中国科学院微生物研究所、中国微生物学会主办
文章信息
- 杨兴盛, 王尚, 何晴, 王朱珺, 张照婧, 姜成英, 马黎萍, 刘贤伟, 胡宝兰, 李咏梅, 邓晔
- Yang Xingsheng, Wang Shang, He Qing, Wang Zhujun, Zhang Zhaojing, Jiang Chengying, Ma Liping, Liu Xianwei, Hu Baolan, Li Yongmei, Deng Ye
- 典型有机固废厌氧消化微生物研究现状与发展方向
- Microorganisms in the typical anaerobic digestion system of organic solid wastes: a review
- 生物工程学报, 2021, 37(10): 3425-3438
- Chinese Journal of Biotechnology, 2021, 37(10): 3425-3438
- 10.13345/j.cjb.210408
-
文章历史
- Received: May 31, 2021
- Accepted: September 3, 2021
2. 中国科学院大学,北京 100049;
3. 山东大学 海洋研究院,山东 青岛 266237;
4. 中国科学院微生物研究所,北京 100101;
5. 华东师范大学 生态与环境科学学院,上海 200241;
6. 中国科技大学 环境科学与工程系,安徽 合肥 230026;
7. 浙江大学 环境与资源学院,浙江 杭州 310058;
8. 同济大学 环境科学与工程学院,上海 200092
2. University of Chinese Academy of Sciences, Beijing 100049, China;
3. Institute of Marine Science and Technology, Shandong University, Qingdao 266237, Shandong, China;
4. Institute of Microbiology, Chinese Academy of Sciences, Beijing 100101, China;
5. School of Ecological and Environmental Sciences, East China Normal University, Shanghai 200241, China;
6. Department of Environmental Science and Engineering, University of Science and Technology of China, Hefei 230026, Anhui, China;
7. College of Environmental & Resource Sciences, Zhejiang University, Hangzhou 310058, Zhejiang, China;
8. College of Environmental Science and Engineering, Tongji University, Shanghai 200092, China
生物处理是目前实现有机固废资源化、能源化、减量化和无害化的重要手段。厌氧消化(Anaerobic digestion) 更是其中的研究热点,已被广泛用于多种有机固废的消减和资源化,如市政污泥、餐厨垃圾、园林和农业废弃物、畜禽粪便等[1-2]。厌氧消化系统的宏观调控较易实现,通常是在遵循微生物组代谢规律的前提下,优化工艺条件以实现特定产物的富集和能源物质的回收[3-5]。然而,以微生物代谢为主体的微观过程却难以实现定向调控。根本原因是有机固废成分的复杂性、环境影响因素的多变性以及它们与菌群交互作用的多样性,限制了对有机固废厌氧消化过程的深入理解。近年来化学分析技术(例如高分辨质谱的应用)、微生物检测技术(例如多组学技术的应用) 以及生物信息技术(例如模型的构建) 的快速发展[6-7],开创了微生物组研究的新方向[8-9],也为更全面地解析有机固废厌氧消化的微观机制提供了新的契机。
1 厌氧消化系统中的微生物以及典型菌群的互作关系厌氧消化的工程调控依然处于“黑箱”操作,随着组学技术的发展,对厌氧消化系统相关微生物分类和功能的认识更加深入,且对其参与的不同阶段的认识也在与时俱进[10-12]。厌氧消化的不同阶段(水解、产酸、产乙酸和产甲烷) 是基于物质流确立的,传统观念认为厌氧消化过程每个阶段的微生物类群具有专一性,然而最新宏基因组学证据表明,微生物类群演替在整个厌氧消化过程具有连续性、无明显的阶梯性变化,例如随着初始底物的变化沿着物质流传递到整个厌氧消化过程时,包括产甲烷菌在内的菌群均会发生演替[13]。参与不同物质转化的微生物被称为相应的功能菌,这种功能菌群的界定是基于微生物的代谢功能而非系统发育信号,所以同一类微生物可能在厌氧消化过程中发挥多重作用。
1.1 参与厌氧消化的微生物类群微生物作为各种物质转化的主要承担者在整个厌氧消化过程中发挥了不可替代的作用。事实上,厌氧消化系统中能够明确鉴定谱系的微生物非常有限,超过90%的微生物无法在属或种的水平进行分类[14]。厌氧消化过程中已知的细菌主要属于厚壁菌门Firmicutes、拟杆菌门Bacteroidetes、变形菌门Proteobacteria[14-15],承担着将复杂有机大分子转化为简单有机物以及CO2、H2等气体成分的一系列任务。相比细菌的高谱系多样性,厌氧消化工程系统中的古菌较为单一,来源于广古菌门Euryarchaeota的产甲烷菌[6, 12]。虽然最新的宏基因组学证据表明深古菌Bathyarchaeota[16]和佛斯特拉古菌Verstraetearchaeota[17-18]等也具有甲烷代谢功能,从而将产甲烷菌谱系扩展到Euryarchaeota之外[19],但目前没有任何非Euryarchaeota的产甲烷菌在厌氧消化系统中被发现。除了细菌和古菌,真菌同样是厌氧消化系统中的重要微生物,参与固体物质的分解和有机大分子的降解与转化过程,然而很少有报道关注真菌在整个系统中的功能和命运[3, 20-21]。正常运行的厌氧消化系统,各类微生物的物种数可能高达上千个[22],这些微生物通常具有高度的功能冗余从而增强了系统应对复杂环境和抵抗环境扰动的能力[23-24]。Yu等认为当功能冗余和群落多样性降低时,菌群间互作网络简单化,系统可能会崩溃[23]。
