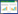中国科学院微生物研究所、中国微生物学会主办
文章信息
- 李玲玲, 刘雪, 邱泽天, 赵广荣
- Li Lingling, Liu Xue, Qiu Zetian, Zhao Guangrong
- 植物多酚的微生物合成
- Microbial synthesis of plant polyphenols
- 生物工程学报, 2021, 37(6): 2050-2076
- Chinese Journal of Biotechnology, 2021, 37(6): 2050-2076
- 10.13345/j.cjb.200747
-
文章历史
- Received: November 24, 2020
- Accepted: February 14, 2021
- Published: February 23, 2021
植物多酚(Plant polyphenols) 是源自苯丙烷代谢途径的一类化合物,含有至少2个酚环[1]。据估计,自然界存在10–20万种植物次级代谢产物,光合作用固定的总碳中有20%流入了苯丙烷途径,形成数量庞大、广泛分布、结构复杂的酚类化合物[2]。按照分子碳骨架的不同,植物多酚大致可分为酚酸类(Polyphenolic acids) ((C6−C3)n)、茋类(Stilbenes) (C6−C2−C6)、姜黄素类(Curcumins) (C6−C7−C6) 和黄酮类(Flavonoids) (C6−C3−C6) (图 1)。其中,黄酮类是最大的一个亚类,根据苯环位置和羟基数目可以细分为黄烷酮(Flavanones)、黄酮(Flavones)、异黄酮(Isoflavones)、黄酮醇(Flavonols)、花青素(Anthocyanidins) 及其衍生物。

|
| 图 1 植物多酚的代表性骨架结构类型 Fig. 1 Representative skeleton structures of plant polyphenols. |
| |
植物多酚类产物的分子结构中存在多个羟基,可与活性氧和活性氮结合[1-3],清除体内过多的自由基,因此植物多酚表现出很强的抗氧化活性。植物多酚还具有扩张脑血管、抗血凝、抗炎、抗肿瘤、抗病毒等作用,可作为治疗这些疾病的药物[4]。丹参素、丹酚酸B、灯盏花素已经被批准用于临床治疗脑栓塞、心肌梗塞和冠心病,而姜黄素、阿克拉定及其衍生物等进入临床抗肿瘤药物的试验阶段。根皮苷具有降血糖作用,以根皮苷为模型药效团,开发上市了一系列治疗糖尿病的化学药物[5]。最近发现丹酚酸C能阻止S蛋白六螺旋束核心的形成,黄芩苷和黄芩素能抑制3-胰凝乳蛋白酶样蛋白酶,是新冠病毒的高效抑制剂[6-7]。
植物多酚在保健品和食品等方面也有着重要的应用[8]。作为营养补充剂,植物多酚能减少机体的氧化性应激、调节免疫系统、延缓衰老。姜黄素、花色苷是食品的增色剂,三叶苷的甜度约为蔗糖的300倍,可作为食品添加剂和商用的甜味剂。植物多酚还可以通过氢键、共价键和配位键等多种相互作用与其他分子结合,作为载体用于构筑各种新型功能材料[9]。
植物多酚在医药、食品、化妆品、化工等领域具有巨大的市场应用前景。预计2026年黄酮类化合物的市场份额将达到12.59亿美元,2027年白藜芦醇和姜黄素的市场份额将分别达到1.49亿美元和1.519亿美元。因此,需要开发大规模生产多酚的方法以满足市场需求。随着合成生物学的技术和工具发展,利用微生物合成天然产物成为一种高效、绿色和经济的方法。文中对丹酚酸、白藜芦醇、姜黄素、黄酮等植物多酚的微生物合成进行综述,以期推动植物源天然产物的工业化发展。
1 丹酚酸类的微生物合成丹参酚酸类是中药丹参中的水溶性活性成分,包括丹参素(Danshensu,salvianic acid A)、迷迭香酸(Rosmarinic acid)、原紫草酸(Prolithospermic acid)、丹酚酸(Salvianolic acids)等。对香豆酸(p-coumaric acid)、咖啡酸(Caffeic acid) 是酚酸类合成的关键前体,丹参素是丹酚酸类产物的另一个苯环的结构来源。丹酚酸可以看成是丹参素和咖啡酸的衍生物或酯化的聚合物。目前丹参素、迷迭香酸的途径已被解析,其他丹参酚酸途径仍然未知。从微生物的芳香氨基酸代谢出发,表达外源基因,可重构关键前体的微生物合成途径,再经过设计改造和表达酚酸类合成基因,可实现丹酚酸类的微生物合成。
1.1 咖啡酸微生物从葡萄糖到咖啡酸有两条合成途径,一条是从酪氨酸的代谢节点切入,表达酪氨酸解氨酶(Tyrosine ammonialyase,TAL)、对香豆酸-3-羟化酶(p-coumarate 3-hydroxylase,C3H)/细胞色素P450还原酶(Cytochrome P450 reductase,CPR),合成咖啡酸;另一条是从苯丙氨酸代谢节点切入,表达苯丙氨酸解氨酶(Phenylalanine ammonialyase,PAL)、反式肉桂酸-4-羟化酶(Trans-cinnamate 4-monooxygenase,C4H)/CPR,合成对香豆酸,进而3′-羟化生成咖啡酸(图 2)。由于该途径涉及的C4H/C3H是细胞色素P450酶类,在真核酿酒酵母Saccharomyces cerevisiae中表达具有一定的优势。在酿酒酵母中,表达TAL、C3H和CPR,强化酪氨酸途径和优化培养基后,酿酒酵母利用葡萄糖合成了29.58 mg/L的对香豆酸和11.3 mg/L咖啡酸[10] (表 1)。虽然TAL能直接将酪氨酸转化为对香豆酸,比从苯丙氨酸出发缩短了合成途径,也规避了C4H的表达,但从目前的研究报道看,酿酒酵母中表达植物咖啡酸途径的效率仍然较低。
|
| 图 2 咖啡酸的微生物合成途径 Fig. 2 Biosynthetic pathway of Caffeic acid. PAL: phenylalanine ammonialyase; C4H: Trans-cinnamate 4-monooxygenase; CPR: cytochrome P450 reductase; TAL: tyrosine ammonia-lyase; C3H: p-coumarate 3-hydroxylase, CYP199A2: cytochrome P450 199A2; HpaBC: 4-hydroxyphenylacetate 3-hydroxylase complexes B and C; TYR: tyrosine; PHE: phenylalanine; CIA: cinnamic acid; PCA: p-coumaric acid; CA: caffeic acid. |
| |
| Microbe | Genes | Substrate | Titer (mg/L) | References |
| Saccharomyces cerevisiae | aro4K229L, aro7G141S, Δpdc5, Δaro10, Rctal, Atc3h, Atcpr1 | Glucose | 11.3 | [10] |
| Rttal, PahpaB, SehpaC | Tyrosine | 289.4 | [17] | |
| Escherichia coli | Rgtal, sam5 | Glucose | 106.0 | [12] |
| Rgtal, cyp199A2, pdr, pux | Tyrosine | 280.0 | [13] | |
| tyrAfbr, ppsA, tktA, aroGfbr, ΔpheLA, ΔtyrA, Rgtal, hpaBC | Glycerol | 766.7 | [16] | |
| Rc: Rhodobacter capsulatus; At: Arabidopsis thaliana; Rt: Rhodosporidium toruloides; Pa: Pseudomonas aeruginosa; Se: Salmonella enterica; Rg: Rhodotorula glutinis; fbr: feedback inhibition resistance. | ||||
由于C4H/C3H在大肠杆菌Escherichia coli中表达较为困难,需要寻找替代酶。目前已经鉴定到西班牙糖丝菌Saccharothrix espanaensis的Sam5[11-12]、沼泽红假单胞菌Rhodopseudomonas palustris的CYP199A2及其突变体[13],大肠杆菌[14]和铜绿假单胞菌Pseudomonas aeruginosa[15]的HpaBC,都可将对香豆酸高效转化为咖啡酸。利用高产酪氨酸大肠杆菌菌株,表达TAL和HpaBC两个酶,以甘油为主要碳源,合成了766.7 mg/L咖啡酸[16]。在酿酒酵母表达铜绿假单胞菌来源的HpaB和肠道沙门氏菌Salmonella enterica来源的HpaC,合成了289.4 mg/L的咖啡酸[17],表明该途径在酵母中也是高效的。
1.2 丹参素笔者课题组采用逆合成策略,从4-羟基苯丙酮酸出发,设计了丹参素的微生物合成途径(图 3A)。在植物中丹参素途径未解析的情况下,通过生物大数据挖掘出两个关键酶,构建出一条丹参素的人工合成途径。该途径以4-羟基苯丙酮酸为底物,大肠杆菌内源性的羟化酶(HpaBC) 催化苯环间位羟基化,植物乳杆菌Lactobacillus pentosus D-乳酸脱氢酶(D-lactate dehydrogenase,D-LDH) 还原酮基,生成丹参素。在这个途径中,酮基还原反应是限速步骤。采用突变体D-LDHY52A,极大地提高了丹参素的产量。利用模块化工程和底盘菌株改造等策略,将丹参素途径与高产酪氨酸的底盘大肠杆菌进行适配化,获得了从葡萄糖发酵合成丹参素的工程菌株(图 3B)[18]。

|
| 图 3 工程大肠杆菌合成丹参素 Fig. 3 Biosynthesis of salvianic acid A in engineered E. coli. (A) SAA retrosynthetic pathway. (B) The artifcial synthetic pathway of SAA from glucose. (C) The targeted integration of three modules into chromosome of engineered E. coli. 6-P-G: 6-phosphate D-glucose; PEP: phosphoenolpyruvate; PYR: pyruvate; DAHP: 3-deoxy-arabino-heptulonate 7-phosphate; DHS: 3-dehydroshikimic acid; SHK: shikimic acid; CHA: chorismic acid; 4HPP: 4-hydroxyphenylpyruvate; DHPP: 3, 4-dihydroxyphenylpyruvate; 4-HPL: 4-hydroxyphenyllactate; TYR: tyrosine; PHE: phenylalanine; SAA: salvianic acid A. |
| |
在上述工作的基础上,笔者课题组采用启动子工程,微调组成型表达丹参素途径。进而采用染色体工程,将酪氨酸途径强化基因模块和丹参素途径合成基因整合到大肠杆菌染色体上,构建了不使用抗生素和诱导剂的高产丹参素菌株(图 3C),补料发酵60 h,丹参素产量达5.6 g/L[19]。该菌株遗传稳定,可放大培养,具备工业化生产的潜力。
除了工程微生物从头合成丹参素外,最近报道了生物转化制备丹参素。江南大学蔡宇杰课题组在大肠杆菌中表达L-氨基酸脱氨酶和D-乳酸脱氢酶,将原料L-多巴转化为丹参素[20-21];同时表达L-谷氨酸脱氢酶或者葡萄糖脱氢酶用于NADH再生,转化得率为95.2%[21]。在此工程菌的基础上,表达酪氨酸酚裂解酶,将丹参素的转化路线推向传统化工原料邻苯二酚和丙酮酸[22],转化得率为59.6%。同济大学Li Chao-课题组构建了D-扁桃酸脱氢酶、苯丙氨酸-4-羟化酶和羟基苯乙酸酯-3-羟化酶的级联酶催化体系,添加NADH和DMPH4双辅因子循环酶和底物,将原料苯丙酮酸转化为丹参素,转化得率为84.9%[23]。这些新工艺路线具有绿色和可持续性,值得进一步优化和改进。
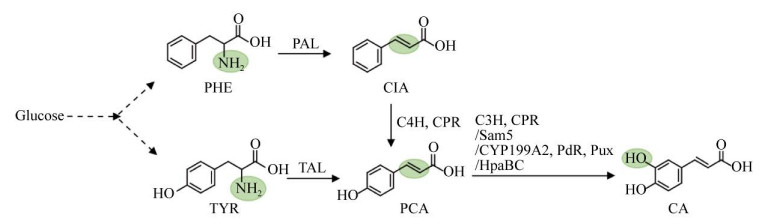
1.3 迷迭香酸迷迭香酸合成酶(Rosmarinic acid synthase,RAS) 催化丹参素和咖啡酰CoA的酯化反应,在酿酒酵母中合成了迷迭香酸[24],表明迷迭香酸合成酶在酵母中具有功能。在酿酒酵母中表达植物迷迭香酸合成途径,包括约氏黄杆菌Flavobacterium johnsoniae FjTAL、拟南芥4-香豆酰CoA连接酶(4-coumarate: coenzyme A ligase) At4CL、褐家鼠Rattus norvegu酪氨酸转氨酶(Tyrosine aminotransferase) RnTAT、彩叶草Plectranthus scutellarioides对羟基苯丙酮酸还原酶(Hydroxyphenylpyruvate reductase,HPPR) CbHPPR、CbRAS、大肠杆菌EcHpaBC、丹参Salivia miltiorrhiza SmCYP98A14和SmCPR (图 4),并通过正交设计优化基因表达强度,利用葡萄糖合成了5.95 mg/L迷迭香酸,同时对羟基肉桂酸、对羟基苯丙酮酸、对羟基-苯乳酸等中间产物大量积累[24],表明植物迷迭香酸途径在酿酒酵母中是低效的。这可能是由于植物中合成迷迭香酸涉及细胞色素P450酶催化的苯环羟化反应,需要氧化还原伴侣酶参与。在大肠杆菌中没有采用植物合成路线,而采用微生物合成丹参素途径(HpaBC和D-LDH)、植物合成咖啡酸途径(PAL和4CL) 和RAS构成的迷迭香酸杂合途径,以葡萄糖为底物合成了0.65 mg/L迷迭香酸[25]。迷迭香酸合成酶是限速因素,一方面该酶活性较低,即使添加底物丹参素和咖啡酸,迷迭香酸产量仍然较低;另一方面,该酶的底物特异性不强,在合成迷迭香酸的同时,还合成了副产物咖啡酰-对-4′-羟基苯乳酸、对香豆酰-苯乳酸[26]。
