺失菌株在10 mmol/L LiCl或0.2 mmol/L NaCl条件下生长缺陷,并且蛋白活性具有pH依赖性,在碱性条件下具有最大活性[48]。通过文库筛选,我们从专性嗜碱微生物Alkalimonas amylolytica N10中分离鉴定出NhaD同源物,该蛋白在碱性条件下才具有Na+-Li+/H+逆向转运活性[49]。进一步研究发现,该菌中另外3种逆向转运蛋白也能够参与碱性pH耐受和高Na+抗性应答过程,如Cpa1家族Aa-NhaP、Cpa2家族Aa-KefB和CaCA家族Aa-CaxA[50]。
霍乱弧菌作为引起霍乱病症的主要病原菌,常发现于水生环境中,能在较宽范围pH和盐度条件下存活。研究发现,霍乱弧菌包括至少3种Nha家族逆向转运蛋白,即NhaA、NhaB和NhaD[8]。NhaA和NhaB蛋白在霍乱弧菌应答高盐环境压力过程中发挥着作用,其中NhaA蛋白和大肠杆菌NhaA蛋白具有相似的活性特征。有趣的是,当NhaA、NhaB和NhaD蛋白同时缺失时,并未显著影响霍乱弧菌在高盐压力下的生长能力,提示存在其他的未鉴定Na+/H+逆向转运蛋白或Na+离子外排机制[51, 52]。基因组序列分析也证实,霍乱弧菌还存在其他Na+/H+逆向转运蛋白,如Mrp家族和NhaC家族等。因此,对这些不同类型Na+/H+逆向转运蛋白相互协同关系的阐释将会进一步提高我们对霍乱弧菌耐压机制的理解。值得注意的是,随着全基因组测序技术和分子生物学技术的发展,除了上文描述的几种常见Na+/H+逆向转运蛋白家族外,微生物中也报道了许多其他类型Na+/H+逆向转运蛋白,如NhaE等[53]。随着基因组学研究的不断深入,同种微生物中发现了多个推测的不同家族Na+/H+逆向转运蛋白,但是其功能分工尚不清楚。因此,对Na+/H+逆向转运蛋白分类学和活性研究将进一步加深我们对该类超家族成员的认识(表 1)。
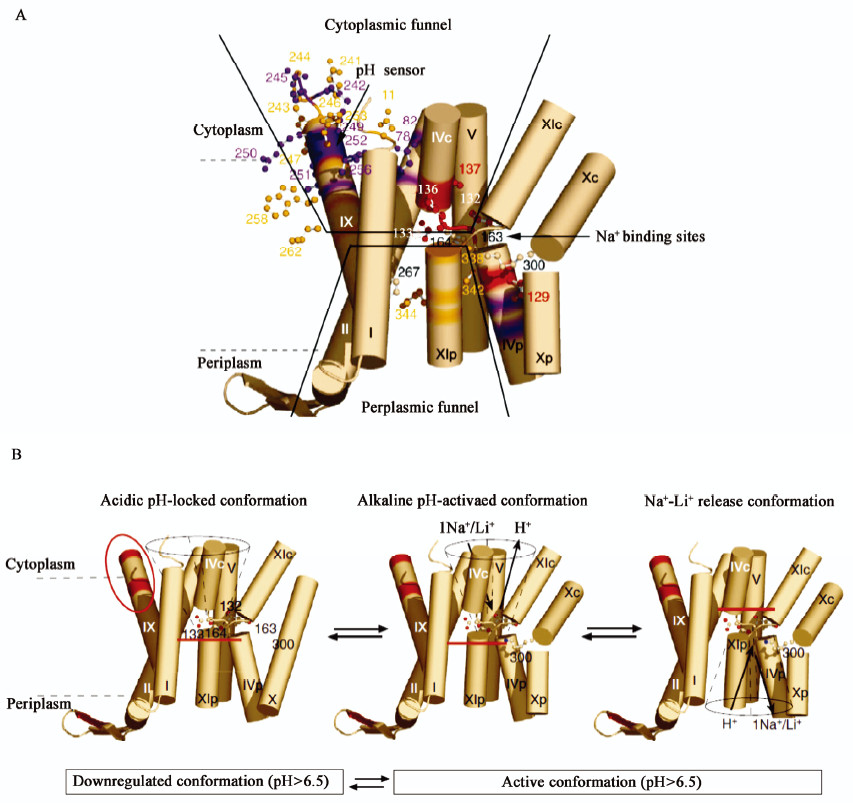
2 Na+/H+逆向转运蛋白的结构和机理Na+/H+逆向转运蛋白是一类高度保守的膜蛋白,其结构和功能研究受到了学术界的广泛关注。自从Mitchell及其同事在大肠杆菌中首次报道Na+/H+逆向转运蛋白后,该蛋白家族在细菌、真菌、植物或动物等整个生命界的细胞膜及内膜系统上被鉴定到[6, 32]。本文以大肠杆菌NhaA Na+/H+逆向转运蛋白为例,通过对其结构阐释,初步揭示Na+/H+逆向转运蛋白发挥作用的分子机制,为其他Na+/H+逆向转运蛋白的功能阐释提供参考[34, 54, 55]。大肠杆菌NhaA Na+/H+逆向转运蛋白由388个氨基酸残基构成,能够形成12个跨膜区域(TMS),其中C端和N端区域都位于胞质内,而且跨膜区VII和VIII明显地短于其他区域[54]。His225、Gly338是NhaA中与pH反应有关的残基,G338S突变导致NhaA失去pH控制,在pH 6.5−9.0时处于激活状态[56, 57]。H225R突变使NhaA催化活性的最适pH向酸性方向偏移,E252C突变使NhaA催化活性的最适pH向碱性方向偏移[57, 58]。研究发现,NhaA蛋白中存在2个漏斗状结构,两者朝向相反并被周质空间中疏水性氨基酸组成的离子屏障所阻隔(图 2A)[54]。胞质漏斗结构由TMS II、Ⅸ、Ⅳc和V跨膜螺旋区组成,富含带负电荷的氨基酸残基;周质漏斗结构由TMS II、Ⅷ和XIp跨膜螺旋区组成,同样富含带负电荷的氨基酸残基。NhaA蛋白pH感应器(pH sensor)位于TMS Ⅸ跨膜螺旋区的N端部位,在胞质漏斗结构开口处,由一系列带负电荷的氨基酸残基构成,能够作为一种离子陷阱捕获阳离子。TMS IV-XI结构由一对不连续的朝向相反的螺旋区域构成,中间被一条延伸链区域阻断。Asp133能够稳定TMS IV-XI结构中带有部分正电荷的N端部位,Lys300也能够抵消TMS IV-XI结构中带有部分负电荷的C端部位,最终在膜中间形成一个电荷平衡区,从而确保NhaA蛋白活性的精细调控。NhaA蛋白中Na+结合位点(Na+ binding sites)位于TMS IV-XI结构的延伸链区域,靠近胞质漏斗结构底部区域,含有高度保守性氨基酸残基,如Asp163、Asp164、Asp133、Thr132等,该区域对维持NhaA蛋白活性是必需的。目前的研究模型表明(图 2B):在酸性条件下,离子转运过程被周质离子屏障阻隔,并且只有Asp164残基能够暴露在胞质漏斗结构底部区域。在碱性条件下,NhaA蛋白pH感应器能够感应环境信号,引起TMS Ⅸ跨膜螺旋区构型的改变,导致TMS IVc、XIp和X跨膜螺旋区的重新定位,并最终完全释放Na+离子结合位点。当NhaA蛋白与底物Na+离子结合后,引起电荷失衡,打开周质离子屏障,将Na+离子结合位点暴露于周质漏斗结构底部区域,最终完成Na+离子释放过程。
| 基因名称 Name | 种属 Species | 定义 definition | 长度 Length (aa) | 底物 substrates | 文献 reference |
| Ec-nhaA | E. coli K12 | Na+: H+ antiporter, NhaA family | 388 | Na+ or Li+ | [31, 32, 33, 34] |
| Ec-nhaB | E. coli K12 | Na+: H+ antiporter, NhaB family | 513 | Na+ or Li+ | [39, 40] |