1.2 厌氧消化系统中典型菌群的互作关系厌氧消化系统中不同菌群在代谢功能上是相互依存的,在维持系统稳定的同时,遵循着物料守恒和能量守恒规律[25]。相互关联的复杂酶系为厌氧消化系统中菌群执行互补或可替代的代谢活动提供了支持[26]。通过鉴定编码关键酶基因的存在和表达是解析系统代谢路径和解构互营关系的重要方法。譬如Hao等结合宏基因组和宏转录组,预测了互营脂肪酸氧化菌互营代谢途径并证实了不同环境下代谢功能存在选择性表达[27]。Zhu等根据参与Wood-Ljungdahl途径和甘氨酸裂解途径关键基因的表达差异,重构了发酵细菌以甘氨酸裂解替代乙酸代谢并耦合氢营养产甲烷的互营模型[24]。厌氧消化前段的发酵细菌与后段的产甲烷菌通过产物交叉喂养构成互营关系,缓解了产物积累造成的压力,并使整个反应在热力学上是有利的。一方面发酵细菌产生的乙酸、H2、CO2、甲基化合物为产甲烷菌提供底物,同时这些细菌的代谢活动消耗了氧、硝酸盐、Fe(Ⅲ) 等氧化性物质,为产甲烷创造适宜的氧化还原条件;另一方面产甲烷菌通过消耗乙酸、H2和CO2,为脂肪酸的氧化解除了产物的抑制作用,提供了能量,同时维持了体系的pH稳定[19]。这种互营依赖于细胞间的电子传递,分为依靠H2、甲酸等小分子进行的种间电子传递(Interspecies electron transfer,IET) 和依靠细胞上的微生物纳米导线或多血红素细胞色素的直接电子传递(Direct interspecies electron transfer,DIET)[28-29]。一直以来,IET被认为是互营菌的主要电子传递方式[30],而Rotaru等证实了DIET在厌氧消化工程系统中的存在,并提出DIET可能在全球产甲烷过程中起重要作用[31]。最新研究显示,电子传递和能量获取策略是构建互营关系的关键决定因素[25]。
厌氧消化各阶段产物汇聚于胞外空间形成一个共享的物质池。除了互营,不同菌群对物质的竞争也广泛存在于整个厌氧消化系统中。例如,互营乙酸氧化菌(Syntrophic acetate oxidation bacteria,SAOB) 和乙酸营养型产甲烷菌构成了对乙酸的竞争,而氢营养产甲烷菌和同型产乙酸菌均以H2和CO2为底物[32]。同型产乙酸菌/乙酸营养产甲烷菌以及互营乙酸氧化菌/氢营养产甲烷菌分别通过产物交叉喂养成为互营伙伴,并由于近似的互营代谢作用而产生生态位的竞争[33] (主要代谢途径见图 1)。微生物的生态位竞争受到其环境适应性的影响,而整个互营代谢的存在往往受限于互营伙伴中更为脆弱的一方。研究表明,互营乙酸氧化(图 1C) 作为Wood-Ljungdahl途径产乙酸(图 1B) 的逆反应在热力学上极为不利,所以SAOB在与乙酸营养型产甲烷菌的竞争中常处于劣势,从而造成整个互营代谢处于不利地位[3, 34]。但温度升高时,乙酸氧化的热力学壁垒会相对降低,从而提升SAOB的竞争能力[3, 35]。此外,相比于发酵细菌,产甲烷古菌一般冗余度较低,具有更低的生态位宽度,更容易受到恶劣条件的影响,进而影响到互营伙伴的代谢乃至危及整个系统的稳态。尤其是乙酸营养型产甲烷菌,相比其他两种类型的产甲烷菌其生态位更窄[36-38],在极端环境如高游离氨、低pH和高盐条件下难以有效存活,造成的生态空位会由耐受性更好的互营乙酸氧化菌/氢营养产甲烷菌填补[36-38]。

|
| 图 1 经典甲烷代谢途径、同型产乙酸途径和互营乙酸氧化途径[5, 19, 34, 41] Fig. 1 Classical metabolic pathway of methanogenesis, homoacetogenesis and SAO[5, 19, 34, 41]. (FWD:甲酰甲烷呋喃脱氢酶;FTR:四氢甲烷蝶呤转移酶;MCH:次甲基四氢甲烷蝶呤环水解酶;MTD:亚甲基四氢甲烷蝶呤脱氢酶;MER:亚甲基四氢甲烷蝶呤还原酶;MTR:四氢甲烷蝶呤甲基转移酶;MCR:甲基辅酶M还原酶;H4MPT:四氢甲烷蝶呤;THF:四氢叶酸;CoFeSP:可啉铁硫蛋白;FDH:甲酸脱氢酶;FTS:甲酸四氢叶酸合成酶;MeTC:亚甲基四氢叶酸环水解酶;MeTD:亚甲基四氢叶酸脱氢酶;MeTR:亚甲基四氢叶酸还原酶;MeTF:乙酰转移酶;ACS:乙酰辅酶A合成酶;CD:CO氧化酶;PTA:磷酸转乙酰酶;AK:乙酸激酶) FWD: formyl-methanofuran dehydrogenase; FTR: formylmeth-anofuran-H4MPT formyltransferase; MCH: methenyl-H4MPT cyclohydrolase; MTD: F420-dependent methylene H4MPT dehydrogenase; MER: F420-dependent methylene-H4MPT reductase; MTR: Na+-translocating methyl-H4MPT-coenzyme-M-methyltransferase; MCR: methyl-coenzyme M reductase; H4MPT: tetrahydromethanopterin; THF: tetrahydrofolate; CoFeSP: corrinoid iron-sulphur protein; FDH: formate dehydrogenase; FTS: formate-THF synthetase; MeTC: methylene-THF cyclohydrolase; MeTD: methylene-THF dehydrogenase; MeTR: methylene-THF reductase; MeTF: methyltransferase; ACS: acetyl-CoA synthase; CD: CO dehydrogenase; PTA: phosphotransacetylase; AK: acetate kinase. |
| |
共享物质池中的某些物质在细胞间频繁作为产物和底物传递,如甲酸、乙醇、CO2、H2等,其浓度会直接影响到相应代谢反应的平衡,从而对菌群演替施加选择压力,其中H2的作用尤为显著[25, 39]。厌氧消化过程中存在能够应对高H2浓度的H2耐受型代谢和在极低H2浓度下就会受到抑制的H2敏感型代谢。葡萄糖降解作为H2耐受型代谢可耐受1 020 Pa的H2分压,而丁酸降解作为H2敏感型代谢仅耐受2.8 Pa[37]。H2敏感型代谢会受到由其他反应生成的H2的抑制,因此厌氧消化系统需要通过耦合消耗H2的互营代谢方式降低H2的积累。而H2的浓度降低不利于细胞间的电子传递,系统需要采取其他途径转移并处置电子,如通过甲酸、细胞色素和纳米导线进行电子传递以及耦合低浓度的氧气代谢作为辅助的电子汇[37]。因此,互营关系的构建以及不同代谢方式对于物质流的竞争受到反应热力学条件的调控,从而使整个系统实现对物质和能量的“精打细算”和“系统最优”[24, 36, 40]。