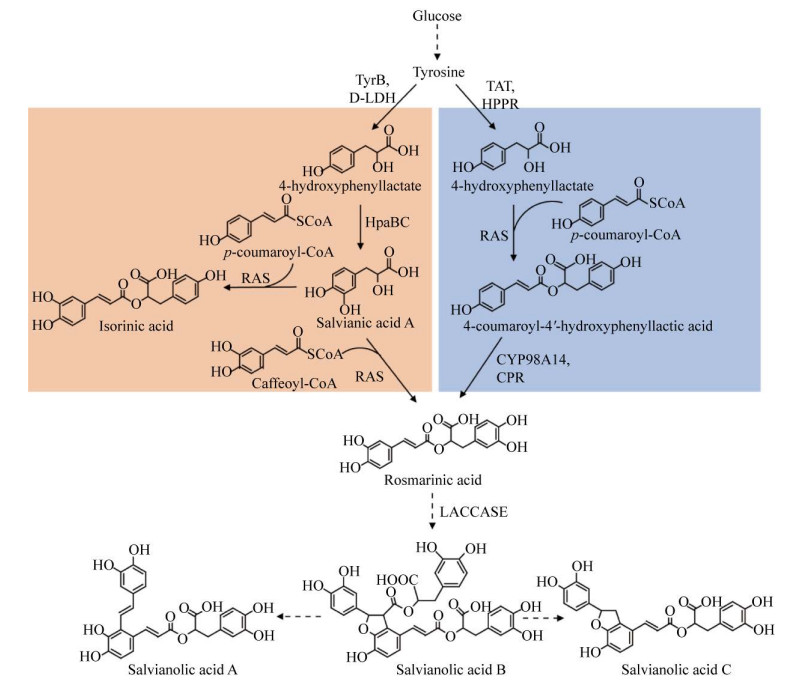
|
| 图 4 迷迭香酸的微生物合成途径 Fig. 4 Biosynthetic pathways of rosmarinic acid. Artifical sythestic pathway is shown in left column and plant-derived pathway in right column. TAT: tyrosine aminotransferase; HPPR: hydroxyphenylpyruvate reductase; HpaBC: 4-hydroxyphenylacetate 3-hydroxylase complexes B and C; RAS: rosmarinic acid synthase; CYP98A14: cytochrome P450 98A14; CPR: cytochrome P450 reductase. |
| |
茋类化合物,又称二苯乙烯类多酚化合物,主要包括白藜芦醇(Resveratrol)及其羟基化衍生物白皮杉醇(Piceatannol)、甲氧基化产物松茋(Pinostilbene) 和紫檀茋(Pterostilbene)、糖基化产物虎杖苷(Polydatin,piceid) (图 5)。
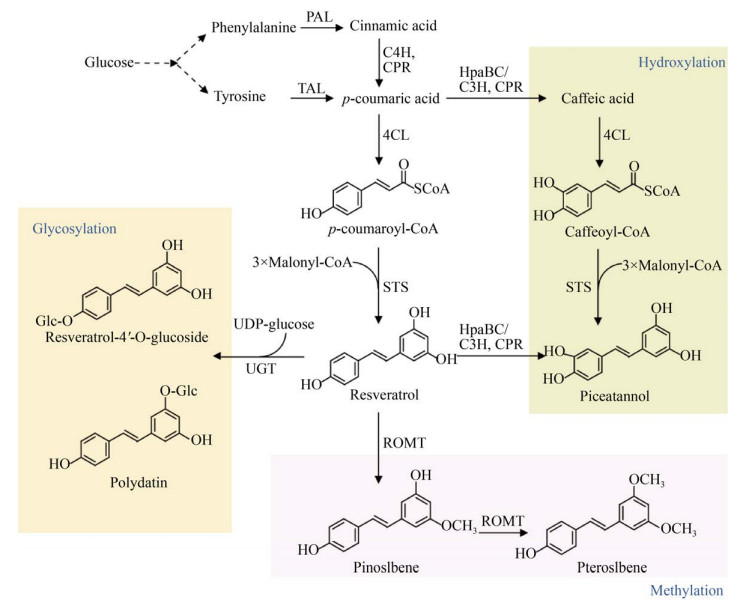
|
| 图 5 茋类的微生物合成途径 Fig. 5 Biosynthetic pathways of stilbenes. 4CL: 4-coumarate-CoA ligase; STS: stilbene synthase; UGT: UDP-glucosyltransferase; ROMT: resveratrol-O-methyltransferase. |
| |
TAL、4CL和白藜芦醇合成酶(Stilbene synthase,STS) 组成了白藜芦醇的合成途径(图 5)。由STS催化对香豆酰CoA和丙二酰CoA生成白藜芦醇,目前已经在大肠杆菌、酿酒酵母、谷氨酸棒杆菌和解脂耶式酵母等微生物中合成(表 2)。
| Microbe | Genes | Substrate | Titer (mg/L) | References |
| Saccharomyces cerevisiae | At4cl-GSG-Vvsts | p-coumaric acid | 5.2 | [27] |
| At4cl-SH3, Vvsts-PDZ, GBD1SH32PDZ4 | p-coumaric acid | 14.4 | [28] | |
| Zif268-4cl, PBSⅡ-sts | p-coumaric acid | > 250 | [29] | |
| At4cl, Vvsts | p-coumaric acid | 262 | [30] | |
| aro4K229L, aro7G141S, acc1S659A, S1157A, Hatal, At4cl, Vvsts | Ethanol | 531.4 | [31] | |
| aro4K229L, aro7G141S, acc1S659A, S1157A, acsL641P, Δaro10, Atpal, Atc4h, Atatr, cyb5, At4cl, Vvsts | Glucose | 812 | [32] | |
| Yarrowia lipolytica | aro4K229L, aroGS180F, aro1, aro2, aro3, aro4, aro5, tkt, Δtrp2, Δtrp3, Δaro8, Δaro9, Δpyk, Δpha2, Rgtal, Pc4cl, Vvsts | Glucose | 12.6 | [33] |
| Fjtal, Vvpal, Atc4h, At4cl, Vvsts | Glycerol | 430 | [34] | |
| aro7G139S, aro4K221L, Δpdc, Δaro10, acc1, Fjtal, At4cl, Vvsts | Glucose | 12 400 | [35] | |
| Escherichia coli | At4cl, Vvsts | p-coumaric acid | 2 340 | [36] |
| ΔtyrR, ΔtrpED, Rgtal, Pc4cl, Vvsts | Glucose | 4.6 | [37] | |
| Setal, Sc4cl, Ahsts | Glucose | 5.2 | [38] | |
| Corynebacterium glutamicum | Δcg0344-47, Δcg2625-40, Δcg1226, Δcg0502, Fjtal, Pc4cl, Ahsts, EcaroH | Glucose | 59 | [39] |
| Vv: Vitis vinifera; Ha: Herpetosiphon aurantiacus; Pc: Petroselinum crispum; Sc: Streptomyces coelicolor; Ah: Arachis hypogaea. | ||||
对白藜芦醇途径中的4CL、STS进行了筛选,包括来源于拟南芥、欧芹Petroselinum crispum、紫草Lithospermum erythrorhizon、烟草Nicotiana tabacum、链霉菌的4CL,葡萄Vitis vinifera、花生Arachis hypogaea、虎杖Polygonum cuspidatum、松叶蕨Psilotum nudum、马尾松Pinus massoniana、北美乔松Pinus strobus、赤松Pinus densiflora的STS,其中拟南芥的4CL、葡萄和花生的STS使用最为广泛。在大肠杆菌和酿酒酵母中表达不同来源的4CL和STS,可以对香豆酸为底物合成白藜芦醇。4CL和STS的不同表达方式直接影响白藜芦醇的产量。用甘氨酸-丝氨酸-甘氨酸(Gly-Ser-Gly,GSG) 接头使4CL和STS的融合表达优于独立表达[27],而用蛋白支架GBD-SH3-PDZ调控4CL和STS的比例,白藜芦醇的产量比融合表达提高了1.7倍[28]。DNA支架也可用于途径的优化,将4CL和STS的基因分别与锌指蛋白结构域Zif268和PBSⅡ融合,通过调整锌指结合位点的重复度、间距和顺序优化4CL和STS表达量,白藜芦醇的产量比融合表达策略提高50倍[29]。这些结果表明蛋白质支架和DNA支架是增强白藜芦醇代谢效率更优秀的策略。
在酿酒酵母合成白藜芦醇的过程中,利用酵母提取物、蛋白胨和葡萄糖的富营养培养基,组成型强启动子HXT7表达At4cl和Vvsts,白藜芦醇的产量达到262 mg/L[30],可见培养基成分对酵母合成白藜芦醇有较大影响。染色体整合多拷贝Hatal、At4cl、Vvsts,同时过表达aro4K229L和aro7G141S强化酪氨酸供给,过表达acc1S659A, S1157A提高胞内丙二酰CoA,乙醇补料发酵110 h,酿酒酵母合成了531 mg/L的白藜芦醇[31]。异源表达PAL、C4H和CPR、4CL、STS,葡萄糖补料发酵,酿酒酵母可通过苯丙氨酸途径合成812 mg/L白藜芦醇[32]。
与酿酒酵母类似,解脂耶式酵母Yarrowia lipolytica可从酪氨酸或苯丙氨酸途径合成白藜芦醇[33]。以葡萄糖为碳源,当单拷贝表达时,PAL-C4H-4CL-STS途径产量略高于TAL-4CL- STS途径;而两拷贝表达时,以甘油为碳源时,采用酪氨酸-苯丙氨酸联合途径(TAL、PAL、C4H、4CL、STS) 效果更优,在5 L发酵罐中合成了430 mg/L白藜芦醇[34]。进一步改造酪氨酸途径、增加基因拷贝数和分批补料培养,发酵90 h,解脂耶式酵母从头合成了12.4 g/L白藜芦醇[35]。
与酵母相比,大肠杆菌合成白藜芦醇的研究相对滞后。虽然以对香豆酸为底物,使用大肠杆菌BW27784,用GAP启动子驱动At4cl和Vvsts的双顺反子表达替代T7启动子驱动的单顺反子表达,白藜芦醇的产量达到2.34 g/L[36]。但无论游离质粒表达tal、4cl、sts基因,还是定点整合到大肠杆菌的染色体上合成白藜芦醇,大肠杆菌从头合成白藜芦醇的效果都不理想[37-38],需要增强酪氨酸和丙二酰CoA供给,从而优化白藜芦醇在大肠杆菌中的合成。
谷氨酸棒杆菌Corynebacterium glutamicum是另一种合成白藜芦醇有潜力的微生物,敲除芳香族化合物的4个降解基因簇,并表达TAL、AroH、4CL和STS,以葡萄糖为碳源,重组菌合成了59 mg/L白藜芦醇[39],与大肠杆菌相比,表现出一定的优越性。
2.2 白藜芦醇衍生物白皮杉醇是白藜芦醇在3′位发生羟基化形成,有两条合成途径:第一条途径是异源表达4CL、STS,从咖啡酸合成白皮杉醇[39-40];第二条途径是异源表达TAL、4CL、C3H或HpaBC和STS,经过对香豆酸合成白皮杉醇[41] (图 5)。
白藜芦醇的甲氧基化产物主要包括松茋和紫檀茋,分别由甲氧基转移酶(O-methyltransferase,OMT) 催化白藜芦醇的5-位和3, 5-二位顺序发生甲氧基化生成。葡萄和高粱的ROMT能以白藜芦醇为底物,在大肠杆菌中生成微量的松茋和紫檀茋[42]。将拟南芥的Atcomt与Setal、Nt4cl2、Vvsts在高产酪氨酸的大肠杆菌中表达,添加L-甲硫氨酸提高胞内甲基供体,从头合成了33.6 mg/L紫檀茋[43] (表 3)。在酿酒酵母中表达葡萄和高粱的Romt与Atpal2、Atc4h、Atatr2、At4cl2和Vvsts1,以葡萄糖为底物,生成了松茋和紫檀茋[32]。
| Products | Microbe | Genes | Substrate | Titer (mg/L) | References |
| Piceatannol | Corynebacterium glutamicum | ΔphdB, ΔpcaF, ΔpobA, Pc4cl, Ahsts | Caffeic acid | 55.0 | [39] |
| Escherichia coli | At4cl, Ahsts | Caffeic acid | 13.2 | [40] | |
| Escherichia coli | Setal, At4cl, Sec3h, Ahsts | Tyrosine | 21.5 | [41] | |
| Pterostilbene | Escherichia coli | Setal, Nt4cl2, Vvsts, Atcomt | Glucose | 33.6 | [43] |
| Saccharomyces cerevisiae | Atpal2, Atc4h, Atatr2, At4cl2, Vvsts1, VvRomt | Glucose | 34.9 | [32] | |
| Pinostilbene | Saccharomyces cerevisiae | Atpal2, Atc4h, Atatr2, At4cl2, Vvsts1, SbRomt | Glucose | 1.9 | [32] |
| Resveratrol-3-O-glucoside | Escherichia coli | Setal, Sc4cl, Ahsts, yjic | Glucose | 2.5 | [44] |
| Resveratrol-4′-O-glucoside | Escherichia coli | Setal, Sc4cl, Ahsts, yjic | Glucose | 7.5 | [44] |
| Nt: Nicotiana tabacum; Sb: Sorghum bicolor. | |||||
白藜芦醇糖苷主要包括虎杖苷(白藜芦醇- 3-O-葡萄糖苷)和白藜芦醇-4′-O-葡萄糖苷。由于地衣芽孢杆菌来源的糖基转移酶YjiC具有非特异性糖基化活性,在大肠杆菌中异源表达Setal、Sc4cl、Ahsts和yjic,催化底物白藜芦醇在3-位和4′位发生糖基化反应,但倾向于合成白藜芦醇-4′-O-葡萄糖苷,而不是虎杖苷[44]。
3 姜黄素类的微生物合成姜黄素类包括姜黄素、去甲氧基姜黄素和双去甲氧基姜黄素等。已经鉴定的姜黄素合成酶有两种,一种是单酶姜黄素类合酶(Curcuminoid synthase,CUS),另一种是双酶,即双酮CoA合酶(Diketide-CoA synthase,DCS) 和姜黄素合酶(Curcumin synthase,CURS) (图 6)。姜黄素类产物异源合成的研究主要集中在大肠杆菌中,最近在解脂耶式酵母中也有报道[45] (表 4)。
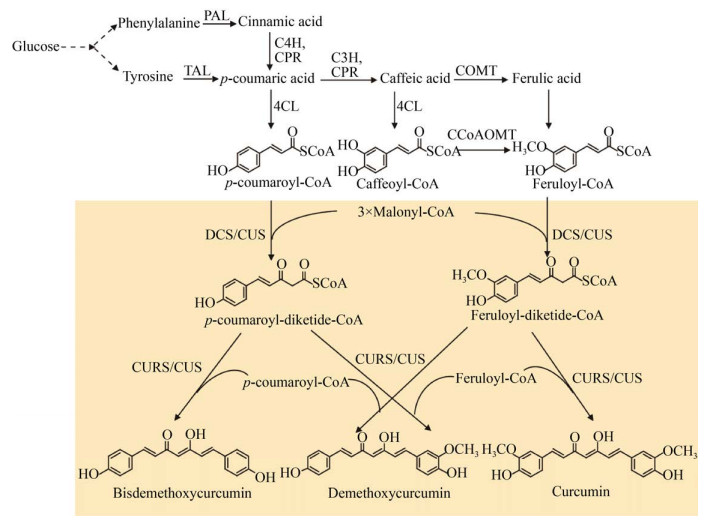
|
| 图 6 姜黄素类的微生物合成途径 Fig. 6 Biosynthetic pathways of Curcuminoids. CUS: curcuminoid synthase; DCS: diketide-CoA synthase; CURS: curcumin synthase; CCoAOMT: caffeoyl-CoA 3-O-methyltransferase. |
| |
| Products | Microbe | Genes | Substrate | Titer (mg/L) | References |
| Bisdemethoxycurcumin | Yarrowia lipolytica | Nt4cl, Oscus, acc1 | 4-coumaric acid | 0.2 | [45] |
| Escherichia coli | Rrpal, Le4cl, Oscus | Tyrosine | 53.4 | [46] | |
| Escherichia coli | ΔtyrR, ΔpheA, Setal, Os4cl, Oscus | Glucose | 4.6 | [47] | |
| Curcumin | Escherichia coli | Le4cl, Oscus | Ferulic acid | 113.0 | [46] |
| Escherichia coli | At4cl, Cldcs, Clcurs | Ferulic acid | 70.0 | [48] | |