| Ec-chaA | E. coli K12 | Ca2+-Na+: H+ antiporter, CaCA family | 366 | Ca2+ or Na+ | [41] |
| Ec-mdfA | E. coli K12 | Multidrug efflux system protein, Mfs family | 410 | Drugs, Na+ or K+ | [30] |
| Ec-yjcE | E. coli K12 | Putative cation: proton antiporter, Cpa1 family | 549 | Na+ | [14] |
| Ec-cvrA (ycgO) | E. coli K12 | Putative cation: proton antiporter, Cpa1 family | 578 | K+ | [14] |
| Ec-KefB | E. coli K12 | K+: H+ antiporter, Cpa2 family | 601 | K+ | [20] |
| Ec-KefC | E. coli K12 | K+: H+ antiporter, Cpa2 family | 620 | K+, Rb+, Li+ or Na+ | [20] |
| Eh-napA | E. hirae ATCC9790 | Na+: H+ antiporter, Cpa2 family | 383 | Na+ | [21] |
| Vc-nhaA | V. cholerae O395 | Na+: H+ antiporter, NhaA family | 382 | Na+ or Li+ | [8, 51] |
| Vc-nhaB | V. cholerae O395 | Na+: H+ antiporter, NhaB family | 530 | Na+ | [8, 51] |
| Vc-nhaD | V. cholerae O395 | Na+: H+ antiporter, NhaD family | 477 | Na+ or Li+ | [8] |
| Vc-nhaC (yqkI) | V. cholerae O395 | Putative Na+: H+ antiporter, NhaC family | 481 | Na+? | This study |
| Vc-nhaP1 | V. cholerae O395 | Na+-K+: H+ antiporter, Cpa1 family | 444 | Na+ or K+ | [18] |
| Vc-nhaP2 | V. cholerae O395 | K+: H+ antiporter, Cpa1 family | 581 | K+, Na+ or Rb+ | [18] |
| Bc-GerN | B. cereus AH187 | Na+-K+: H+-K+ antiporter, Cpa2 family | 375 | Na+ | [22] |
| Bh-mrp (mnh, sha) | B. halodurans C-125 | Monovalent cation: H+ antiporter A-G, Cpa3 family | 95−804 | Na+ | [23, 25] |
| Bs-mrp (mnh, pha) | B. subtilis 168 | Monovalent cation: H+ antiporter A-G, Cpa3 family | 94−801 | Na+, Li+, not K+, Ca2+ | [23, 26] |
| Bs-nhaC (yheL) | B. subtilis 168 | Na+: H+ antiporter, NhaC family | 453 | Na+ | [46] |
| Bs-mleN (yqkI) | B. subtilis 168 | Na+-lactate: malate-2H+ antiporter, NhaC family | 468 | malate, lactate, Na+ | [45] |
| Bs-nhaK (yvgP) | B. subtilis 168 | Monovalent cation: H+ antiporter, Cpa1 family | 670 | Na+, Li+, K+ or Rb+ | [16] |
| Bs-tetB | B. subtilis 168 | Tetracycline-divalent cation efflux protein, Mfs family | 458 | Tet-metal, Na+ or K+ | [8] |
| Bp-mrp | B. pseudofirmus OF4 | Monovalent cation: H+ antiporter A-G, Cpa3 family | 91−805 | Na+ or Li+, not K+ | [24, 27, 29] |
| Bp-nhaC1 | B. pseudofirmus OF4 | Putative Na+: H+ antiporter, NhaC family | 439 | Na+? | This study |