正是由于厌氧消化系统中存在独特的物质-环境-微生物互作关系,前人研究未能在该类系统中发现普遍存在的“核心功能菌”[14]。因此,深入解析菌群间的互作关系以及菌群-物质的代谢耦合关系,有助于发现优化构效关系的关键环境因子及微生物类群,对于生物转化定向调控策略的制定及转化效率的提高至关重要。
2 典型有机固废厌氧消化存在的问题以及微生物在其中发挥的作用有机固废同时具备污染属性和资源属性,其组成复杂,富含多糖、蛋白质、脂质等大分子成分,可以转化生成生物沼气、脂肪酸、羟基酸、氨基酸、醇类、植物化合物等多种产物[42-44]。丰富的有机成分为微生物提供了多样化的代谢底物和物质环境,相应的微生物群落结构和多样性与特定基质的组成也表现出高度相关[14]。例如有报道称,纤维素的降解和梭菌属解纤维梭菌Clostridium cellulolyticum的水平呈正相关[45],脂类的降解和互营单胞菌属Syntrophomonas水平呈正相关[46],梭菌纲Clostridia和软壁菌门Tenericutes特异性地出现在粪便的厌氧消化池[47],高蛋白的食物废物容易造成氨氮的大量积累,使SAOB在生态位上占优势[3, 48]。同时某些成分或中间产物也可能对部分微生物有胁迫或者毒害作用[3, 49]。下面针对3种典型有机固废厌氧消化过程中存在的问题及微生物的作用进行简述。
2.1 市政污泥市政污泥是市政污水处理的副产物,具有含水率高、有机质含量低、产量大、水解难度大等特点。剩余活性污泥中包含大量的微生物细胞,其中约80%是活细胞[7]。实际运行过程中有机物和微生物细胞形成稳定的絮状污泥限制了有机成分的释放,而大量存在的腐殖质也难以被生物利用[7]。因此,常采用预处理手段来提高有机质的可生化性[50-51]。Liu等[52]通过热水解提高了污泥的厌氧消化降解能力,发现Firmicutes和Bacteroidetes等发酵细菌丰度显著增加,而产甲烷菌组成变化不大,这可能是由于产甲烷菌以细菌代谢的中间产物为营养源而不受水解过程初始底物的影响[53]。除了预处理,向消化池投加酶和化学试剂也可以通过生物和化学效应改善水解条件[54-56]。外源酶的投加直接降低了系统水解菌群的产酶需求,人为破除了水解阶段对整个厌氧消化过程的速率限制[54-55]。化学试剂CaO2的投加可以提供羟基自由基从而有效去除难降解的有机污染物,并在生物效应上表现为水解和产酸酶系的活性相对提高[57]。最新的研究发现,市政污泥厌氧消化中CaO2的投加能够富集10余种脂肪酸生物合成的关键基因并促进溶菌酶的作用和多种单糖的代谢[58]。
市政污泥中存在的大量活细胞是病原微生物和抗生素抗性基因持续输入的来源[59-60]。温度被认为是影响厌氧消化对病原体和抗性基因去除能力的重要因素,通过调节营养物质和温度,选择性地恶化目标微生物的生活环境,能够在厌氧消化系统中实现对它们的有效去除[60-61]。对不同温度厌氧消化系统的宏基因组分析表明,高温(55 ℃) 能够有效降低抗生素外排泵编码基因的丰度。此外,菌群多样性的降低可以消减抗性基因的宿主以及潜在宿主,从而抑制了抗性基因的垂直转移;同时,菌群代谢速率的显著提高则可以加速对抗性基因水平转移载体的降解[62]。除了风险微生物,市政污泥富集了来自污水处理厂的各种污染物,针对当前层出不穷的新兴污染物,微生物作用可以有效削减某些污染物的数量和毒性[63-64]。同时这些新兴污染物也可能诱导微生物产生新的特异性代谢机制,需要持续予以关注[63-64]。
2.2 餐厨垃圾餐厨垃圾是餐饮垃圾和厨余垃圾的总称,具有高油、高盐、高含水率、高有机含量等特点[65-66]。在餐厨垃圾厌氧消化过程中大量有机成分往往能高效水解、酸化转化为脂肪酸,但大部分脂肪酸的进一步转化属于H2敏感型代谢而速率不高,转化速率的巨大差异容易造成酸的积累(尤其是丙酸)[39],进而导致体系pH降低,造成产甲烷菌活性降低,因为大部分产甲烷菌在pH < 6.7时就已停止代谢[12]。Sun等发现当pH低至5.1,几乎90%的乙酸营养和氢营养的产甲烷菌在6 d内就会从培养物中消失[67]。尽管脂肪酸同样可以作为系统产出的产品,由于过低pH不利于有机酸的解离,持续的酸化会对微生物造成普遍的危害最终同样会影响挥发性脂肪酸(Volatile fatty acids,VFAs)的产生,导致整个体系的崩溃[68]。工程上从宏观效应的角度出发,通过投加碱性物质的方式来调节体系pH,从而保证体系性能和产出效率[69]。然而,为了更有效地调控有机固废的生物转化过程,需要从微观机制如电子传递过程进行精准调控。研究表明,通过建立和强化直接电子传递可以提高VFAs利用速率和甲烷产率[70-71]。目前调控电子传递主要有两种途径:电子传递源控制和电子传递过程控制[28, 71]。电子传递源控制是以乙醇为产乙酸的前体,这在热力学上更为有利,而且已证实伴随着乙醇的氧化,厌氧消化菌群建立了DIET来促进甲烷的生成[29, 31]。现有研究中,Li等[71]和Gao等[72]分别通过乙醇型预发酵和原位投加酵母菌实现了以乙醇为电子传递源的DIET强化。电子传递过程控制一般是通过投加诸如颗粒活性炭、生物炭、纳米零价铁或铁氧化物等导电材料强化产甲烷菌和其他细菌间的电子和能量传递[28]。Lim等发现添加生物炭后,体系富集了梭状芽胞杆菌属Clostridia等电活性微生物,促进了脂肪酸的降解和乙酸营养型产甲烷过程,最终使得甲烷产率提升了18%[73]。Dang等同样证实了碳布和颗粒活性炭促进了体系电子传递从而增加了微生物降解复杂有机质的能力[70]。此外,通过施加电压也可以加速系统中的电子传递,例如微生物电解池(MEC) 的应用可以显著促进厌氧消化效率[74]。
餐厨垃圾常伴随着高油脂,而高浓度的油脂会限制底物和产物的运输,进而影响到乙酸降解菌和产甲烷菌的活性[75]。传统方法通过油脂分离的方式避免其干扰。事实上,厌氧消化微生物群具备一定程度上抵抗油脂抑制作用的潜力。Amha等发现虽然油脂的添加在初期造成了餐厨垃圾厌氧消化的代谢滞后和VFAs的积累,但随着时间推移微生物群可以通过种间相互作用,转向更有利的代谢途径从而恢复体系功能[76]。此外,研究发现长期的生物驯化能够富集Firmicutes、Bacteroidetes、互养菌门Synergistetes和Euryarchaeota等微生物,从而使厌氧消化系统具备快速降解长链脂肪酸的能力[77]。虽然将微生物逐渐暴露于不利环境下可以使群落内部发生适应性变化[78],但微生物耐受这种极端环境胁迫的分子机制尚不明确。