| Escherichia coli | At4cl, Cldcs, Clcurs | Ferulic acid | 353.4 | [49] | |
| Escherichia coli | Rgtal, At4cl, Setal, CCoAomt, Cldcs, Clcurs | Tyrosine | 0.2 | [48] | |
| Rr: Rhodotorula rubra; Le: Lithospermum erythrorhizon; Cl: Curcuma longa; Os: Oryza sativa. | |||||
通过异源表达红酵母Rhodotorula rubra RrPAL、紫草Le4CL、水稻OsCUS,以酪氨酸为底物,合成双去甲氧基姜黄素[46];通过敲除大肠杆菌酪氨酸途径负调控基因,提高酪氨酸前体供应,从头合成了双去甲氧基姜黄素[47]。
在大肠杆菌中异源表达4CL、CUS或DCS和CURS,可将阿魏酸转化为姜黄素[46, 48]。使用大肠杆菌BL21(DE3) 为宿主优于K12,对诱导条件、培养基成分和碳源浓度等参数进行优化,以阿魏酸为底物合成353.4 mg/L的姜黄素[49]。由于At4CL对阿魏酸的催化效率低,表达PAL、C3H和咖啡酰CoA-3- O-甲基转移酶(Caffeoyl-CoA 3-O-methyltransferase,CCoAOMT),可绕过阿魏酸,外源添加酪氨酸底物,合成了姜黄素[48],但产量仍然很低。
4 黄酮类的微生物合成根据黄酮类的生物合成途径,查耳酮合成酶(Chalcone synthase,CHS) 催化1分子对香豆酰CoA和3分子丙二酰CoA发生克莱森缩合反应,生成了黄酮类的核心骨架结构,即黄烷酮。然后黄烷酮经过修饰转化生成黄酮、异黄酮、黄酮醇及其衍生物。
4.1 黄烷酮、异黄酮和黄酮黄烷酮、异黄酮、黄酮等产物(图 7) 已经在大肠杆菌、酿酒酵母、解脂耶式酵母和链霉菌中合成(表 5)。

|
| 图 7 黄烷酮、异黄酮、黄酮的微生物合成途径 Fig. 7 Biosynthetic pathways of flavanones, isoflavones and flavones. CHS: chalcone synthase; CHI: chalcone isomerase; CHR: chalcone reductase; DBR: double bond reductase; F3′H: flavonoid 3′-hydroxylase; IFS: isoflavone synthase; FNS: flavone synthase. |
| |
| Products | Microbe | Genes | Substrate | Titer (mg/L) | References |
| Naringenin | Escherichia coli | Rstal, Sc4cl, Atchs, Plchi | Glucose | 29 | [50] |
| Escherichia coli | aroGfbr, tyrAfbr, ΔtyrR, Tctal, Pc4cl, Phchs, Mschi | Glucose | 588 | [51] | |
| Saccharomyces cerevisiae | Pc4cl, Phchs, Mschi | p-coumaric acid | 1 210 | [53] | |
| Saccharomyces cerevisiae | Pdppal, Gmc4h, Pdpcpr, Gm4cl, Gmchs, Gmchi | Glucose | 15.6 | [54] | |
| Saccharomyces cerevisiae | aro4G226S, Δaro3, Δaro10, Δpdc5, Δpdc6, Atpal, Atc4h, Atcpr, At4cl3, Atchs3, Atchi | Glucose | 108 | [55] | |
| Saccharomyces cerevisiae | aro4K229L, Δaro10, Δpdc5, Δtrp2, Δpha2, Fjtal, At4cl, Hachs, Mschi | Glucose | ~90 | [56] | |
| Saccharomyces cerevisiae | aro4K229L, aro7G141S, Δpdc1, Δpdc5, Δpdc6, Δdpp1, Δtsc13, acc1, Mdecr, pdh, Fjtal, At4cl, Atchs, Atchi | Glucose | 144.1 | [57] | |
| Streptomyces venezuelae | Sc4cl, Atchs, Atchi, matBC | p-coumaric acid | 35.6 | [58] | |
| Yarrowia lipolytica | aro1, acs, acc1, Rttal, Pc4cl, Phchs, Mschi | Glucose | 252.4 | [59] | |
| Yarrowia lipolytica | aro4K221L, pex10, acc1, Setal, Nt4cl, Sesam8, Hspks | Glucose | 898 | [45] | |
| Eriodictyol | Saccharomyces cerevisiae | Pc4cl, Phchs, Phchi | Caffeic acid | 6.5 | [60] |
| Escherichia coli | Rgtal, Pc4cl, Phchs, Mschi, Ghf3'h-Crcpr, acc, acs, ΔackA | Tyrosine | 107 | [61] | |
| Streptomyces albus | Rctal, Sc4cl, Gmchs, Gmchi, Atf3'h | Glucose | 0.002 | [62] | |
| Yarrowia lipolytica | aro1, acs, acc1, Rttal, Pc4cl, Phchs, Mschi, Ghf3'h, Crcpr | Glucose | 134.2 | [59] | |
| Liquiritigenin | Escherichia coli | Rstal, Pc4cl, Phchs-Amchr, Mschi | Tyrosine | 1.84 | [63] |
| Saccharomyces cerevisiae | Gupal, Guc4h, Gu4cl, Guchs, Guchi | Glucose | 0.3 | [64] | |
| Saccharomyces cerevisiae | Fjtal, Pc4cl, Phchs, Amchr, Mschi | Glucose | 5.3 | [65] | |
| Phloretin | Saccharomyces cerevisiae | Atpal2, Amc4h, Sccpr1, At4cl2, Hachs, tsc13 | Glucose | 42.7 | [66] |
| Genistein | Saccharomyces cerevisiae | Pdppal, Pdpcpr, Gmc4h, Gm4cl, Gmchs, Gmchi, Gmifs | Glucose | 0.1 | [54] |
| Escherichia coli | Os4cl, Pechs, Tpifs, Oscpr | p-coumaric acid | 35 | [68] | |
| Apigenin | Saccharomyces cerevisiae | Pc4cl, Phchs, Phchi, Amfns, Sccpr1 | p-coumaric acid | 2 | [69] |
| Escherichia coli | Rrpal, Sc4cl, Gechs, Plchi, Pcfns, Cgacc | Tyrosine | 13 | [70] | |
| Streptomyces albus | Rctal, Sc4cl, Gmchs, Gmchi, Pcfns | Glucose | 0.08 | [62] | |
| Luteolin | Escherichia coli | Pc4cl, Phchs, Mschi, Pcfns | Caffeic acid | 4 | [71] |
| Saccharomyces cerevisiae | Pc4cl, Phchs, Phchi, Pcfns | Caffeic acid | 2 | [69] | |
| Streptomyces albus | Rctal, Sc4cl, Gmchs, Gmchi, Atf3'h, Pcfns | Glucose | 0.09 | [62] | |
| Rs: Rhodobacter sphaeroides; Pl: Pueraria lobata; Tc: Trichosporon cutaneum; Ph: Petunia X hybrida; Ms: Medicago sativa; Pdp: Poplar hybrid Populus trichocarpa P. deltoides; Gm: Glycine max; Md: Malus x domestica; Hs: Huperiza serrata; Cr: Catharanthus roseus; Gh: Gerbera hybrid; Am: Astragalus mongholicus; Gu: Glycyrrhiza uralensis; Pe: Populus euramericana; Tp: Trifolium pretense. | |||||
黄烷酮主要包括柚皮素(Naringenin)、圣草酚(Eriodictyol)、甘草素(Liquiritigenin) 和根皮素(Phloretin) 等产物。PAL具有一定的底物泛杂性,它对酪氨酸也具有氨基裂解的催化活性。大肠杆菌表达PAL、4CL、查耳酮合成酶(Chalcone synthase,CHS) (图 7) 生成查耳酮,查耳酮在水相中自发形成柚皮素。引入查耳酮异构酶(Chalcone isomerase,CHI) 催化查耳酮立体异构化闭环,用高产酪氨酸的底盘大肠杆菌细胞,则实现了葡萄糖发酵合成柚皮素[50]。通过在大肠杆菌中建立柚皮素动态调控系统、优化前体酪氨酸通量和培养条件优化,无需诱导剂和前体添加,合成了588 mg/L柚皮素[51]。
与在大肠杆菌中类似,酿酒酵母表达植物来源的基因4cl、chs和chi,采用启动子工程策略,构建和筛选一系列不同强度的天然启动子优化这三步基因的表达,从对香豆酸合成了1.21 g/L柚皮素[52-53]。虽然在酿酒酵母表达P450酶比大肠杆菌表达具有一定的优势,表达PAL、C4H和CPR、4CL、CHS和CHI,经过苯丙氨酸途径,能合成柚皮素[54-55],但合成能力不及酪氨酸途径[56-57]。
除了大肠杆菌和酿酒酵母,委内瑞拉链霉菌Streptomyces venezuelae和解脂耶式酵母也被开发为柚皮素合成平台。在委内瑞拉链霉菌中表达Sc4cl、Atchs、Atchi和matBC,从对香豆酸合成柚皮素[58]。产油酵母体内具有丰富的乙酰CoA和丙二酰CoA以及疏水性环境,在合成黄酮产物上具有显著的优势。将柚皮素途径引入解脂耶式酵母底盘[59],通过筛选高活性酶、提高限速步骤CHS的表达拷贝和前体代谢流,在3 L发酵罐中,柚皮素产量达898 mg/L[45]。
从结构上看,圣草酚(Eriodictyol)是3′-羟基-柚皮素。有两种策略实现圣草酚的微生物合成。第一种策略是以咖啡酸为前体,在酿酒酵母[60]中表达4CL、CHS、CHI,合成圣草酚(图 7),但产量较低。第二种策略是在柚皮素途径基础上再表达黄酮-3′-羟化酶(Flavonoid 3′-hydroxylase,F3′H)和CPR,构成圣草酚途径(图 7)。由于途径中F3′H是细胞色素P450酶,采用截短跨膜区域和融合表达策略,在大肠杆菌中表达F3′H和CPR的融合蛋白,显著提高了F3′H在细胞内环境中的活性,以酪氨酸为底物合成出圣草酚[61]。此外,在解脂耶式酵母[59]和白色链霉菌Streptomyces albus[62]中也实现了从头合成圣草酚。由于白色链霉菌是原核细胞,直接表达F3′H存在困难,产量很低。因此,选择真核宿主酵母和加强F3′H的表达效率有利于圣草酚产量提高。
与合成圣草酚的思路相反,在柚皮素途径中表达查耳酮还原酶(Chalcone reductase,CHR),则可构成甘草素(5-去羟基-柚皮素) 途径(图 7)。由于CHS催化生成的四酮中间体在水溶液中极易转化为柚皮素,需要加快中间体向CHR的传递。在大肠杆菌中构建TAL、4CL、CHS、CHR、CHI的代谢途径,将CHS和CHR融合表达,甘草素产量提高了约4倍[63]。对酪氨酸途径改造,利用酵母内源辅因子,从头合成了甘草素,但产量仍然很低,副产物柚皮素积累严重[64-65]。甘草素途径在大肠杆菌和酿酒酵母中都表现较低的效率,主要问题是生成了副产物柚皮素,需要进一步筛选高活性的CHR酶,优化整个途径中酶的表达方式。
根皮素的合成途径与前4种产物不同,需要表达双键还原酶(Double bond reductase,DBR) 催化对香豆酰CoA,生成p-二氢香豆酰CoA,然后在CHS作用下生成根皮素,而无需CHI催化闭环反应(图 7)。根皮素的合成主要在酿酒酵母中,鲜见在大肠杆菌中合成的报道。酵母内源性的TSC13具有DBR活性,表达AtPAL2、AmC4H、CPR1、At4CL2、浆果金丝桃Hypericum androsaemum HaCHS,实现了从头合成根皮素[66]。根皮素合成途径中的异源酶比柚皮素和圣草酚途径少,但产量却较低,主要问题是生成副产物柚皮素和脱轨产物二氢-双去甲基-洋高宁(Dihydro-bisnoryangonin,2H-BNY)、二氢-香豆酰三乙酸内酯(Dihydro- coumaroyl triaceticacid lactones,2H-CTAL)。对8种植物来源的CHS筛选表明,金丝桃的CHS合成根皮素的产量最高,苹果和梨的CHS倾向于合成柚皮素而不是根皮素[66]。对14种植物来源的CHS进行筛选结果表明[67],灯盏花的CHS合成根皮素的能力最高,降低了脱轨产物的比例。