| Bp-nhaC2 | B. pseudofirmus OF4 | Putative Na+: H+ antiporter, NhaC family | 515 | Na+? | This study |
| Bp-nhaC3 | B. pseudofirmus OF4 | Putative Na+: H+ antiporter, NhaC family | 251 | Na+? | This study |
| Bp-nhaC4 | B. pseudofirmus OF4 | Putative Na+: H+ antiporter, NhaC family | 220 | Na+? | This study |
| Bp-nhaC5 | B. pseudofirmus OF4 | Na+: H+ antiporter, NhaC family | 462 | Na+ | [47] |
| Bp-nhaC6 | B. pseudofirmus OF4 | Putative Na+: H+ antiporter, NhaC family | 436 | Na+? | This study |
| Bp-nhaP | B. pseudofirmus OF4 | Putative Na+-K+: H+ antiporter, Cpa1 family | 494 | Na+ or K+? | This study |
| Bp-nhaP2 | B. pseudofirmus OF4 | Putative Na+-K+: H+ antiporter, Cpa1 family | 501 | Na+ or K+? | This study |
| Bp-napA | B. pseudofirmus OF4 | Na+: H+ antiporter, Cpa2 family | 494 | Na+? | This study |
| Sa-mnh1 | S. aureus N315 | Monovalent cation: H+ antiporter A-G, Cpa3 family | 97−801 | Na+ or K+? | This study |
| Sa-mnh2 | S. aureus N315 | Monovalent cation: H+ antiporter A-G, Cpa3 family | 100−800 | K+? | This study |
| Pa-nhaP | P. aeruginosa PAO1 | Na+: H+ antiporter, Cpa1 family | 424 | Na+ or Li+ | [17, 18] |
| Aa-nhaD | A. amylolytica N10 | Na+-Li+: H+ antiporter, NhaD family | 483 | Na+ or Li+ | [49] |
Na+/H+逆向转运蛋白是在生物界普遍存在的负责Na+/H+交换的一种跨膜转运蛋白[8, 59, 60]。在高等植物中,主要通过降低Na+离子吸收、促进Na+离子外排和Na+离子区隔化3种策略,维持胞内pH稳态和离子动态平衡,从而消除Na+离子毒害作用。其中,Na+离子外排和区隔化是次级主动运输过程,主要依赖于质膜和液泡膜上的Na+/H+逆向转运蛋白来完成[60]。研究发现,植物Na+/H+逆向转运蛋白在进化上可能具有多样性,液泡型Na+/H+逆向转运蛋白基因间具有高度同源性,质膜型Na+/H+逆向转运蛋白基因也具有高度同源性,但是两者间的同源性相对较低,而且细菌、真菌和动物中Na+/H+逆向转运蛋白与植物Na+/H+逆向转运蛋白的同源性更低[61]。由于大多数原核生物没有恒定的内膜系统,系统发育分析揭示细菌Na+/H+逆向转运蛋白在进化上可能仅与植物质膜型Na+/H+逆向转运蛋白间具有一定同源性[60, 62]。
有趣的是,嗜盐或嗜碱等极端微生物能在高Na+离子或pH环境条件下生长,但是其胞内Na+离子并不高,提示其膜系统具有较强的Na+/H+逆向转运蛋白活性[7]。近年来,尽管细菌Na+/H+逆向转运蛋白的研究取得了一定进展,鉴定和描述出许多新型Na+/H+逆向转运蛋白,为发掘功能强大的Na+/H+逆向转运蛋白基因资源奠定了基础,但是关于细菌Na+/H+逆向转运蛋白功能特征和应答调控机制的研究仍处于起步阶段。另一方面,越来越多的细菌Na+/H+逆向转运蛋白基因被相继克隆和鉴定,也为其在农业或工业上的应用提供了可能。在番茄细胞中异源表达嗜碱菌Alkalimonas amylolytica Na+/H+逆向转运蛋白后,显著提高转基因植物在盐碱土壤中的生长能力[63];在水稻中异源表达大肠杆菌NhaA Na+/H+逆向转运蛋白后,同样明显增强转基因水稻的耐盐和耐旱特性,有效提高农作物产量[64]。利用基因工程技术进行的类似尝试,将为培育抗盐碱农作物提供了新的思路。但是,由于菌株的耐盐特性是多基因协同作用的结果,单一基因的过表达具有较大的局限性,通常而言并不会影响菌株的耐盐性。在大肠杆菌中,尽管过表达NhaA和NhaR后并没有显著提高菌株的耐盐性,但却改变了菌株的中心碳代谢途径,显著增加了菌株发酵过程中的乳酸积累量[65]。因此,继续挖掘和研究耐盐碱菌高活性的Na+/H+逆向转运蛋白,将为工业菌株改造和抗盐碱农作物培育提供新的工具和手段。