2.3 农业废弃物农业生产中主要会产生畜禽粪便和以秸秆为代表的植物纤维这两类有机固废。粪便和植物纤维分别是低碳氮比(C/N) 和高C/N的典型案例。C/N是衡量厌氧消化底物营养平衡的重要参数,厌氧消化过程中微生物消耗碳的速度是氮的25–30倍,为实现碳的最大转换效率,体系碳氮比需达到20︰1–30︰1的要求[12]。高含氮底物厌氧消化中随着游离氨的积累会显著抑制微生物代谢。Bi等单独以鸡粪为底物进行厌氧消化时,高浓度的氨态氮抑制乙酸转化和产甲烷过程,与去除氨的处理相比,甲烷产率低约30%[79]。而Zhang等发现秸秆的添加有效缓解了体系的氨抑制[80]。这是因为将富氮(如粪便) 和富碳(如植物纤维废物) 的固体废弃物进行共消化恰好平衡了两个体系对营养源组分的需求,同时解除了抑制效应。此外,多个来源的有机固废的共消化可以增加微生物代谢的多样性[14]。Liu等发现单独以玉米秸秆为底物的厌氧消化过程中物种总丰富度较低,古菌多样性不足[81]。而Oladejo等研究表明,粪便的添加为厌氧消化提供了优质的产甲烷菌物种库,混合基质能滋养高丰富度和多样性的微生物组,从而显著提高系统的稳定性和产出效率[82]。共消化以一种极其简便的方式实现了对物质环境和微生物群落结构的调节,使它们彼此契合进行有机固废的高效资源化转化[83]。
物质环境和微生物群落不相适应是厌氧消化系统出现问题的普遍原因,每个厌氧消化系统独特的环境-微生物互作关系导致了不同研究中可能出现迥然不同的结论[84-85],但在相似的系统中确定存在着“核心功能菌”以及同样的微生物群落构建机制[14],这样的特征使得基于大数据建模来调控厌氧消化过程成为可能。目前已经能够在宏观上通过改造物质环境(如预热水解) 和强化微生物功能(如投加导电材料强化细胞间电子传递)来提升厌氧消化系统性能,而更精细的定向调控需要依靠解析复杂体系中微生物组代谢机制,这无疑对分析检测方法提出了更高的要求,相关研究依然任重道远。
3 组学技术推动下有机固废微生物组研究发展方向目前大部分关于厌氧消化的研究已经由单纯的系统性能研究转为同时关注与系统性能密切相关的微生物机制[41, 86-88]。自上世纪末以来,组学技术蓬勃发展并逐渐应用到厌氧消化微生物组的研究中(图 2),尤其是高通量测序(指扩增子测序)的应用极大地丰富了厌氧消化菌群的生物多样性[6, 89]。但扩增子测序存在一些不可避免的缺陷,例如引物覆盖度不足等因素造成遗传信息的缺失或偏差,不具备单独用于识别新物种的能力等[6]。宏基因组的应用会克服这些缺陷,尽管2008年就有研究者对厌氧消化菌群进行了鸟枪宏基因组测序,但由于分析门槛较高,价格相对昂贵,所以宏基因组的应用远不及扩增子测序[21, 90-91]。通过宏蛋白组和宏转录组能够获得体系中具有代谢活性的微生物的转录本表达情况和蛋白分子组成情况,评估当前环境下微生物的基因表达情况和菌群的代谢潜力[92-93]。代谢组的应用通过直接检测小分子代谢产物的方法,可以识别关键代谢途径的信息细节,进一步推断微生物组潜在的代谢机制[92]。一些新兴的技术如单细胞测序技术[94]、基于单细胞分离的细胞内融合基因技术[59] (epicPCR) 在厌氧消化系统中还未见有应用。多组学技术的联用为进一步揭示物种代谢能力和趋向提供了新的方向[95],如通过培养组学和现有组学技术的结合来解密厌氧消化体系中大量的未培养微生物、通过高分辨质谱技术与组学技术的结合揭示菌群-物质的匹配与互作。
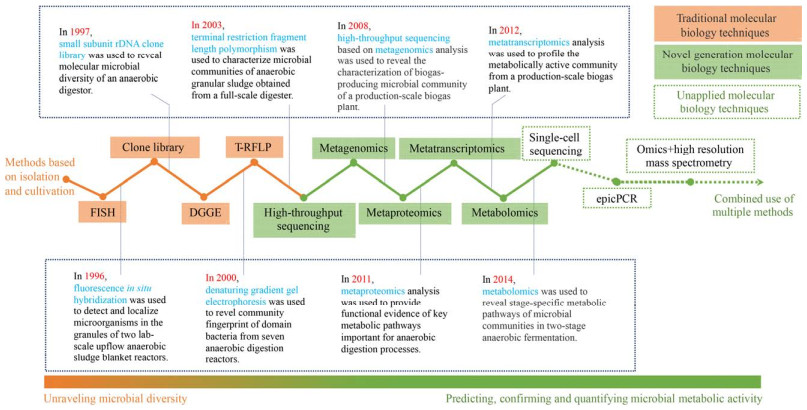
|
| 图 2 现代组学方法应用于厌氧消化系统的时间轴[90, 96-102] Fig. 2 Timeline for the development of omics-based technologies applied to anaerobic digestion[90, 96-102]. (FISH:荧光原位杂交;DGGE:变性梯度凝胶电泳;T-RFLP:末端限制性片段长度多态性分析;eqicPCR:细胞内融合基因技术) FISH: fluorescence in situ hybridization; DGGE: denaturing gradient gel electrophoresis; T-RFLP: terminal restriction fragment length polymorphism; eqicPCR: emulsion, paired isolation, and concatenation polymerase chain reaction. |
| |
菌群-物质的代谢偶联是解析有机固废生物转化体系中微生物学机制的核心任务,可为污染物的定向转化提供重要的科学依据。由于厌氧消化系统中物质种类繁多、物种多样性高,造成了代谢产物与物种相匹配的困难,生态网络分析[103]等信息学方法的引入为大量有机分子与微生物相匹配带来了新的机会。基于大数据分析构建分子生态网络,一方面可以揭示高度复杂微生物群落中的关键物种以及有机物之间、菌群之间、有机物和菌群的互作关系,另一方面通过引入更为详细的体系运行参数和环境参数,可以建立确定性因素主导下的环境-菌群互作模式。此外,目前有机固废厌氧消化的微生物组研究还处于发展阶段,还没有一个能够荟萃全球厌氧消化微生物组研究成果的数据库[22]。已有的数据分散在不同的数据存储机构,缺乏统一的存储协议和对厌氧消化微生物组信息专项的管理,亟需建立一个全球性的厌氧消化微生物组数据集和相应的数据库。综上所述,深入研究厌氧消化过程中的微生物组将为提升系统的稳定性、增加能源回收效率提供有效的理论支持和科学依据,是面向未来有机固废减量化、无害化和资源化的必由之路。
| [1] |
Guo MX, Song WP, Buhain J. Bioenergy and biofuels: history, status, and perspective. Renew Sustain Energy Rev, 2015, 42: 712-725. DOI:10.1016/j.rser.2014.10.013
|
| [2] |
郑琳珂, 戴晓虎. 污泥协同消化微生物的研究进展. 广州化工, 2020, 48(3): 20-22, 34. Zheng LK, Dai XH. Research progress on microorganism in sludge co-digestion. Guangzhou Chem Ind, 2020, 48(3): 20-22, 34 (in Chinese). |
| [3] |
HattiKaul R, Mamo G, Mattiasson B. Anaerobes in Biotechnology. Berlin: Springer-verlag, 2016, 195-234.