从黄烷酮途径出发,在微生物中异源表达一步异黄酮合成酶(Isoflavone synthase,IFS) 和CPR,则生成异黄酮;异源表达黄酮合成酶(Flavone synthase,FNS),则合成黄酮产物(图 7)。
IFS属于细胞色素P450酶,催化黄烷酮B环从C2向C3迁移。在酿酒酵母中,将整条染料木素(Genistein) 途径(PAL、C4H、4CL、CHS、CHI、IFS、CPR) 导入酵母,从头合成染料木素的产量极低,即使培养基添加苯丙氨酸、对香豆酸或柚皮素前体,也只能检测到0.1–7.7 mg/L的产物[54] (表 5)。在大肠杆菌中表达IFS和CPR,染料木素的产量较低,这与大肠杆菌中P450酶表达差是一致的。为增强IFS的催化效率,将N端跨膜域替换为哺乳动物来源的亲水性序列(MALLLAVF),截除CPR的跨膜域,在大肠杆菌中融合表达IFS-CPR,提高了染料木素产量[68]。虽然IFS催化甘草素生成大豆苷元(Daidzein),但未见从葡萄糖合成大豆苷元的报道。
由FNS分别催化柚皮素和圣草酚,生成黄酮产物芹菜素(Apigenin) 和木犀草素(Luteolin)。FNS酶有两种类型,一类是α-酮戊二酸双加氧酶,另一类是NADPH依赖的P450单加氧酶。这两类酶在酵母菌和大肠杆菌中,都能合成芹菜素[69-70],但α-酮戊二酸双加氧酶类FNS更适合黄酮途径的异源构建。木犀草素既可以经过圣草酚合成[71],也可以从芹菜素合成[62, 69]。
4.2 黄酮醇黄酮醇产物包括山萘酚(Kaempferol)、槲皮素(Quercetin)、杨梅黄酮(Myricetin) 和菲瑟酮(Fisetin) 等。从黄烷酮出发,经过一步或几步黄烷酮羟化酶(F3H、F3′H、F3′5′H) 催化生成二氢黄酮醇,然后在黄烷酮合酶(Flavonol synthase,FLS) 作用下合成黄酮醇(图 8)。其中F3′H和F3′5′H属于细胞色素P450酶。
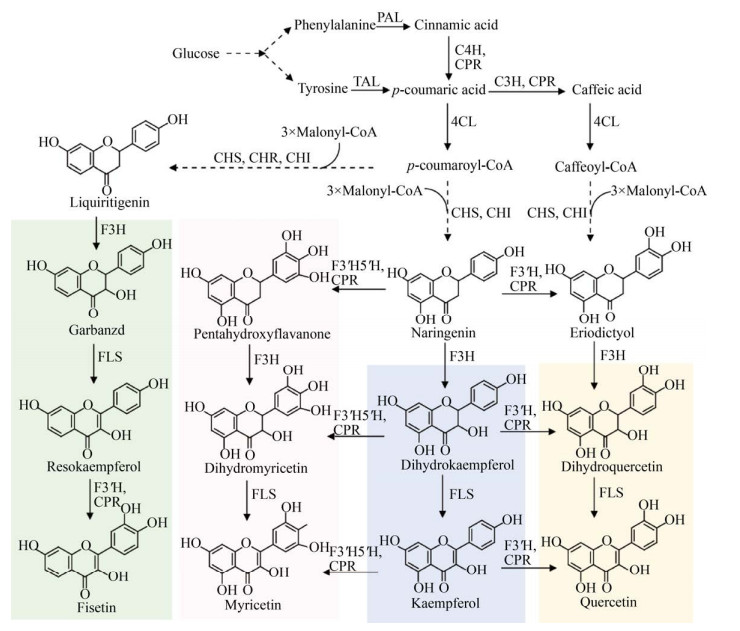
|
| 图 8 黄酮醇的微生物合成途径 Fig. 8 Biosynthetic pathways of flavonols. F3H: Flavanone-3-hydroxylase; F3′5′H: Flavonoid-3′, 5′-hydroxylase; FLS: Flavonol synthase. |
| |
在柚皮素途径基础上再表达F3H、FLS两步酶,则生成山萘酚。在大肠杆菌中,表达4CL、CHS、CHI、F3H、FLS,添加对香豆酸底物,合成了产物山萘酚[72] (表 6)。然而检测到大量中间产物二氢山萘酚的积累,可见FLS是限制性因素。通过筛选F3H和FLS酶、设计融合蛋白、增加基因拷贝数和优化发酵参数,以柚皮素为前体,合成了1.18 g/L山萘酚[73]。在酿酒酵母中,筛选5种植物来源的FLS,其中美洲黑杨、拟南芥、柑橘的FLS兼具F3H功能,从对香豆酸合成山萘酚,同时减少了副产物的积累[74]。改造乙酸利用途径,或强化酪氨酸途径,异源表达山萘酚途径,实现了酵母中从葡萄糖合成山萘酚[65, 74]。筛选F3H和FLS表达组合,敲除苯乙醇竞争途径、添加前体以及优化碳源等策略[74],可进一步提高山萘酚的产量。
| Products | Microbe | Genes | Substrate | Titer (mg/L) | References |
| Kaempferol | Escherichia coli | Pc4cl, Phchs, Phchi, Mdf3h, Atfls | p-coumaric acid | 0.3 | [72] |
| Escherichia coli | Csf3h, Cufls | Naringenin | 1 180 | [73] | |
| Saccharomyces cerevisiae | Ebpal, Ebc4h, Eb4cl, Ebchs, Ebchi, Atf3h, Pdfls, adh2, ald6, acs, acc1 | Glucose | 66.2 | [74] | |
| Saccharomyces cerevisiae | Fjtal, Pc4cl, Phchs, Mschi, Amf3h, Atfls | Glucose | 26.5 | [65] | |
| Streptomyces albus | Rctal, Sc4cl, Gmchs, Gmchi, Atfls, Pcf3h | Glucose | 0.06 | [75] | |
| Quercetin | Escherichia coli | Pc4cl, Phchs, Phchi, Mdf3h, Atfls, Crf3'h, Crcpr | p-coumaric acid | 0.05 | [72] |
| Saccharomyces cerevisiae | Ppdpal, Ppdcpr, Gmc4h, Gm4cl, Gmchs, Gmchi, Gmf3h, GmF3′H, Stfls | Glucose | 0.3 | [54] | |
| Saccharomyces cerevisiae | Pc4cl, Phchs, Phchi, Phf3h, Pdfls | Caffeic acid | 10 | [74] | |
| Saccharomyces cerevisiae | Fjtal, Pc4cl, Phchs, Mschi, Amf3h, Atfls, Phf3'h, Crcpr | Glucose | 20.3 | [65] | |
| Streptomyces coelicolor | Rctal, Sc4cl, Gmchs, Gmchi, Atfls, Atf3'h, Pcf3h | Glucose | 0.03 | [75] | |
| Streptomyces albus | Rctal, Sc4cl, Gmchs, Gmchi, Atfls, Atf3'h, Pcf3h | Glucose | 0.10 | [75] | |
| Myricetin | Escherichia coli | Pc4cl, Phchs, Phchi, Mdf3h, Atfls, Crf3'h, Crcpr | Naringenin | 0.01 | [72] |
| Streptomyces coelicolor | Rctal, Sc4cl, Gmchs, Gmchi, Atfls, Pcf3h, Phf3'h | Glucose | 0.03 | [75] | |
| Streptomyces albus | Rctal, Sc4cl, Gmchs, Gmchi, Atfls, Pcf3h, Phf3'h | Glucose | 0.04 | [75] | |
| Fisetin | Escherichia coli | Rstal, Pc4cl, Phchs, Amchr, Mschi, Atf3h, Phf3'h, Atfls, Crcpr | Tyrosine | 0.3 | [63] |
| Saccharomyces cerevisiae | Fjtal, Pc4cl, Phchs, Mschi, Amchr, Amf3h, Phf3'h, Atfls, Crcpr | Glucose | 2.3 | [54] | |
| Cs: Citrus sinensis; Cu: Citrus unshiu; St: Solanum tuberosum; Eb: Erigeron breviscapus; Pd: Populus deltoides. | |||||
槲皮素是山萘酚的3′-羟基化产物。在山萘酚途径中,表达F3′H酶和CPR,以柚皮素或对香豆酸为底物或前体,酿酒酵母可合成槲皮素。也可以咖啡酸为中间产物,构建工程酿酒酵母,合成槲皮素[74]。将槲皮素途径引入高产酪氨酸的酵母底盘中,融合表达F3′H和CPR,从葡萄糖合成了槲皮素[54]。此外,已报道白色链霉菌和天蓝色链霉菌Streptomyces coelicolor从头合成槲皮素[75],但效率远远低于酵母菌。
杨梅黄酮是山萘酚的3′-和5′-二羟基化产物,由F3′5′H催化山萘酚生成。F3′5′H已经在大肠杆菌、白色链霉菌和天蓝色链霉菌中实现功能性表达,合成了杨梅黄酮[72, 75],但未见在酿酒酵母中的报道。
在甘草素途径基础上,依次经过F3H、FLS和F3′H催化反应,大肠杆菌利用酪氨酸底物,合成了菲瑟酮[63];而酿酒酵母则实现了以葡萄糖为底物合成菲瑟酮[54]。
4.3 黄酮衍生物黄酮类经后修饰反应,包括糖基化、异戊烯基化、甲基化等形成黄酮衍生物(图 9)。在大肠杆菌和酿酒酵母中合成黄酮糖苷、异戊二烯柚皮素和甲基柚皮素等产物(表 7)。

|
| 图 9 黄酮糖苷的微生物合成途径 Fig. 9 Biosynthetic pathways of flavonoid glycosides. F6H: flavone-6-hydroxylase; F7GAT: flavonoid-7-O- glucuronosyltransferase; DFR: dihydroflavonol-4-reductase; ANS: anthocyanidin synthase; LAR: leucoanthocyanidin reductase; 3GT: anthocyanidin-3-O-glycosyltransferase; OGT: O-UDP-glycosyltransferase; CGT: C-glycosyltransferase. |
| |
| Products | Microbe | Genes | Substrate | Titer (mg/L) | References |
| Glycosylation | |||||
| Phlorizin | Saccharomyces cerevisiae | Atpal, Amc4h, Sccpr, At4cl, Hachs, Sctsc13, Pycugt88F2 | Glucose | 65 | [66] |
| Nothofagin | Saccharomyces cerevisiae | Atpal, Amc4h, Sccpr, At4cl, Hachs, Sctsc13, Oscgt | Glucose | 59 | [66] |
| Trilobatin | Saccharomyces cerevisiae | Atpal, Amc4h, Sccpr, At4cl, Hachs, Sctsc13, Atugt73B2 | Glucose | 32.8 | [66] |
| Pelargonidin-3-O-glucoside | Saccharomyces cerevisiae | Atpal Amc4h, Sccpr, Hachs, Mschi, At4cl, Mdf3h, Aadfr, Phans, Dca3gt | Glucose | 0.8 | [77] |
| Cyanidin-3-O-glucoside | Saccharomyces cerevisiae | Atpal, Amc4h, Sccpr, Hachs, Mschi, At4cl, Mdf3h, Phf3'h, Ptdfr, Phans, Faa3gt | Glucose | 1.5 | [77] |
| Delphinidin-3-O-glucoside | Saccharomyces cerevisiae | Atpal, Amc4h, Sccpr, Hachs, Mschi, At4cl, Mdf3h, Slf3'5'h, Ihdfr, Phans, Faa3gt | Glucose | 1.8 | [77] |
| Apigenin-7-O-glucuronide | Saccharomyces cerevisiae | Ebpal, Ebc4h, Eb4cl, Ebchs, Ebchi, Ebfns, Ebf7gat | Glucose | 185 | [78] |
| Scutellarin | Saccharomyces cerevisiae | Ebpal, Ebc4h, Eb4cl, Ebchs, Ebchi, Ebfns, Ebf6h, Ebf7gat | Glucose | 105 | [78] |
| Liquiritin | Saccharomyces cerevisiae | Gupal, Guc4h, Gu4cl, Guchs, Guchr, Guchi, Guugt | Glucose | 0.4 | [64] |
| Prenylation | |||||
| 8-prenylnaringenin | Saccharomyces cerevisiae | Atpal, Atc4h, Atcpr, At4cl, Atchs, Atchi, Sffpt | Glucose | 0.1 | [79] |
| Methylation | |||||
| Ponciretin | Escherichia coli | Setal, Os4cl, Pechs, Gmsomt | Glucose | 42.5 | [80] |
| Sakuranetin | Escherichia coli | Setal, Os4cl, Pechs, Osnomt | Glucose | 40.1 | [80] |
| Pyc: Pyrus communis; Aa: Anthurium andraeanum; Pt: Populus trichocarpa; Ih: Iris hollandica; Sl: Solanum lycopersicum; Dc: Dianthus caryophyllus; Fa: Fragaria ×ananassa; Sf: Sophora flavescens. | |||||
黄酮糖基化产物是目前研究最多的一类黄酮衍生物,由糖基转移酶催化活化的糖基从供体转移到黄酮苷元上形成,如槲皮素糖苷、根皮素糖苷和花色苷、芹菜素糖苷。槲皮素含有5个羟基(3-OH、5-OH、7-OH、3′-OH、4′-OH),均可发生O-糖基化反应,大肠杆菌表达糖基转移酶,以槲皮素为底物,已经实现了多种槲皮素糖苷的合成[76]。这里主要介绍根皮素糖苷、花色苷、芹菜素糖苷和甘草苷的微生物合成。