通过异源表达策略,以大肠杆菌Na+/H+逆向转运蛋白基因缺失菌株KNabc为宿主系统,分离和鉴定出许多新的Na+/H+逆向转运蛋白,是研究细菌Na+/H+逆向转运蛋白的最主要方法之一[42]。需要注意的是,尽管许多微生物,尤其是嗜碱菌和嗜盐菌,都存在多种Na+/H+逆向转运蛋白,但是这些Na+/H+逆向转运蛋白间的内在关系和调控机制尚不清楚,仍有许多Na+/H+逆向转运蛋白需要进行功能注释。因此,利用高通量筛选和分子生物学方法,发现更多的Na+/H+逆向转运蛋白,并在分子和细胞水平上阐释不同Na+/H+逆向转运蛋白的应答机理和互作关系,将为我们更好地了解和应用Na+/H+逆向转运蛋白提供重要的理论依据和指导意义。
| [1] |
Xu N, Dong Y, Cheng X, et al. Cellular iron homeostasis mediated by the Mrs4-Ccc1-Smf3 pathway is essential for mitochondrial function, morphogenesis and virulence in |
| [2] |
Xu N, Cheng X, Yu Q, et al. Aft2, a novel transcription regulator, is required for iron metabolism, oxidative stress, surface adhesion and hyphal development in |
| [3] |
Xu N, Cheng XX, Yu QL, et al. Research advances of iron homeostasis regulatory networks in |
| [4] |
Yu QL, Xu N, Li MC. Calcium homeostasis systems and calcium signaling pathways in |
| [5] | Kempf B, Bremer E. Uptake and synthesis of compatible solutes as microbial stress responses to high-osmolality environments[J]. Archives of Microbiology, 1998, 170(5): 319-330 |
| [6] | Padan E, Venturi M, Gerchman Y, et al. Na+/H+ antiporters[J]. Biochimica Biophysica Acta, 2001, 1505(1): 144-157 |
| [7] | Krulwich TA. Alkaliphiles: ‘basic’ molecular problems of pH tolerance and bioenergetics[J]. Molecular microbiology, 1995, 15(3): 403-410 |
| [8] | Padan E, Bibi E, Ito M, et al. Alkaline pH homeostasis in bacteria: new insights[J]. Biochimica Biophysica Acta, 2005, 1717(2): 67-88 |
| [9] | Yang LF. Cloning and functional study on Na+ extrusion related genes in Gram-positive moderately halophilic bacteria[D]. Beijing: Doctoral Dissertation of China Agricultural University, 2006 (in Chinese)杨礼富. 革兰氏阳性中度嗜盐菌钠离子输出相关基因的克隆与功能研究[D]. 北京: 中国农业大学博士学位论文, 2006 |
| [10] | Du LX, Dong KH, Zhu HS. Research progess on Na+/H+ antiporter of plant[J]. Grassland and Turf, 2012(2): 82-86 (in Chinese)杜利霞, 董宽虎, 朱慧森. 植物Na+/H+逆向转运蛋白研究进展[J]. 草原与草坪, 2012(2): 82-86 |
| [11] | Saier MH Jr, Reddy VS, Tamang DG, et al. The transporter classification database[J]. Nucleic Acids Research, 2014, 42: D251-258 |
| [12] | Krulwich TA, Hicks DB, Ito M. Cation/proton antiporter complements of bacteria: why so large and diverse?[J]. Molecular microbiology, 2009, 74(2): 257-260 |
| [13] | Saier MH Jr, Eng BH, Fard S, et al. Phylogenetic characterization of novel transport protein families revealed by genome analyses[J]. Biochimica Biophysica Acta, 1999, 1422(1): 1-56 |