|
| [4] |
Sarkar O, Butti SK, Venkata Mohan S. Acidogenesis driven by hydrogen partial pressure towards bioethanol production through fatty acids reduction. Energy, 2017, 118: 425-434. DOI:10.1016/j.energy.2016.12.017
|
| [5] |
Lyu Z, Shao NN, Akinyemi T, et al. Methanogenesis. Curr Biol, 2018, 28(13): 727-732. DOI:10.1016/j.cub.2018.05.021
|
| [6] |
李叶青, 景张牧, 江皓, 等. 微生物组学及其在厌氧消化中的研究进展. 生物技术通报, 2021, 37(1): 90-101. Li YQ, Jing ZM, Jiang H, et al. Microbiome and its research progress of anaerobic digestion. Biotechnol Bull, 2021, 37(1): 90-101 (in Chinese). |
| [7] |
余泽晖, 梁志伟, 李浩聪, 等. 厌氧消化污泥微生物组研究方法及应用. 微生物学通报, 2019, 46(8): 2053-2068. Yu ZH, Liang ZW, Li HC, et al. Anaerobic sludge digestion microbiome—analytical methods and applications. Microbiol China, 2019, 46(8): 2053-2068 (in Chinese). |
| [8] |
刘双江, 施文元, 赵国屏. 中国微生物组计划: 机遇与挑战. 中国科学院院刊, 2017, 32: 241-250. Liu SJ, Shi WY, Zhao GP. China microbiome initiative: opportunity and challenges. Bull Chin Acad Sci, 2017, 32: 241-250 (in Chinese). |
| [9] |
Marchesi JR, Ravel J. The vocabulary of microbiome research: a proposal. Microbiome, 2015, 3(1): 31. DOI:10.1186/s40168-015-0094-5
|
| [10] |
郝鑫. 处理黑臭水体疏浚底泥的混合物料仿生干发酵工艺研究[D]. 重庆: 重庆交通大学, 2019. Hao X. Study on bionic dry fermentation process of mixture for treatment of black stinky water body dredged sediment[D]. Chongqing: Chongqing Jiaotong University, 2019 (in Chinese). |
| [11] |
Bryant MP. Microbial methane product-teheoretical aspects. J Anim Sci, 1979, 48(1): 193-201. DOI:10.2527/jas1979.481193x
|
| [12] |
Laiq Ur Rehman M, Iqbal A, Chang CC, et al. Anaerobic digestion. Water Environ Res, 2019, 91(10): 1253-1271. DOI:10.1002/wer.1219
|
| [13] |
Nabi M, Liang JS, Zhang PY, et al. Anaerobic digestion of sewage sludge pretreated by high pressure homogenization using expanded granular sludge blanket reactor: feasibility, operation optimization and microbial community. J Environ Chem Eng, 2021, 9(1): 104720. DOI:10.1016/j.jece.2020.104720
|
| [14] |
Campanaro S, Treu L, Rodriguez-R LM, et al. New insights from the biogas microbiome by comprehensive genome-resolved metagenomics of nearly 1600 species originating from multiple anaerobic digesters. Biotechnol Biofuels, 2020, 13: 25. DOI:10.1186/s13068-020-01679-y
|
| [15] |
Jiang F, Wang S, Zhang Y, et al. Variation of metagenome from feedstock to digestate in full-scale biogas plants. Front Microbiol, 2021, 12: 660225. DOI:10.3389/fmicb.2021.660225
|
| [16] |
Evans PN, Parks DH, Chadwick GL, et al. Methane metabolism in the archaeal phylum Bathyarchaeota revealed by genome-centric metagenomics. Science, 2015, 350(6259): 434-438. DOI:10.1126/science.aac7745
|
| [17] |
Vanwonterghem I, Evans PN, Parks DH, et al. Methylotrophic methanogenesis discovered in the archaeal phylum Verstraetearchaeota. Nat Microbiol, 2016, 1: 16170. DOI:10.1038/nmicrobiol.2016.170
|
| [18] |
周雷, 刘来雁, 刘鹏飞, 等. 佛斯特拉古菌门(Verstraetearchaeota)研究进展. 生物资源, 2020, 42(5): 515-521. Zhou L, Liu LY, Liu PF, et al. Research progress of the archaeal phylum Verstraetearchaeota. Biotic Resour, 2020, 42(5): 515-521 (in Chinese). |
| [19] |
Evans PN, Boyd JA, Leu AO, et al. An evolving view of methane metabolism in the archaea. Nat Rev Microbiol, 2019, 17(4): 219-232. DOI:10.1038/s41579-018-0136-7
|
| [20] |
Vinzelj J, Joshi A, Insam H, et al. Employing anaerobic fungi in biogas production: challenges & opportunities. Bioresour Technol, 2020, 300: 122687. DOI:10.1016/j.biortech.2019.122687
|
| [21] |
Qin S, Wainaina S, Awasthi SK, et al. Fungal dynamics during anaerobic digestion of sewage sludge combined with food waste at high organic loading rates in immersed membrane bioreactors. Bioresour Technol, 2021, 335: 125296. DOI:10.1016/j.biortech.2021.125296
|
| [22] |
Jiang C, Peces M, Andersen MH, et al. Characterizing the growing microorganisms at species level in 46 anaerobic digesters at Danish wastewater treatment plants: a six-year survey on microbial community structure and key drivers. Water Res, 2021, 193: 116871. DOI:10.1016/j.watres.2021.116871
|
| [23] |
Yu JJ, Tang SN, Lee PKH. Microbial communities in full-scale wastewater treatment systems exhibit deterministic assembly processes and functional dependency over time. Environ Sci Technol, 2021, 55(8): 5312-5323. DOI:10.1021/acs.est.0c06732
|
| [24] |
Zhu XY, Campanaro S, Treu L, et al. Novel ecological insights and functional roles during anaerobic digestion of saccharides unveiled by genome-centric metagenomics. Water Res, 2019, 151: 271-279. DOI:10.1016/j.watres.2018.12.041
|
| [25] |