根皮素的糖苷包括根皮苷(Phlorizen)、诺瑟法根(Nothofagin) 和三叶苷(Trilobatin),其糖基转移酶已被鉴定出来。在合成根皮素的酿酒酵母菌中[66],表达苹果Malus x domestica MdUGT88A1或西洋梨Pyrus communis PcUGT88F2,催化根皮素2′位发生O-糖基化,合成根皮苷;表达水稻OsCGT,催化3′位发生C-糖基化,合成诺瑟法根;表达拟南芥AtUGT73B2,催化4′位O-糖基化,合成了三叶苷。
花色苷是花青素的糖基化形式,通过拓展黄酮醇合成途径,异源表达F3H、二氢黄酮醇-4-还原酶(Dihydroflavonol-4-reductase,DFR)、花青素合成酶(Anthocyanidin synthase,ANS) 和花青素-3-O-葡萄糖基转移酶(Anthocyanidin-3-O- glycosyltransferase,A3GT),可在微生物中重构花色苷合成途径。另一条合成途径由F3H、DFR、无色花色素还原酶(Leucoanthocyanidin reductase,LAR)、ANS、3GT组成,经过黄烷-3-醇合成花色苷。由于花青素合成过程中经历多步CYP450酶,ANS的底物谱宽泛,花青素及其糖苷在酸性条件下稳定(pH≤5),在中性条件下会迅速降解,因此多以酿酒酵母为宿主合成花色苷。表达花青素人工途径,测试了2种F3′H、4种F3′5′H、5种DHF、9种糖基转移酶,对6步途径酶筛选优化的基础上,从头合成了微量的天竺葵素-3-O-葡萄糖苷(Pelargonidin-3-O-glucoside)、矢车菊素-3- O-葡萄糖苷(Cyanidin-3-O-glucoside) 和飞燕草素-3-O-葡萄糖苷(Delphinidin-3-O-glucoside) 等花色苷[77]。
灯盏花素(Scutellarin) 是灯盏花(Erigeron breviscapus) 中药的主要活性成分,临床上用于治疗脑供血不足。采用基因组、转录组分析和高通量筛选技术,从灯盏花中挖掘和鉴定了灯盏花素合成途径的两步关键酶:黄酮-6-羟化酶(Flavone-6- hydroxylase,F6H) 和黄酮-7-O-葡萄糖醛酸转移酶(Flavonoid-7-O-glucuronosyltransferase,F7GAT),在工程酿酒酵母中,从葡萄糖合成了芹菜素-7-O-糖苷(Apigenin-7-O-glucuronide) 和灯盏花素[78]。甘草苷(Liquiritin) 是濒危药材甘草的活性成分,其微生物合成备受关注。在甘草素途径基础上,表达糖基转移酶,酿酒酵母合成了甘草苷[64]。
相对于黄酮糖苷,异戊烯基化和甲基化黄酮产物较少,而且大多数的转移酶未被鉴定出来,限制其在微生物中的合成。目前,在合成柚皮素的酿酒酵母中,表达苦参Sophora flavescens的异戊二烯基转移酶(Flavonoid prenyltransferase) SfFPT,从头合成了8-异戊二烯柚皮素(8-prenylnaringenin)[79]。在大肠杆菌中,通过改造莽草酸途径和柚皮素途径,表达两种甲基转移酶,分别合成了樱花素(7-O-甲基柚皮素,Sakuranetin)和异樱花素(7′-O-甲基柚皮素,Ponciretin)[80]。
5 微生物合成多酚类的调控工程 5.1 前体代谢工程芳香氨基酸是微生物异源合成多酚产物的前体之一。芳香氨基酸代谢途径清楚,已经建立了多种策略,在大肠杆菌和酿酒酵母等底盘细胞中实现了酪氨酸和苯丙氨酸的高效合成[16, 18, 51, 52],因此不再赘述。这里讨论多酚合成的另一个底物丙二酰CoA的代谢调控。
在黄酮、白藜芦醇、姜黄素等生物合成过程中,都需要3分子丙二酰CoA作为二碳单元的链延伸底物。微生物细胞内丙二酰CoA浓度低,是多酚合成的一个主要限速因子。丙二酰CoA由乙酰CoA羧化酶(Acetyl-CoA carboxylase,ACC) 催化乙酰-CoA合成,在细胞中主要用于脂肪酸合成。通过开源节流的方式提高胞内丙二酰CoA的水平,进而达到优化多酚合成途径的目的(表 8)。
| Products | Microbe | Pathway genes | Substrate | Strategies | Titer (mg/L) | References |
| Resveratrol | Escherichia coli | Le4cl, Ahsts | p-coumaric acid | Cgacc | 171 | [81] |
| Escherichia coli | At4cl, Vvsts | p-coumaric acid | OptForce | 1 600 | [88] | |
| Naringenin | Escherichia coli | Rrpal, Sc4cl, Gechs, Plchi | Tyrosine | Cgacc | 60 | [82] |
| Escherichia coli | Pc4cl, Phchs, Phchi | p-coumaric acid | Placc, PlBirA and acs | 119 | [83] | |
| Escherichia coli | Rgtal, Pc4cl, Phchs, Mschi | Glucose | matBC | 84 | [50] | |
| Escherichia coli | Pc4cl, Phchs, Mschi | p-coumaric acid | matBC, adding cerulenin | 186 | [71] | |
| Escherichia coli | Rgtal, Pc4cl, Phchs, Mschi | Tyrosine | matBC, anti-fabB/fabF | 391 | [84] | |
| Escherichia coli | Rgtal, Pc4cl, Phchs, Mschi | Tyrosine | CRISPRi regulation | 421 | [85] | |
| Escherichia coli | Pc4cl, Phchs, Mschi | p-coumaric acid | OptForce | 474 | [87] | |
| Eriodictyol | Escherichia coli | Pc4cl, Phchs, Phchi | Caffeic acid | Placc, PlBirA and acs | 52 | [83] |
| Escherichia coli | Rgtal, Pc4cl, Phchs, Mschi, Ghf3'h, Crcpr | Tyrosine | Acc, Acs, ΔAckA | 107 | [61] | |
| Escherichia coli | Pc4cl, Phchs, Mschi | Caffeic acid | matBC, adding cerulenin | 54 | [71] | |
| Phloretin | Saccharomyces cerevisiae | Ha4cl, Ebchs | Dihydro-p-coumaric acid | adh2, ald6, acs, acc1, matBC | 619.5 | [67] |
开源丙二酰CoA的方式是增加其供应。第一种策略是过表达ACC。大肠杆菌的ACC由4个亚基组成,且其表达受酰基载体蛋白的反馈抑制。而谷氨酸棒杆菌的CgACC只有两个亚基(AccBC和DtsR1),最常用来提高多酚产量如白藜芦醇、柚皮素等[81-82]。ACC是一种生物素依赖酶,将ACC和生物素连接酶BirA在大肠杆菌中共表达,使得黄烷酮的产量提高了11.6倍[83]。另一种策略是改造乙酸同化途径,包括削弱乙酸和乙醇积累、干扰柠檬酸循环、强化葡萄糖到丙酮酸的代谢流等。协同过表达脂酰CoA合酶(Acyl-CoA synthetase,Acs)、敲除乙酸激酶(Acetate kinase,AckA) 和过表达CgACC,使得大肠杆菌胞内丙二酰CoA浓度提高了16.3倍[61]。第三种策略是构建丙二酸转化途径,包括丙二酰合酶(Malonate synthetase,MatB) 基因和丙二酸转运蛋白(Malonate carrier protein,MatC) 基因,把培养基中的丙二酸物转运到胞内,并转化为丙二酰CoA,用于多酚的合成。采用此策略,大肠杆菌合成柚皮素和圣草酚产量分别提高了2.69倍和3.55倍[71]。在合成根皮素的过程中,联合两种策略,将乙醇脱氢酶adh2 (Alcohol dehydrogenase)、乙醛脱氢酶ald6 (Aldehyde dehydrogenase)、acs和acc1整合到酵母染色体上,同时质粒过表达matBC,根皮素产量提高约1.3倍[67]。第四种策略是利用β-氧化途径。过表达与过氧化物酶体的形成相关蛋白PEX10,促进脂肪酸的β-氧化形成乙酰CoA,提高了解脂耶式酵母合成柚皮素和白藜芦醇的能力[45]。
节流丙二酰CoA的方式是弱化脂肪酸合成途径,将丙二酰CoA导流到多酚合成途径。第一种策略是培养基中添加浅蓝菌素(Cerulenin),浅蓝菌素可有效抑制脂肪酸合酶FabB和FabF (β-ketoacyl-ACP synthase Ⅰ/Ⅱ) 的活性。谷氨酸棒杆菌在最适浅蓝菌素剂量下的白藜芦醇产量提升了13倍[39]。第二种策略是利用反义RNA (anti-RNA) 的基因沉默技术,微调脂肪酸代谢途径,平衡细胞生长和黄酮类产物合成对丙二酰CoA需求[84],大肠杆菌的柚皮素产量提高了4.31倍。第三种策略是CRISPRi系统,它调控范围广,涉及多个内源性基因,效应更显著。用CRISPRi系统调控乙酰CoA途径的17个基因和脂肪酸途径的7个基因,抑制ppsA、eno、adhE、mdh、fumC、sdhA、sucC、citE、fabH、fabB、fabF和fabI等12个靶标基因,不显著影响细胞生长的同时提高细胞内丙二酰CoA的水平,使柚皮素产量提高了7.4倍,达到421 mg/L[85]。综合表达CgACC、anti-RNA、促进产物外排,提高了白藜芦醇的产量[86]。
随着计算生物学的发展,利用微生物代谢预测算法指导敲除或过表达靶标基因,提高胞内丙二酰CoA的水平。运用OptForce模型,在大肠杆菌中过表达了acc、pgk、pdh基因,同时敲除fumC、suuC基因,与野生型菌株相比,丙二酰CoA水平提高了3.7倍,柚皮素产量达到474 mg/L[87]。该模型也成功用于白藜芦醇途径调节和合成[88]。
5.2 途径动态调控工程简单地过表达途径中的基因,可以有效提高细胞的生产效能,但这种静态调控策略无法实时调控基因的表达水平,且易受发酵环境的干扰。动态调控策略可以使微生物根据细胞中代谢物积累水平自动调节途径的表达强度,从而适应不断变化的细胞内、外环境。动态调控一般由信号输入、生物传感器、信号输出3部分组成,构建生物传感器是实现动态调控的关键。目前已经开发了多种响应代谢物的生物传感器,用于改善合成途径中关键酶的活性或获取高效重组菌。
以恶臭假单胞菌的调节蛋白TtgR作为白藜芦醇的传感器,设计了TtgR结合的启动子,以红色荧光蛋白为效应信号,构建4CL随机突变文库,筛选获得4CL变体,使工程大肠杆菌合成了556.4 mg/L白藜芦醇,提高了4.7倍。该突变体还适用于优化柚皮素的产量,大肠杆菌合成了225 mg/L柚皮素[89]。RppA催化5分子的丙二酰CoA生成红色的淡黄霉素,可用作胞内丙二酰CoA的丰度传感器,且具有较宽的动态响应范围。将RppA与大肠杆菌基因组规模的anti-RNA文库结合起来,筛选出了14个增强丙二酰CoA的靶基因,用于指导优化工程大肠杆菌构建,白藜芦醇的产量为51.8 mg/L,提高了4.2倍[90]。类似地,基于柚皮素与AlCl3的黄色反应建立生物传感器,通过迭代高通量筛选组成型启动子,对柚皮素生物合成途径基因表达进行优化,工程大肠杆菌合成191.9 mg/L柚皮素[91]。
偶联细胞生长的动态调节是一种较先进的模式,可用于优化多酚合成。积累的柚皮素激活转录调节激活子FdeR,进而抑制酰基载体蛋白基因的表达,减少丙二酰CoA进入脂肪酸途径;当对香豆酸积累时,阻遏物PadR失活,开启acs和acc基因表达,从而增强丙二酰CoA的合成供应;这样建立了柚皮素-对香豆酸-丙二酰CoA的动态调控系统,并通过定向进化启动子,细胞生长得到改善,柚皮素产量达到251.4 mg/L,提高了8.4倍[92]。
群体效应(Quorum-sensing,QS) 是一种依赖细胞自身信号的传感系统,也可用于代谢途径调控。在大肠杆菌中表达两种细菌QS,设计两个开关:Lux动态沉默脂肪酸途径基因以增加胞内丙二酰CoA的水平,Esa动态延迟TAL-4CL的表达以解除对香豆酰CoA对TAL的毒性作用;通过双重QS系统调控,柚皮素产量为125.9 mg/L,提高了10倍[93]。
5.3 共培养工程在自然界,每种微生物之间形成劳动分工的相互作用关系,构成稳定生态群落。植物多酚的生物合成途径较长,可模仿自然生态群落,将多酚合成途径分配到不同的菌株中,从而构建多菌组成的共培养系统,不仅可克服途径长、碳流效率低和还原力不足等问题,而且也可分段优化、提高构建效率。这些策略已经应用于优化白藜芦醇、姜黄素、黄酮、迷迭香酸等的微生物合成(表 9)。
| Products | Coculture system and genes | Substrate | Titer (mg/L) | References |
| Resveratrol | Escherichia coli 1: tktAfbr, aroGfbr, ΔpheA, Rgtal; Escherichia coli 2: Sc4cl, Ahsts | Glycerol | 22.6 | [94] |
| Escherichia coli 1: Δpgi, ΔtyrR, tyrAfbr, aroGfbr, Rgtal; Escherichia coli 2: Δzwf, Cgacc, Pc4cl, Ahsts | Glucose | 55.7 | [95] | |