| [14] |
Verkhovskaya ML, Barquera B, Wikstrom M. Deletion of one of two |
| [15] |
Gouda T, Kuroda M, Hiramatsu T, et al. |
| [16] |
Fujisawa M, Kusumoto A, Wada Y, et al. NhaK, a novel monovalent cation/H+ antiporter of |
| [17] |
Kuroda T, Fujita N, Utsugi J, et al. A major Li+ extrusion system NhaB of |
| [18] | Resch CT, Winogrodzki JL, Hase CC, et al. Insights into the biochemistry of the ubiquitous NhaP family of cation/H+ antiporters[J]. Biochemistry and Cell Biology, 2011, 89(2): 130-137 |
| [19] |
Janto B, Ahmed A, Ito M, et al. Genome of alkaliphilic |
| [20] | Fujisawa M, Ito M, Krulwich TA. Three two-component transporters with channel-like properties have monovalent cation/proton antiport activity[J]. Proceedings of the National Academy of Sciences of the United States of America, 2007, 104(33): 13289-13294 |
| [21] |
Waser M, Hess-Bienz D, Davies K, et al. Cloning and disruption of a putative NaH-antiporter gene of |
| [22] |
Southworth TW, Guffanti AA, Moir A, et al. GerN, an endospore germination protein of |
| [23] | Swartz TH, Ikewada S, Ishikawa O, et al. The Mrp system: a giant among monovalent cation/proton antiporters?[J]. Extremophiles, 2005, 9(5): 345-354 |
| [24] |
Morino M, Natsui S, Ono T, et al. Single site mutations in the hetero-oligomeric Mrp antiporter from alkaliphilic |
| [25] |
Hamamoto T, Hashimoto M, Hino M, et al. Characterization of a gene responsible for the Na+/H+ antiporter system of alkalophilic |
| [26] |
Ito M, Guffanti AA, Oudega B, et al. mrp, a multigene, multifunctional locus in |
| [27] | Morino M, Suzuki T, Ito M, et al. Purification and functional reconstitution of a seven-subunit mrp-type Na+/H+ antiporter[J]. Journal of Bacteriology, 2014, 196(1): 28-35 |
| [28] |
Yamaguchi T, Tsutsumi F, Putnoky P, et al. pH-dependent regulation of the multi-subunit cation/proton antiporter Pha1 system from |
| [29] |
Swartz TH, Ito M, Ohira T, et al. Catalytic properties of |
| [30] | Fluman N, Adler J, Rotenberg S A, et al. Export of a single drug molecule in two transport cycles by a multidrug efflux pump[J]. Nature Communications, 2014, 5: 4615 |
| [31] |
Padan E, Tzubery T, Herz K, et al. NhaA of |
| [32] |
West IC, Mitchell P. Proton/sodium ion antiport in |
| [33] |