Zhu XY, Campanaro S, Treu L, et al. Metabolic dependencies govern microbial syntrophies during methanogenesis in an anaerobic digestion ecosystem. Microbiome, 2020, 8(1): 22. DOI:10.1186/s40168-019-0780-9
|
| [26] |
Sorokin DY, Makarova KS, Abbas B, et al. Discovery of extremely halophilic, methyl-reducing euryarchaea provides insights into the evolutionary origin of methanogenesis. Nat Microbiol, 2017, 2: 17081. DOI:10.1038/nmicrobiol.2017.81
|
| [27] |
Hao L, Michaelsen TY, Singleton CM, et al. Novel syntrophic bacteria in full-scale anaerobic digesters revealed by genome-centric metatranscriptomics. ISME J, 2020, 14(4): 906-918. DOI:10.1038/s41396-019-0571-0
|
| [28] |
Shen L, Zhao QC, Wu XE, et al. Interspecies electron transfer in syntrophic methanogenic consortia: from cultures to bioreactors. Renew Sustain Energy Rev, 2016, 54: 1358-1367. DOI:10.1016/j.rser.2015.10.102
|
| [29] |
Summers ZM, Fogarty HE, Leang C, et al. Direct exchange of electrons within aggregates of an evolved syntrophic coculture of anaerobic bacteria. Science, 2010, 330(6009): 1413-1415. DOI:10.1126/science.1196526
|
| [30] |
Hamilton JJ, Calixto Contreras M, Reed JL. Thermodynamics and H2 transfer in a methanogenic, syntrophic community. PLoS Comput Biol, 2015, 11(7): e1004364. DOI:10.1371/journal.pcbi.1004364
|
| [31] |
Rotaru AE, Shrestha PM, Liu FH, et al. A new model for electron flow during anaerobic digestion: direct interspecies electron transfer to Methanosaeta for the reduction of carbon dioxide to methane. Energy Environ Sci, 2014, 7(1): 408-415. DOI:10.1039/C3EE42189A
|
| [32] |
Hattori S. Syntrophic acetate-oxidizing microbes in methanogenic environments. Microbes Environ, 2008, 23(2): 118-127. DOI:10.1264/jsme2.23.118
|
| [33] |
Westerholm M, Dolfing J, Schnürer A. Growth characteristics and thermodynamics of syntrophic acetate oxidizers. Environ Sci Technol, 2019, 53(9): 5512-5520. DOI:10.1021/acs.est.9b00288
|
| [34] |
Timmers PHA, Vavourakis CD, Kleerebezem R, et al. Metabolism and occurrence of methanogenic and sulfate-reducing syntrophic acetate oxidizing communities in haloalkaline environments. Front Microbiol, 2018, 9: 3039. DOI:10.3389/fmicb.2018.03039
|
| [35] |
Dolfing J. Thermodynamic constraints on syntrophic acetate oxidation. Appl Environ Microbiol, 2014, 80(4): 1539-1541. DOI:10.1128/AEM.03312-13
|
| [36] |
Lu F, Hao L, Guan D, et al. Synergetic stress of acids and ammonium on the shift in the methanogenic pathways during thermophilic anaerobic digestion of organics. Water Res, 2013, 47(7): 2297-2306. DOI:10.1016/j.watres.2013.01.049
|
| [37] |
Nobu MK, Narihiro T, Mei R, et al. Catabolism and interactions of uncultured organisms shaped by eco-thermodynamics in methanogenic bioprocesses. Microbiome, 2020, 8(1): 111. DOI:10.1186/s40168-020-00885-y
|
| [38] |
De Vrieze J, Christiaens MER, Walraedt D, et al. Microbial community redundancy in anaerobic digestion drives process recovery after salinity exposure. Water Res, 2017, 111: 109-117. DOI:10.1016/j.watres.2016.12.042
|
| [39] |
Smith DP, McCarty PL. Energetic and rate effects on methanogenesis of ethanol and propinate in perturbed CATRs. Biotechnol Bioeng, 1989, 34(1): 39-54. DOI:10.1002/bit.260340106
|
| [40] |
Dyksma S, Jansen L, Gallert C. Syntrophic acetate oxidation replaces acetoclastic methanogenesis during thermophilic digestion of biowaste. Microbiome, 2020, 8(1): 105. DOI:10.1186/s40168-020-00862-5
|
| [41] |
Schuchmann K, Müller V. Autotrophy at the thermodynamic limit of life: a model for energy conservation in acetogenic bacteria. Nat Rev Microbiol, 2014, 12(12): 809-821. DOI:10.1038/nrmicro3365
|
| [42] |
Zamri MFMA, Hasmady S, Akhiar A, et al. A comprehensive review on anaerobic digestion of organic fraction of municipal solid waste. Renew Sustain Energy Rev, 2021, 137: 110637. DOI:10.1016/j.rser.2020.110637
|
| [43] |
戴晓虎. 我国污泥处理处置现状及发展趋势. 科学, 2020, 72(6): 30-34, 4. Dai XH. Applications and perspectives of sludge treatment and disposal in China. Science, 2020, 72(6): 30-34, 4 (in Chinese). |
| [44] |
Hunter SM, Blanco E, Borrion A. Expanding the anaerobic digestion map: a review of intermediates in the digestion of food waste. Sci Total Environ, 2021, 767: 144265. DOI:10.1016/j.scitotenv.2020.144265
|
| [45] |
Sun L, Liu T, Müller B, et al. The microbial community structure in industrial biogas plants influences the degradation rate of straw and cellulose in batch tests. Biotechnol Biofuels, 2016, 9: 128. DOI:10.1186/s13068-016-0543-9
|
| [46] |
Ziels RM, Beck DA, Marti M, et al. Monitoring the dynamics of syntrophic β-oxidizing bacteria during anaerobic degradation of oleic acid by quantitative PCR. FEMS Microbiol Ecol, 2015, 91(4): fiv028.