| Escherichia coli: ΔtyrR, aroGfbr, tyrAfbr, Tctal; Saccharomyces cerevisiae: acc1S659A, S1157A, At4cl, Vvsts | Glucose | 36.0 | [96] | |
| Curcumin | Escherichia coli 1: Rgtal, Sec3h, Atcomt; Escherichia coli 2: At4cl, Cldcs, Clcurs | Tyrosine | 15.9 | [97] |
| Afzelechin | Escherichia coli 1: ΔsucC, ΔfumC, At4cl, Phchs, Cmchi; Escherichia coli 2: Csf3h, Fadfr, DuLAR | p-coumaric acid | 40.7 | [98] |
| Naringenin | Escherichia coli 1: Rgtal; Escherichia coli 2: Rgtal, Pc4cl, Phchs, Mschi | Glucose | 41.5 | [99] |
| Escherichia coli: ΔptsG, ΔtyrR, ΔpykA, ΔpykF, ΔpheA; Saccharomyces cerevisiae: Rttal, Pc4cl, Phchs, Phchi | Xylose | 21.2 | [100] | |
| Genistein | Escherichia coli: Rrpal, Sc4cl, Gechs, Plchi, Cgacc; Saccharomyces cerevisiae: Geifs | Tyrosine | 5.8 | [101] |
| Kaempferol | Saccharomyces cerevisiae 1: aro4K229L, Fjtal, At4cl, Hachs, Phchi; Saccharomyces cerevisiae 2: Atfls, Csf3h | Glucose | 200.0 | [102] |
| Saccharomyces cerevisiae 1: Δaro10, Δpdc5, Δtsc13, aro4K229L, aro7G141S, acc1, Ecpdh, Fjtal, At4cl, Atchs, Atchi; Saccharomyces cerevisiae 2: Ntf3h, Atfls | Glucose | 168.1 | [57] | |
| Quercetin | Saccharomyces cerevisiae 1: Δaro10, Δpdc5, Δtsc13, aro4K229L, aro7G141S, acc1, Ecpdh, Fjtal, At4cl, Atchs, Atchi; Saccharomyces cerevisiae 2: Ntf3h, Phf3'h, Atfls | Glucose | 154.2 | [57] |
| Myricetin | Saccharomyces cerevisiae 1: Δaro10, Δpdc5, Δtsc13, aro4K229L, aro7G141S, acc1, Ecpdh, Fjtal, At4cl, Atchs, Atchi; Saccharomyces cerevisiae 2: Ntf3h, slf3’5’h, Phf3'h, Atfls | Glucose | 145.0 | [57] |
| Pelargonidin | Saccharomyces cerevisiae 1: Δaro10, Δpdc5, Δtsc13, aro4K229L, aro7G141S, acc1, Ecpdh, Fjtal, At4cl, Atchs, Atchi; Saccharomyces cerevisiae 2: Ntf3h, Aadfr, Ghans | Glucose | 33.3 | [57] |
| Cyanidin | Saccharomyces cerevisiae 1: Δaro10, Δpdc5, Δtsc13, aro4K229L, aro7G141S, acc1, Ecpdh, Fjtal, At4cl, Atchs, Atchi; Saccharomyces cerevisiae 2: Ntf3h, Phf3'h, Aadfr, Ghans | Glucose | 31.7 | [57] |
| Delphinidin | Saccharomyces cerevisiae 1: Δaro10, Δpdc5, Δtsc13, Fjtal, At4cl, Atchs, Atchi, acc1, Ecpdh, aro4K229L, aro7G141S; Saccharomyces cerevisiae 2: Ntf3h, Phf3'h, slf3’5’h, Aadfr, Ghans | Glucose | 26.1 | [57] |
| Rosmarinic acid | Escherichia coli 1: ΔpheA, ΔtyrR, tyrAfbr, aroGfbr, ΔptsH, ΔptsI, Δcrr, ΔaroE, ΔydiB, Rgtal, hpaBC; Escherichia coli 2: ΔxylA, aroE, aroL, aroA, aroC, tyrAfbr, aroGfbr, hpaBC, Lpd-ldh; Escherichia coli 3: ΔpheA, ΔtyrR, tyrAfbr, aroGfbr, ΔptsH, ΔptsI, Δcrr, ΔaroE, ΔydiB, Pc4cl, MoRAS | Glucose | 172.0 | [103] |
| Pelargonidin-3- O-glucoside | Escherichia coli 1: tyrAfbr, aroGfb, Rgtal; Escherichia coli 2: ΔsumC, ΔfumC, ΔpheA, ΔtyrR, tyrAfbr, aroGfb, At4CL, Phchs, Cmchi, RtmatBC; Escherichia coli 3: Csf3h, Fadfr, DuLAR; Escherichia coli 4: Phans, At3gt | Glucose | 9.5 | [104] |
白藜芦醇合成途径中,一株大肠杆菌以葡萄糖为底物合成对香豆酸,另一株大肠杆菌负责供应丙二酰CoA,合成了白藜芦醇[94]。也可将异源途径与内源途径偶联起来,一株菌通过磷酸戊糖途径合成对香豆酸,第二株菌通过糖酵解途径提供丙二酰CoA,平衡柠檬酸循环和丙二酰CoA供给,合成白藜芦醇[95]。除了同物种共培养系统,大肠杆菌合成的对香豆酸,转运到酿酒酵母中合成白藜芦醇,由大肠杆菌-酿酒酵母组成的共培养系统实现了无诱导剂的白藜芦醇从头合成[96]。
姜黄素异源合成途径包括6步反应,两株大肠杆菌分别表达阿魏酸模块和姜黄素模块,总姜黄素和姜黄素的产量与单一菌株相比,分别提高6.6倍和2.6倍[97]。将阿福豆素(Afzelechin) 合成途径分解为依赖丙二酰CoA的上游模块(4CL、CHS、CHI) 和依赖NADPH的下游模块(F3H、DFR、LAR),利用大肠杆菌-大肠杆菌共培养策略,阿福豆素产量是单菌培养的970倍[98]。
途径中不同基因表达需要不同的微环境,选择不同种微生物进行共培养,可提供多元化的酶反应场所。以葡萄糖为碳源,两株大肠杆菌共培养合成柚皮素时存在竞争[99],不同微生物的混合培养是一种新策略。以木糖为碳源,在大肠杆菌和酿酒酵母之间建立起共存的关系,大肠杆菌合成酪氨酸并分泌乙酸,为利用乳酸酿酒酵母提了供前体和碳源,大肠杆菌-酿酒酵母混菌与单一酵母菌合成柚皮素相比产量提高了8倍[100]。在染料木素合成中,将柚皮素途径在大肠杆菌中表达,P450酶IFS分配到酿酒酵母中表达,两种菌株共转化酪氨酸底物,改善了染料木素的合成效率[101]。
在两株酿酒酵母共培养系统中,第一个工程菌负责从葡萄糖合成柚皮素,第二个工程菌负责从柚皮素合成山萘酚,较好地解决了中间产物积累问题[102]。类似地,让第二个酵母表达柚皮素的下游模块,该共培养系统也成功应用于槲皮素、杨梅黄酮、天竺葵素和矢车菊素、飞燕草素等5种黄酮类产物的合成[57]。
三菌共培养系统可用于平衡非线性-汇聚式的代谢途径。在迷迭香酸异源合成途径中,丹参素途径和咖啡酸途径同时竞争酪氨酸前体,且途径酶存在底物交叉反应,单菌合成效率很低。将迷迭香酸合成途径划分为3个模块,咖啡酸模块、丹参素模块和迷迭香酸模块,分别置于3个独立的大肠杆菌中,共培养的迷迭香酸产量较单菌培养提高了38倍,达到172 mg/L[103]。
四菌共培养适合于较长的代谢途径。矢车菊素-3-O-葡萄糖苷的途径包括9个外源基因,酶催化效率低下且表达多个基因给细胞带来严重的代谢负担。将花青素途径分成4个模块,分别是酪氨酸模块、柚皮素模块、黄烷醇模块和花青素模块,构建了4株大肠杆菌的共培养体系,首次在大肠杆菌中实现了花青素的异源从头合成[104]。
6 总结与展望植物多酚的微生物合成研究,早期是培养基中添加咖啡酸、肉桂酸、对香豆酸等,从植物中间代谢物出发合成产物。随后是衔接到微生物代谢节点,如酪氨酸和苯丙氨酸,打通了植物多酚的微生物合成途径。葡萄糖是发酵工业的廉价原料,经过十几年的发展,终于在模式微生物大肠杆菌和酿酒酵母中建立了植物多酚骨架结构的从头合成途径,先后实现了咖啡酸、丹参素、迷迭香酸、白藜芦醇、姜黄素、黄烷酮、黄酮醇、异黄酮等的合成。同时部分多酚的糖基化、甲基化、异戊烯基化等衍生物也被合成,为深度后修饰多酚产物奠定了基础。
近年来,植物多酚的异源合成宿主已经拓展到谷氨酸棒杆菌、链霉菌、解脂酵母等微生物,期待着未来深入开发和应用。谷氨酸棒杆菌是生产氨基酸的优质微生物,基因组相对较小;而链霉菌具有强大的丙二酰CoA供给能力,但芳香氨基酸合成途径较弱,今后可改造提高酪氨酸途径通量,应用于多酚的合成。这些革兰氏阳性工业细菌没有脂多糖和内毒素的困扰,从生物工程下游制备产品的角度看,它们优于革兰氏阴性细菌大肠杆菌。解脂酵母的芳香氨基酸合成能力与酿酒酵母相当,而丙二酰CoA供给能力优于酿酒酵母,柚皮素等产物的合成能力似乎超过了模式微生物,是极具潜力的多酚合成宿主。毕赤酵母Pichia pastoris是表达异源蛋白的良好宿主,具有高密度发酵能力,而且没有乙醇抑制效应,有待开发成为多酚产物合成宿主。
在黄酮类产物的微生物合成中,由于CHS的底物非特异性,催化缩合不同数量丙二酰CoA生成副产物和脱轨产物,成为CHS酶催化反应中共同性的问题。植物细胞合成黄酮的研究结果表明,CHIL (CHI-like) 蛋白能够与CHS发生相互作用,形成复酶体,增强催化效率[105],同时纠正CHS的功能,减少副产物产生[106]。深入研究CHIL-CHS之间的适配性,将是减少副产物和脱轨产物生成的一种策略。
由于多酚的从头合成途径较长,异源途径优化和底盘适配是今后提升多酚产量的主要策略。除了常规的启动子工程、模块化工程、融合表达、前体工程用于优化合成代谢途径外,近年来发展的动态调控和共培养系统为植物多酚的微生物合成提供了新策略。目前的共培养研究仍局限于大肠杆菌和酿酒酵母。可扩大共培养微生物的种类和培养方式,根据各种微生物的生理生化特点和培养特性,对代谢途径进行合理分工,深入研究异源途径和内源代谢的相互作用,有望延长异源合成路线、扩大合成的产物范围,从头高效合成更多的植物多酚。细胞器工程已经在化学品和其他天然产物的微生物合成中展现优势[107-108],这种区域化定位表达,有可能提高酶的催化活性,协调各反应步骤之间的平衡;同时隔离非特异性底物与酶的接触,有望大幅度减少副产物和脱轨产物的生成,提高多酚产物的合成能力。
| [1] |
Quideau S, Deffieux D, Douat-Casassus C. Plant polyphenols: chemical properties, biological activities and synthesis. Angew Chem Int Ed Engl, 2011, 50(3): 586-621. DOI:10.1002/anie.201000044
|
| [2] |
Pereira DM, Valentão P, Pereira JA, et al. Phenolics: from chemistry to biology. Molecules, 2009, 14(6): 2202-2211. DOI:10.3390/molecules14062202
|
| [3] |
Yahfoufi N, Alsadi N, Jambi M, et al. The immunomodulatory and anti-inflammatory role of polyphenols. Nutrients, 2018, 10(11): 1618-1640. DOI:10.3390/nu10111618
|
| [4] |
Niedzwiecki A, Roomi MW, Kalinovsky T, et al. Anticancer efficacy of polyphenols and their combinations. Nutrients, 2016, 8(9): 552-568. DOI:10.3390/nu8090552
|
| [5] |
Bays H. Sodium glucose co-transporter type 2 (SGLT2) inhibitors: targeting the kidney to improve glycemic control in diabetes mellitus. Diabetes Ther, 2013, 4(2): 195-220. DOI:10.1007/s13300-013-0042-y
|
| [6] |
Yang C, Pan XY, Xu XF, et al. Salvianolic acid C potently inhibits SARS-CoV-2 infection by blocking the formation of six-helix bundle core of spike protein. Signal Transduct Ter, 2020, 5(1): 220.