Mager T, Rimon A, Padan E, et al. Transport mechanism and pH regulation of the Na+/H+ antiporter NhaA from |
| [34] | Padan E. Functional and structural dynamics of NhaA, a prototype for Na+ and H+ antiporters, which are responsible for Na+ and H+ homeostasis in cells[J]. Biochimica Biophysica Acta, 2014, 1837(7): 1047-1062 |
| [35] |
Padan E, Schuldiner S. Molecular physiology of the Na+/H+ antiporter in |
| [36] |
Rahav-Manor O, Carmel O, Karpel R, et al. NhaR, a protein homologous to a family of bacterial regulatory proteins (LysR), regulates nhaA, the sodium proton antiporter gene in |
| [37] |
Lentes CJ, Mir SH, Boehm M, et al. Molecular characterization of the Na+/H+-antiporter NhaA from |
| [38] | Calinescu O, Danner E, Bohm M, et al. Species differences in bacterial NhaA Na+/H+ exchangers[J]. FEBS Letters, 2014, 588(17): 3111-3116 |
| [39] |
Pinner E, Padan E, Schuldiner S. Kinetic properties of NhaB, a Na+/H+ antiporter from |
| [40] |
Pinner E, Kotler Y, Padan E, et al. Physiological role of NhaB, a specific Na+/H+ antiporter in |
| [41] |
Ohyama T, Igarashi K, Kobayashi H. Physiological role of the chaA gene in sodium and calcium circulations at a high pH in |
| [42] |
Nozaki K, Inaba K, Kuroda T, et al. Cloning and sequencing of the gene for Na+/H+ antiporter of |
| [43] | Wang X, Xu F, Chen S. Metagenomic cloning and characterization of Na+/H+ antiporter genes taken from sediments in Chaerhan Salt Lake in China[J]. Biotechnology Letters, 2013, 35(4): 619-624 |
| [44] |
Meng L, Hong S, Liu H, et al. Cloning and identification of Group 1 mrp operon encoding a novel monovalent cation/proton antiporter system from the moderate halophile |
| [45] |
Wei Y, Guffanti AA, Ito M, et al. |
| [46] |
Pragai Z, Eschevins C, Bron S, et al. |
| [47] |
Ito M, Guffanti AA, Zemsky J, et al. Role of the nhaC-encoded Na+/H+ antiporter of alkaliphilic |
| [48] |
Nozaki K, Kuroda T, Mizushima T, et al. A new Na+/H+ antiporter, NhaD, of |
| [49] |
Liu J, Xue Y, Wang Q, et al. The activity profile of the NhaD-type Na+-Li+/H+ antiporter from the soda Lake Haloalkaliphile |
| [50] |
Wei Y, Liu J, Ma Y, et al. Three putative cation/proton antiporters from the soda lake alkaliphile |
| [51] |
Herz K, Vimont S, Padan E, et al. Roles of NhaA, NhaB, and NhaD Na+/H+ antiporters in survival of |
| [52] |