|
| [47] |
Zhang W, Werner JJ, Agler MT, et al. Substrate type drives variation in reactor microbiomes of anaerobic digesters. Bioresour Technol, 2014, 151: 397-401. DOI:10.1016/j.biortech.2013.10.004
|
| [48] |
Westerholm M, Moestedt J, Schnürer A. Biogas production through syntrophic acetate oxidation and deliberate operating strategies for improved digester performance. Appl Energy, 2016, 179: 124-135. DOI:10.1016/j.apenergy.2016.06.061
|
| [49] |
吴远远. 基于厌氧产酸发酵的新型果蔬垃圾厌氧处理工艺研究[D]. 北京: 清华大学, 2016. Wu YY. Anaerobic digestion of fruit and vegetable waste based on acidogenic fermentation[D]. Beijing: Tsinghua University, 2016 (in Chinese). |
| [50] |
唐兴. 腐殖质对污泥厌氧消化的影响及其屏蔽方法研究[D]. 北京: 北京建筑大学, 2017. Tang X. Study on effect of humus on anaerobic digestion and shielding method of negative impact[D]. Beijing: Beijing University of Civil Engineering and Architecture, 2017 (in Chinese). |
| [51] |
Chen JH, Liu SH, Wang YM, et al. Effect of different hydrolytic enzymes pretreatment for improving the hydrolysis and biodegradability of waste activated sludge. Water Sci Technol, 2018, 2017(2): 592-602. DOI:10.2166/wst.2018.185
|
| [52] |
Liu XG, Wang Q, Tang YZ, et al. Hydrothermal pretreatment of sewage sludge for enhanced anaerobic digestion: resource transformation and energy balance. Chem Eng J, 2021, 410: 127430. DOI:10.1016/j.cej.2020.127430
|
| [53] |
Walter A, Probst M, Franke-Whittle IH, et al. Microbiota in anaerobic digestion of sewage sludge with and without co-substrates. Water Environ J, 2019, 33(2): 214-222. DOI:10.1111/wej.12392
|
| [54] |
郝晓地, 唐兴, 李季, 等. 金属离子屏蔽腐殖酸对污泥厌氧消化抑制作用实验研究. 环境科学学报, 2018, 38(9): 3539-3545. Hao XD, Tang X, Li J, et al. Experimental study on shielding the inhibition effect of humic acid on anaerobic digestion of excess sludge by metals. Acta Sci Circumstant, 2018, 38(9): 3539-3545 (in Chinese). |
| [55] |
Li J, Hao X, van Loosdrecht MCM, et al. Relieving the inhibition of humic acid on anaerobic digestion of excess sludge by metal ions. Water Res, 2021, 188: 116541. DOI:10.1016/j.watres.2020.116541
|
| [56] |
Xin XD, He JG, Feng JH, et al. Solubilization augmentation and bacterial community responses triggered by co-digestion of a hydrolytic enzymes blend for facilitating waste activated sludge hydrolysis process. Chem Eng J, 2016, 284: 979-988. DOI:10.1016/j.cej.2015.09.060
|
| [57] |
Li YM, Wang J, Zhang A, et al. Enhancing the quantity and quality of short-chain fatty acids production from waste activated sludge using CaO2 as an additive. Water Res, 2015, 83: 84-93. DOI:10.1016/j.watres.2015.06.021
|
| [58] |
Ping Q, Zheng M, Dai XH, et al. Metagenomic characterization of the enhanced performance of anaerobic fermentation of waste activated sludge with CaO2 addition at ambient temperature: fatty acid biosynthesis metabolic pathway and CAZymes. Water Res, 2020, 170: 115309. DOI:10.1016/j.watres.2019.115309
|
| [59] |
Wei ZY, Feng K, Wang ZJ, et al. High-throughput single-cell technology reveals the contribution of horizontal gene transfer to typical antibiotic resistance gene dissemination in wastewater treatment plants. Environ Sci Technol, 2021, 55(17): 11824-11834. DOI:10.1021/acs.est.1c01250
|
| [60] |
Yun H, Liang B, Ding Y, et al. Fate of antibiotic resistance genes during temperature-changed psychrophilic anaerobic digestion of municipal sludge. Water Res, 2021, 194: 116926. DOI:10.1016/j.watres.2021.116926
|
| [61] |
Chen Z, Li W, Qin WT, et al. Long-term performance and microbial community characteristics of pilot-scale anaerobic reactors for thermal hydrolyzed sludge digestion under mesophilic and thermophilic conditions. Sci Total Environ, 2020, 720: 137566. DOI:10.1016/j.scitotenv.2020.137566
|
| [62] |
Xu R, Zhang YR, Xiong WP, et al. Metagenomic approach reveals the fate of antibiotic resistance genes in a temperature-raising anaerobic digester treating municipal sewage sludge. J Clean Prod, 2020, 277: 123504. DOI:10.1016/j.jclepro.2020.123504
|
| [63] |
Paulo LM, Stams AJM, Sousa DZ. Methanogens, sulphate and heavy metals: a complex system. Rev Environ Sci Bio-Technol, 2015, 14(4): 537-553. DOI:10.1007/s11157-015-9387-1
|
| [64] |
Hammer L, Palmowski L. Fate of selected organic micropollutants during anaerobic sludge digestion. Water Environ Res, 2021, wer.1603.
|
| [65] |
Uçkun Kiran E, Trzcinski AP, Ng WJ, et al. Bioconversion of food waste to energy: a review. Fuel, 2014, 134: 389-399. DOI:10.1016/j.fuel.2014.05.074
|
| [66] |
Braguglia CM, Gallipoli A, Gianico A, et al. Anaerobic bioconversion of food waste into energy: a critical review. Bioresour Technol, 2018, 248(pt a): 37-56.