|
| [7] |
Su HX, Yao S, Zhao WF, et al. Anti-SARS-CoV-2 activities in vitro of Shuanghuanglian preparations and bioactive ingredients. Acta Pharmacol Sin, 2020, 41(9): 1167-1177. DOI:10.1038/s41401-020-0483-6
|
| [8] |
Fraga CG, Croft KD, Kennedy DO, et al. The effects of polyphenols and other bioactives on human health. Food Funct, 2019, 10(2): 514-528. DOI:10.1039/C8FO01997E
|
| [9] |
崔可建, 蔡超, 朱才镇. 基于植物多酚构筑新型功能材料. 材料导报, 2018, 32(5): 755-764, 771. Cui KJ, Cai C, Zhu CZ. Creating novel functional materials based on plant polyphenols: a review. Mater Rev, 2018, 32(5): 755-764, 771 (in Chinese). |
| [10] |
Li YZ, Mao JW, Liu QL, et al. De novo biosynthesis of caffeic acid from glucose by engineered Saccharomyces cerevisiae. ACS Synth Biol, 2020, 9(4): 756-765. DOI:10.1021/acssynbio.9b00431
|
| [11] |
Berner M, Krug D, Bihlmaier C, et al. Genes and enzymes involved in caffeic acid biosynthesis in the actinomycete Saccharothrix espanaensis. J Bacteriol, 2006, 188(7): 2666-2673. DOI:10.1128/JB.188.7.2666-2673.2006
|
| [12] |
Zhang HR, Stephanopoulos G. Engineering E. coli for caffeic acid biosynthesis from renewable sugars. Appl Microbiol Biotechnol, 2013, 97(8): 3333-3341. DOI:10.1007/s00253-012-4544-8
|
| [13] |
Furuya T, Arai Y, Kino K. Biotechnological production of caffeic acid by bacterial cytochrome P450 CYP199A2. Appl Environ Microbiol, 2012, 78(17): 6087-6094. DOI:10.1128/AEM.01103-12
|
| [14] |
Lin YH, Yan YJ. Biosynthesis of caffeic acid in Escherichia coli using its endogenous hydroxylase complex. Microb Cell Fact, 2012, 11: 42-50. DOI:10.1186/1475-2859-11-42
|
| [15] |
Furuya T, Kino K. Catalytic activity of the two-component flavin-dependent monooxygenase from Pseudomonas aeruginosa toward cinnamic acid derivatives. Appl Microbiol Biotechnol, 2014, 98(3): 1145-1154. DOI:10.1007/s00253-013-4958-y
|
| [16] |
Huang Q, Lin YH, Yan YJ. Caffeic acid production enhancement by engineering a phenylalanine over-producing Escherichia coli strain. Biotechnol Bioeng, 2013, 110(12): 3188-3196. DOI:10.1002/bit.24988
|
| [17] |
Liu LQ, Liu H, Zhang W, et al. Engineering the biosynthesis of caffeic acid in Saccharomyces cerevisiae with heterologous enzyme combinations. Engineering, 2019, 5(2): 287-295. DOI:10.1016/j.eng.2018.11.029
|
| [18] |
Yao YF, Wang CS, Qiao J, et al. Metabolic engineering of Escherichia coli for production of salvianic acid A via an artificial biosynthetic pathway. Metab Eng, 2013, 19: 79-87. DOI:10.1016/j.ymben.2013.06.001
|
| [19] |
Zhou L, Ding Q, Jiang GZ, et al. Chromosome engineering of Escherichia coli for constitutive production of salvianic acid A. Microb Cell Fact, 2017, 16(1): 84-94. DOI:10.1186/s12934-017-0700-2
|
| [20] |
Xiong TZ, Jiang J, Bai YJ, et al. Redox self-sufficient biocatalyst system for conversion of 3, 4-dihydroxyphenyl-L-alanine into (R)- or (S)-3, 4-dihydroxyphenyllactic acid. J Ind Microbiol Biotechnol, 2019, 46(8): 1081-1090. DOI:10.1007/s10295-019-02200-7
|
| [21] |
Xiong TZ, Jiang J, Bai YJ, et al. Biosynthesis of D-danshensu from L-DOPA using engineered Escherichia coli whole cells. Appl Microbiol Biotechnol, 2019, 102(15): 6097-6105. DOI:10.1007/s00253-019-09947-0
|
| [22] |
Xiong TZ, Jia P, Jiang J, et al. One-pot, three-step cascade synthesis of D-danshensu using engineered Escherichia coli whole cells. J Biotechnol, 2019, 300: 48-54. DOI:10.1016/j.jbiotec.2019.05.008
|
| [23] |
Li C, Zhang ZD, Wang J. A thermophilic biofunctional multienzyme cascade reaction for cell-free synthesis of salvianic acid A and 3, 4-dihydroxymandelic acid. ACS Sustainable Chem Eng, 2019, 7(22): 18247-18253. DOI:10.1021/acssuschemeng.9b05496
|
| [24] |
Babaei M, Zamfir GMB, Chen X, et al. Metabolic engineering of Saccharomyces cerevisiae for rosmarinic acid production. ACS Synth Biol, 2020, 9(8): 1978-1988. DOI:10.1021/acssynbio.0c00048
|
| [25] |
Bloch SE, Schmidt-Dannert C. Construction of a chimeric biosynthetic pathway for the de novo biosynthesis of rosmarinic acid in Escherichia coli. ChemBioChem, 2014, 15(16): 2393-2401. DOI:10.1002/cbic.201402275
|
| [26] |
Jiang JJ, Bi HP, Zhuang YB, et al. Engineered synthesis of rosmarinic acid in Escherichia coli resulting production of a new intermediate, caffeoyl-phenyllactate. Biotechnol Lett, 2016, 38(1): 81-88. DOI:10.1007/s10529-015-1945-7
|
| [27] |
Zhang YS, Li SZ, Li J, et al. Using unnatural protein fusions to engineer resveratrol biosynthesis in yeast and mammalian cells. J Am Chem Soc, 2006, 128(40): 13030-13031. DOI:10.1021/ja0622094
|
| [28] |
Wang YC, Yu O. Synthetic scaffolds increased resveratrol biosynthesis in engineered yeast cells. J Biotechnol, 2012, 157(1): 258-260. DOI:10.1016/j.jbiotec.2011.11.003
|
| [29] |
Conrado RJ, Wu GC, Boock JT, et al. DNA-guided assembly of biosynthetic pathways promotes improved catalytic efficiency. Nucleic Acids Res, 2012, 40(4): 1879-1889. DOI:10.1093/nar/gkr888
|
| [30] |
Sydor T, Schaffer S, Boles E. Considerable increase in resveratrol production by recombinant industrial yeast strains with use of rich medium. Appl Environ Microbiol, 2010, 76(10): 3361-3363. DOI:10.1128/AEM.02796-09
|
| [31] |
Li MJ, Kildegaard KR, Chen Y, et al. De novo production of resveratrol from glucose or ethanol by engineered Saccharomyces cerevisiae. Metab Eng, 2015, 32: 1-11. DOI:10.1016/j.ymben.2015.08.007
|
| [32] |
Li MJ, Schneider K, Kristensen M, et al. Engineering yeast for high-level production of stilbenoid antioxidants. Sci Rep, 2016, 6: 36827. DOI:10.1038/srep36827
|
| [33] |
Gu Y, Ma JB, Zhu YJ, et al. Engineering Yarrowia lipolytica as a chassis for de novo synthesis of five aromatic-derived natural products and chemicals. ACS Synth Biol, 2020, 9(8): 2096-2106. DOI:10.1021/acssynbio.0c00185
|
| [34] |
He Q, Szczepańska P, Yuzbashev T, et al. De novo production of resveratrol from glycerol by engineering different metabolic pathways in Yarrowia lipolytica. Metab Eng Commun, 2020, 11: e00146. DOI:10.1016/j.mec.2020.e00146
|
| [35] |
Sáez-Sáez J, Wang GK, Marella ER, et al. Engineering the oleaginous yeast Yarrowia lipolytica for high-level resveratrol production. Metab Eng, 2020, 62: 51-61. DOI:10.1016/j.ymben.2020.08.009
|
| [36] |
Lim CG, Fowler ZL, Hueller T, et al. High-yield resveratrol production in engineered Escherichia coli. Appl Environ Microbiol, 2011, 77(10): 3451-3460. DOI:10.1128/AEM.02186-10
|
| [37] |
Liu XL, Lin J, Hu HF, et al. De novo biosynthesis of resveratrol by site-specific integration of heterologous genes in Escherichia coli. FEMS Microbiol Lett, 2016, 363(8): fnw061. DOI:10.1093/femsle/fnw061
|
| [38] |
Kang SY, Lee JK, Choi O, et al. Biosynthesis of methylated resveratrol analogs through the construction of an artificial biosynthetic pathway in E. coli. BMC Biotechnol, 2014, 14: 67-77. DOI:10.1186/1472-6750-14-67
|
| [39] |
Kallscheuer N, Vogt M, Stenzel A, et al. Construction of a Corynebacterium glutamicum platform strain for the production of stilbenes and (2S)-flavanones. Metab Eng, 2016, 38: 47-55. DOI:10.1016/j.ymben.2016.06.003
|
| [40] |
Watts KT, Lee PC, Schmidt-Dannert C. Biosynthesis of plant-specific stilbene polyketides in metabolically engineered Escherichia coli. BMC Biotechnol, 2006, 6: 22-33.
|
| [41] |
Wang SY, Zhang SW, Xiao AF, et al. Metabolic engineering of Escherichia coli for the biosynthesis of various phenylpropanoid derivatives. Metab Eng, 2015, 29: 153-159. DOI:10.1016/j.ymben.2015.03.011
|
| [42] |
Jeong YJ, An CH, Woo SG, et al. Production of pinostilbene compounds by the expression of resveratrol O-methyltransferase genes in Escherichia coli. Enzyme Microb Technol, 2014, 54: 8-14. DOI:10.1016/j.enzmictec.2013.09.005
|
| [43] |
Heo KT, Kang SY, Hong YS. De novo biosynthesis of pterostilbene in an Escherichia coli strain using a new resveratrol O-methyltransferase from Arabidopsis. Microb Cell Fact, 2017, 16(1): 30-37. DOI:10.1186/s12934-017-0644-6
|
| [44] |
Choi O, Lee JK, Kang SY, et al. Construction of artificial biosynthetic pathways for resveratrol glucoside derivatives. J Microbiol Biotechnol, 2014, 24(5): 614-618. DOI:10.4014/jmb.1401.01031
|
| [45] |
Palmer CM, Miller KK, Nguyen A, et al. Engineering 4-coumaroyl-CoA derived polyketide production in Yarrowia lipolytica through a β-oxidation mediated strategy. Metab Eng, 2020, 57: 174-181. DOI:10.1016/j.ymben.2019.11.006
|
| [46] |
Katsuyama Y, Matsuzawa M, Funa N, et al. Production of curcuminoids by Escherichia coli carrying an artificial biosynthesis pathway. Microbiology (Reading), 2008, 154(9): 2620-2628. DOI:10.1099/mic.0.2008/018721-0
|
| [47] |
Kim EJ, Cha MN, Kim BG, et al. Production of curcuminoids in engineered Escherichia coli. J Microbiol Biotechnol, 2017, 27(5): 975-982. DOI:10.4014/jmb.1701.01030
|
| [48] |
Rodrigues JL, Araújo RG, Prather KLJ, et al. Production of curcuminoids from tyrosine by a metabolically engineered Escherichia coli using caffeic acid as an intermediate. Biotechnol J, 2015, 10(4): 599-609. DOI:10.1002/biot.201400637
|
| [49] |
Couto MR, Rodrigues JL, Rodrigues LR. Optimization of fermentation conditions for the production of curcumin by engineered Escherichia coli. J Roy Soc Interface, 2017, 14(133): 20170470. DOI:10.1098/rsif.2017.0470
|
| [50] |
Santos CNS, Koffas M, Stephanopoulos G. Optimization of a heterologous pathway for the production of flavonoids from glucose. Metab Eng, 2011, 13(4): 392-400. DOI:10.1016/j.ymben.2011.02.002
|
| [51] |
Zhou SH, Hao TT, Zhou JW. Fermentation and metabolic pathway optimization to de novo synthesize (2S)-naringenin in Escherichia coli. J Microbiol Biotechnol, 2020, 30(10): 1574-1582. DOI:10.4014/jmb.2008.08005
|
| [52] |
Gao S, Lyu YB, Zeng WZ, et al. Efficient biosynthesis of (2S)-naringenin from p-coumaric acid in Saccharomyces cerevisiae. J Agric Food Chem, 2020, 68(4): 1015-1021. DOI:10.1021/acs.jafc.9b05218
|
| [53] |
Gao S, Zhou HR, Zhou JW, et al. Promoter-library-based pathway optimization for efficient (2S)-naringenin production from p-coumaric acid in Saccharomyces cerevisiae. J Agric Food Chem, 2020, 68(25): 6884-6891. DOI:10.1021/acs.jafc.0c01130
|
| [54] |
Trantas E, Panopoulos N, Ververidis F. Metabolic engineering of the complete pathway leading to heterologous biosynthesis of various flavonoids and stilbenoids in Saccharomyces cerevisiae. Metab Eng, 2009, 11(6): 355-366. DOI:10.1016/j.ymben.2009.07.004
|
| [55] |
Koopman F, Beekwilder J, Crimi B, et al. De novo production of the flavonoid naringenin in engineered Saccharomyces cerevisiae. Microb Cell Fact, 2012, 11: 155-169. DOI:10.1186/1475-2859-11-155
|
| [56] |
Lyu XM, Ng KR, Lee JL, et al. Enhancement of naringenin biosynthesis from tyrosine by metabolic engineering of Saccharomyces cerevisiae. J Agric Food Chem, 2017, 65(31): 6638-6646. DOI:10.1021/acs.jafc.7b02507
|
| [57] |
Du Y, Yang BR, Yi ZQ, et al. Engineering Saccharomyces cerevisiae coculture platform for the production of flavonoids. J Agric Food Chem, 2020, 68(7): 2146-2154. DOI:10.1021/acs.jafc.9b07916
|
| [58] |
Park SR, Ahn MS, Han AR, et al. Enhanced flavonoid production in Streptomyces venezuelae via metabolic engineering. J Microbiol Biotechnol, 2011, 21(11): 1143-1146. DOI:10.4014/jmb.1108.08012
|
| [59] |
Lv YK, Marsafari M, Koffas M, et al. Optimizing oleaginous yeast cell factories for flavonoids and hydroxylated flavonoids biosynthesis. ACS Synth Biol, 2019, 8(11): 2514-2523. DOI:10.1021/acssynbio.9b00193
|
| [60] |
Yan YJ, Kohli A, Koffas MAG. Biosynthesis of natural flavanones in Saccharomyces cerevisiae. Appl Environ Microbiol, 2005, 71(9): 5610-5613. DOI:10.1128/AEM.71.9.5610-5613.2005
|
| [61] |
Zhu SL, Wu JJ, Du GC, et al. Efficient synthesis of eriodictyol from L-tyrosine in Escherichia coli. Appl Environ Microbiol, 2014, 80(10): 3072-3080. DOI:10.1128/AEM.03986-13
|
| [62] |
Marín L, Gutiérrez-Del-Río I, Yagüe P, et al. De novo biosynthesis of apigenin, luteolin, and eriodictyol in the actinomycete Streptomyces albus and production improvement by feeding and spore conditioning. Front Microbiol, 2017, 8: 921-932. DOI:10.3389/fmicb.2017.00921
|
| [63] |
Stahlhut SG, Siedler S, Malla S, et al. Assembly of a novel biosynthetic pathway for production of the plant flavonoid fisetin in Escherichia coli. Metab Eng, 2015, 31: 84-93. DOI:10.1016/j.ymben.2015.07.002
|
| [64] |
Yin Y, Li YP, Jiang D, et al. De novo biosynthesis of liquiritin in Saccharomyces cerevisiae. Acta Pharm Sin B, 2020, 10(4): 711-721. DOI:10.1016/j.apsb.2019.07.005
|
| [65] |
Rodriguez A, Strucko T, Stahlhut SG, et al. Metabolic engineering of yeast for fermentative production of flavonoids. Bioresour Technol, 2017, 245: 1645-1654. DOI:10.1016/j.biortech.2017.06.043
|
| [66] |
Eichenberger M, Lehka BJ, Folly C, et al. Metabolic engineering of Saccharomyces cerevisiae for de novo production of dihydrochalcones with known antioxidant, antidiabetic, and sweet tasting properties. Metab Eng, 2017, 39: 80-89. DOI:10.1016/j.ymben.2016.10.019
|
| [67] |
Jiang CM, Liu XN, Chen XQ, et al. Raising the production of phloretin by alleviation of by-product of chalcone synthase in the engineered yeast. Sci China Life Sci, 2020, 63(11): 1734-1743. DOI:10.1007/s11427-019-1634-8
|
| [68] |
Kim BG. Biological synthesis of genistein in Escherichia coli. J Microbiol Biotechnol, 2019, 30(5): 770-776.