Steuber J, Halang P, Vorburger T, et al. Central role of the Na+-translocating NADH:quinone oxidoreductase (Na+-NQR) in sodium bioenergetics of |
| [53] |
Sousa PM, Videira MA, Vorburger T, et al. The novel NhaE-type Na+/H+ antiporter of the pathogenic bacterium |
| [54] | Padan E, Kozachkov L, Herz K, et al. NhaA crystal structure: functional-structural insights[J]. Journal of Experimental Biology, 2009, 212(Pt 11): 1593-1603 |
| [55] | Padan E. The enlightening encounter between structure and function in the NhaA Na+/H+ antiporter[J]. Trends in Biochemical Sciences, 2008, 33(9): 435-443 |
| [56] |
Rimon A, Gerchman Y, Kariv Z, et al. A point mutation (G338S) and its suppressor mutations affect both the pH response of the NhaA-Na+/H+ antiporter as well as the growth phenotype of |
| [57] |
Olami Y, Rimon A, Gerchman Y, et al. Histidine 225, a residue of the NhaA Na+/H+ antiporter of |
| [58] |
Tzubery T, Rimon A, Padan E. Mutation E252C increases drastically the Km value for Na+ and causes an alkaline shift of the pH dependence of NhaA Na+/H+ antiporter of |
| [59] | Reguera M, Bassil E, Blumwald E. Intracellular NHX-type cation/H+ antiporters in plants[J]. Molecular Plant, 2014, 7(2): 261-263 |
| [60] | Rodriguez-Rosales MP, Galvez FJ, Huertas R, et al. Plant NHX cation/proton antiporters[J]. Plant Signal Behavior, 2009, 4(4): 265-276 |
| [61] | Brett CL, Donowitz M, Rao R. Evolutionary origins of eukaryotic sodium/proton exchangers[J]. American Journal of Physiology Cell Physiology, 2005, 288(2): C223-239 |
| [62] | Chen GP, Wang HZ, Shi NN, et al. Na+/H+ antiporter and its relationship with plant salt tolerance[J]. China Biotechnology, 2006, 26(5): 101-106 (in Chinese)陈观平, 王慧中, 施农农, 等. Na+/H+逆向转运蛋白与植物耐盐性关系研究进展[J]. 中国生物工程杂志, 2006, 26(5): 101-106 |
| [63] | Zhong N, Han L, Wu X, et al. Ectopic expression of a bacterium NhaD-type Na+/H+ antiporter leads to increased tolerance to combined salt/alkali stresses[J]. Journal of Integrative Plant Biology, 2012, 54(6): 412-421 |
| [64] | Wu L, Fan Z, Guo L, et al. Over-expression of the bacterial nhaA gene in rice enhances salt and drought tolerance[J]. Plant Science, 2005, 168(2): 297-302 |
| [65] |
Wu X, Altman R, Eiteman MA, et al. Effect of overexpressing nhaA and nhaR on sodium tolerance and lactate production in |
 2015, Vol. 42
2015, Vol. 42