|
| [69] |
Leonard E, Yan YJ, Lim KH, et al. Investigation of two distinct flavone synthases for plant-specific flavone biosynthesis in Saccharomyces cerevisiae. Appl Environ Microbiol, 2005, 71(12): 8241-8248. DOI:10.1128/AEM.71.12.8241-8248.2005
|
| [70] |
Miyahisa I, Funa N, Ohnishi Y, et al. Combinatorial biosynthesis of flavones and flavonols in Escherichia coli. Appl Microbiol Biotechnol, 2006, 71(1): 53-58. DOI:10.1007/s00253-005-0116-5
|
| [71] |
Leonard E, Yan YJ, Fowler ZL, et al. Strain improvement of recombinant Escherichia coli for efficient production of plant flavonoids. Mol Pharmaceutics, 2008, 5(2): 257-265. DOI:10.1021/mp7001472
|
| [72] |
Leonard E, Yan YJ, Koffas MAG. Functional expression of a P450 flavonoid hydroxylase for the biosynthesis of plant-specific hydroxylated flavonols in Escherichia coli. Metab Eng, 2006, 8(2): 172-181. DOI:10.1016/j.ymben.2005.11.001
|
| [73] |
Pei JJ, Chen AN, Dong P, et al. Modulating heterologous pathways and optimizing fermentation conditions for biosynthesis of kaempferol and astragalin from naringenin in Escherichia coli. J Ind Microbiol Biotechnol, 2019, 46(2): 171-186. DOI:10.1007/s10295-018-02134-6
|
| [74] |
Duan LJ, Ding WT, Liu XN, et al. Biosynthesis and engineering of kaempferol in Saccharomyces cerevisiae. Microb Cell Fact, 2017, 16(1): 165-174. DOI:10.1186/s12934-017-0774-x
|
| [75] |
Marín L, Gutiérrez-Del-Río I, Entrialgo-Cadierno R, et al. De novo biosynthesis of myricetin, kaempferol and quercetin in Streptomyces albus and Streptomyces coelicolor. PLoS One, 2018, 13(11): e0207278. DOI:10.1371/journal.pone.0207278
|
| [76] |
封悦洋, 王颖, 姚明东, 等. 生物合成槲皮素糖苷类衍生物的研究进展. 化工学报, 2020, 71(7): 2945-2955. Feng YY, Wang Y, Yao MD, et al. Advances in biosynthesis of quercetin glycoside derivatives. J Chem Ind Eng (China), 2020, 71(7): 2945-2955 (in Chinese). |
| [77] |
Eichenberger M, Hansson A, Fischer D, et al. De novo biosynthesis of anthocyanins in Saccharomyces cerevisiae. FEMS Yeast Res, 2018, 18(4). DOI:10.1093/femsyr/foy046
|
| [78] |
Liu XN, Cheng J, Zhang GH, et al. Engineering yeast for the production of breviscapine by genomic analysis and synthetic biology approaches. Nat Commun, 2018, 9(1): 448-457. DOI:10.1038/s41467-018-02883-z
|
| [79] |
Levisson M, Araya-Cloutier C, De Bruijn WJC, et al. Toward developing a yeast cell factory for the production of prenylated flavonoids. J Agric Food Chem, 2019, 67(49): 13478-13486. DOI:10.1021/acs.jafc.9b01367
|
| [80] |
Kim MJ, Kim BG, Ahn JH. Biosynthesis of bioactive O-methylated flavonoids in Escherichia coli. Appl Microbiol Biotechnol, 2013, 97(16): 7195-7204. DOI:10.1007/s00253-013-5020-9
|
| [81] |
Katsuyama Y, Funa N, Miyahisa I, et al. Synthesis of unnatural flavonoids and stilbenes by exploiting the plant biosynthetic pathway in Escherichia coli. Chem Biol, 2007, 14(6): 613-621. DOI:10.1016/j.chembiol.2007.05.004
|
| [82] |
Miyahisa I, Kaneko M, Funa N, et al. Efficient production of (2S)-flavanones by Escherichia coli containing an artificial biosynthetic gene cluster. Appl Microbiol Biotechnol, 2005, 68(4): 498-504. DOI:10.1007/s00253-005-1916-3
|
| [83] |
Leonard E, Lim KH, Saw PN, et al. Engineering central metabolic pathways for high-level flavonoid production in Escherichia coli. Appl Environ Microbiol, 2007, 73(12): 3877-3886. DOI:10.1128/AEM.00200-07
|
| [84] |
Wu JJ, Yu O, Du GC, et al. Fine-tuning of the fatty acid pathway by synthetic antisense RNA for enhanced (2S)-naringenin production from L-tyrosine in Escherichia coli. Appl Environ Microbiol, 2014, 80(23): 7283-7292. DOI:10.1128/AEM.02411-14
|
| [85] |
Wu JJ, Du GC, Chen J, et al. Enhancing flavonoid production by systematically tuning the central metabolic pathways based on a CRISPR interference system in Escherichia coli. Sci Rep, 2015, 5: 13477. DOI:10.1038/srep13477
|
| [86] |
Zhao Y, Wu BH, Liu ZN, et al. Combinatorial optimization of resveratrol production in engineered E. coli. J Agric Food Chem, 2018, 66(51): 13444-13453. DOI:10.1021/acs.jafc.8b05014
|
| [87] |
Xu P, Ranganathan S, Fowler ZL, et al. Genome-scale metabolic network modeling results in minimal interventions that cooperatively force carbon flux towards malonyl-CoA. Metab Eng, 2011, 13(5): 578-587. DOI:10.1016/j.ymben.2011.06.008
|
| [88] |
Bhan N, Xu P, Khalidi O, et al. Redirecting carbon flux into malonyl-CoA to improve resveratrol titers: proof of concept for genetic interventions predicted by OptForce computational framework. Chem Eng Sci, 2013, 103: 109-114. DOI:10.1016/j.ces.2012.10.009
|
| [89] |
Xiong DD, Lu SK, Wu JY, et al. Improving key enzyme activity in phenylpropanoid pathway with a designed biosensor. Metab Eng, 2017, 40: 115-123. DOI:10.1016/j.ymben.2017.01.006
|
| [90] |
Yang D, Kim WJ, Yoo SM, et al. Repurposing type Ⅲ polyketide synthase as a malonyl-CoA biosensor for metabolic engineering in bacteria. Proc Natl Acad Sci USA, 2018, 115(40): 9835-9844. DOI:10.1073/pnas.1808567115
|
| [91] |
Zhou SH, Lyu YB, Li HZ, et al. Fine-tuning the (2S)-naringenin synthetic pathway using an iterative high-throughput balancing strategy. Biotechnol Bioeng, 2019, 116(6): 1392-1404. DOI:10.1002/bit.26941
|
| [92] |
Zhou SH, Yuan SF, Nair PH, et al. Development of a growth coupled dynamic regulation network balancing malonyl-CoA node to enhance (2S)-naringenin synthesis in E. coli. BioRxiv Synth Biol, 2020. DOI:10.1101/2020.07.07.192633
|
| [93] |
Dinh CV, Prather KLJ. Development of an autonomous and bifunctional quorum-sensing circuit for metabolic flux control in engineered Escherichia coli. Proc Natl Acad Sci USA, 2019, 116(51): 25562-25568. DOI:10.1073/pnas.1911144116
|
| [94] |
Camacho-Zaragoza JM, Hernández-Chávez G, Moreno-Avitia F, et al. Engineering of a microbial coculture of Escherichia coli strains for the biosynthesis of resveratrol. Microb Cell Fact, 2016, 15(1): 163-173. DOI:10.1186/s12934-016-0562-z
|
| [95] |
Hong J, Im DK, Oh MK. Investigating E. coli coculture for resveratrol production with 13C metabolic flux analysis. J Agric Food Chem, 2020, 68(11): 3466-3473. DOI:10.1021/acs.jafc.9b07628
|
| [96] |
Yuan SF, Yi XN, Johnston TG, et al. De novo resveratrol production through modular engineering of an Escherichia coli-Saccharomyces cerevisiae co-culture. Microb Cell Fact, 2020, 19(1): 143-154. DOI:10.1186/s12934-020-01401-5
|
| [97] |
Rodrigues JL, Gomes D, Rodrigues LR. A combinatorial approach to optimize the production of curcuminoids from tyrosine in Escherichia coli. Front Bioeng Biotechnol, 2020, 8: 59-73. DOI:10.3389/fbioe.2020.00059
|
| [98] |
Jones JA, Vernacchio VR, Sinkoe AL, et al. Experimental and computational optimization of an Escherichia coli co-culture for the efficient production of flavonoids. Metab Eng, 2016, 35: 55-63. DOI:10.1016/j.ymben.2016.01.006
|
| [99] |
Ganesan V, Li ZH, Wang XN, et al. Heterologous biosynthesis of natural product naringenin by co-culture engineering. Synth Syst Biotechnol, 2017, 2(3): 236-242. DOI:10.1016/j.synbio.2017.08.003
|
| [100] |
Zhang W, Liu H, Li X, et al. Production of naringenin from D-xylose with co-culture of E. coli and S. cerevisiae. Eng Life Sci, 2017, 17(9): 1021-1029. DOI:10.1002/elsc.201700039
|
| [101] |
Katsuyama Y, Miyahisa I, Funa N, et al. One-pot synthesis of genistein from tyrosine by coincubation of genetically engineered Escherichia coli and Saccharomyces cerevisiae cells. Appl Microbiol Biotechnol, 2007, 73(5): 1143-1149. DOI:10.1007/s00253-006-0568-2
|
| [102] |
Lyu XM, Zhao GL, Ng KR, et al. Metabolic engineering of Saccharomyces cerevisiae for de novo production of kaempferol. J Agric Food Chem, 2019, 67(19): 5596-5606. DOI:10.1021/acs.jafc.9b01329
|
| [103] |
Li ZH, Wang XN, Zhang HR. Balancing the non-linear rosmarinic acid biosynthetic pathway by modular co-culture engineering. Metab Eng, 2019, 54: 1-11. DOI:10.1016/j.ymben.2019.03.002
|
| [104] |
Jones JA, Vernacchio VR, Collins SM, et al. Complete biosynthesis of anthocyanins using E. coli polycultures. mBio, 2017, 8(3): e00621-17.
|
| [105] |
Ban ZN, Qin H, Mitchell AJ, et al. Noncatalytic chalcone isomerase-fold proteins in Humulus lupulus are auxiliary components in prenylated flavonoid biosynthesis. Proc Natl Acad Sci USA, 2018, 115(22): E5223-E5232. DOI:10.1073/pnas.1802223115
|
| [106] |
Waki T, Mameda R, Nakano T, et al. A conserved strategy of chalcone isomerase-like protein to rectify promiscuous chalcone synthase specificity. Nat Commun, 2020, 11(1): 870-883. DOI:10.1038/s41467-020-14558-9
|
| [107] |
Hammer SK, Avalos JL. Harnessing yeast organelles for metabolic engineering. Nat Chem Biol, 2017, 13(8): 823-832. DOI:10.1038/nchembio.2429
|
| [108] |
Wei SP, Qian ZG, Hu CF, et al. Formation and functionalization of membraneless compartments in Escherichia coli. Nat Chem Biol, 2020, 16(10): 1143-1148. DOI:10.1038/s41589-020-0579-9
|
 2021, Vol. 37
2021, Vol. 37