|
| [67] |
Sun M, Liu B, Yanagawa K, et al. Effects of low pH conditions on decay of methanogenic biomass. Water Res, 2020, 179: 115883. DOI:10.1016/j.watres.2020.115883
|
| [68] |
Wainaina S, Lukitawesa, Kumar Awasthi M, et al. Bioengineering of anaerobic digestion for volatile fatty acids, hydrogen or methane production: a critical review. Bioengineered, 2019, 10(1): 437-458. DOI:10.1080/21655979.2019.1673937
|
| [69] |
左剑恶, 胡纪萃, 陆正禹, 等. 厌氧消化过程中的酸碱平衡及pH控制的研究. 中国沼气, 1998, 16(1): 3-7. Zuo JE, Hu JC, Lu ZY, et al. Study on acid-base balance and pH control in anaerobic digestion. China Biogas, 1998, 16(1): 3-7 (in Chinese). |
| [70] |
Dang Y, Holmes DE, Zhao Z, et al. Enhancing anaerobic digestion of complex organic waste with carbon-based conductive materials. Bioresour Technol, 2016, 220: 516-522. DOI:10.1016/j.biortech.2016.08.114
|
| [71] |
Li Y, Tang YP, Xiong P, et al. High-efficiency methanogenesis via kitchen wastes served as ethanol source to establish direct interspecies electron transfer during anaerobic co-digestion with waste activated sludge. Water Res, 2020, 176: 115763. DOI:10.1016/j.watres.2020.115763
|
| [72] |
Gao M, Zhang S, Ma XX, et al. Effect of yeast addition on the biogas production performance of a food waste anaerobic digestion system. R Soc Open Sci, 2020, 7(8): 200443. DOI:10.1098/rsos.200443
|
| [73] |
Lim EY, Tian HL, Chen YY, et al. Methanogenic pathway and microbial succession during start-up and stabilization of thermophilic food waste anaerobic digestion with biochar. Bioresour Technol, 2020, 314: 123751. DOI:10.1016/j.biortech.2020.123751
|
| [74] |
Li Y, Chen YG, Wu J. Enhancement of methane production in anaerobic digestion process: a review. Appl Energy, 2019, 240: 120-137. DOI:10.1016/j.apenergy.2019.01.243
|
| [75] |
Long JH, Aziz TN, Reyes FLDL Ⅲ, et al. Anaerobic co-digestion of fat, oil, and grease (FOG): a review of gas production and process limitations. Process Saf Environ Prot, 2012, 90(3): 231-245. DOI:10.1016/j.psep.2011.10.001
|
| [76] |
Amha YM, Sinha P, Lagman J, et al. Elucidating microbial community adaptation to anaerobic co-digestion of fats, oils, and grease and food waste. Water Res, 2017, 123: 277-289. DOI:10.1016/j.watres.2017.06.065
|
| [77] |
Kurade MB, Saha S, Kim JR, et al. Microbial community acclimatization for enhancement in the methane productivity of anaerobic co-digestion of fats, oil, and grease. Bioresour Technol, 2020, 296: 122294. DOI:10.1016/j.biortech.2019.122294
|
| [78] |
Okoro-Shekwaga CK, Ross AB, Camargo-Valero MA. Enhanced in-situ biomethanation of food waste by sequential inoculum acclimation: energy efficiency and carbon savings analysis. Waste Manag, 2021, 130: 12-22. DOI:10.1016/j.wasman.2021.04.053
|
| [79] |
Bi SJ, Qiao W, Xiong LP, et al. Improved high solid anaerobic digestion of chicken manure by moderate in situ ammonia stripping and its relation to metabolic pathway. Renew Energy, 2020, 146: 2380-2389. DOI:10.1016/j.renene.2019.08.093
|
| [80] |
Zhang T, Mao CL, Zhai NN, et al. Influence of initial pH on thermophilic anaerobic co-digestion of swine manure and maize stalk. Waste Manag, 2015, 35: 119-126. DOI:10.1016/j.wasman.2014.09.004
|
| [81] |
Liu C, Wachemo AC, Tong H, et al. Biogas production and microbial community properties during anaerobic digestion of corn stover at different temperatures. Bioresour Technol, 2018, 261: 93-103. DOI:10.1016/j.biortech.2017.12.076
|
| [82] |
Oladejo OS, Dahunsi SO, Adesulu-Dahunsi AT, et al. Energy generation from anaerobic co-digestion of food waste, cow dung and piggery dung. Bioresour Technol, 2020, 313: 123694. DOI:10.1016/j.biortech.2020.123694
|
| [83] |
Noonari AA, Mahar RB, Sahito AR, et al. Anaerobic co-digestion of canola straw and banana plant wastes with buffalo dung: effect of Fe3O4 nanoparticles on methane yield. Renewe Energy, 2019, 133: 1046-1054. DOI:10.1016/j.renene.2018.10.113
|
| [84] |
Peces M, Astals S, Jensen PD, et al. Deterministic mechanisms define the long-term anaerobic digestion microbiome and its functionality regardless of the initial microbial community. Water Res, 2018, 141: 366-376. DOI:10.1016/j.watres.2018.05.028
|
| [85] |
Liu T, Sun L, Nordberg A, et al. Substrate-induced response in biogas process performance and microbial community relates back to inoculum source. Microorganisms, 2018, 6(3): 80. DOI:10.3390/microorganisms6030080
|
| [86] |
Xu R, Yang ZH, Zheng Y, et al. Organic loading rate and hydraulic retention time shape distinct ecological networks of anaerobic digestion related microbiome. Bioresour Technol, 2018, 262: 184-193. DOI:10.1016/j.biortech.2018.04.083
|
| [87] |
Chen H, Hao S, Chen Z, et al. Mesophilic and thermophilic anaerobic digestion of aqueous phase generated from hydrothermal liquefaction of cornstalk: molecular and metabolic insights. Water Res, 2020, 168: 115199. DOI:10.1016/j.watres.2019.115199
|
| [88] |
Inaba T, Aoyagi T, Hori T, et al. Long-term acclimatization of sludge microbiome for treatment of high-strength organic solid waste in anaerobic membrane bioreactor. Biochem Eng J, 2020, 154: 107461. DOI:10.1016/j.bej.2019.107461
|
| [89] |
Dueholm MS, Andersen KS, McIlroy SJ, et al. Generation of comprehensive ecosystem-specific reference databases with species-level resolution by high-throughput full-length 16S rRNA gene sequencing and automated taxonomy assignment (AutoTax). BioRxiv, 2020, 10.
|
| [90] |
Schlüter A, Bekel T, Diaz NN, et al. The metagenome of a biogas-producing microbial community of a production-scale biogas plant fermenter analysed by the 454-pyrosequencing technology. J Biotechnol, 2008, 136(1/2): 77-90.
|
| [91] |
Nguyen AQ, Wickham R, Nguyen LN, et al. Impact of anaerobic co-digestion between sewage sludge and carbon-rich organic waste on microbial community resilience. Environ Sci: Water Res Technol, 2018, 4(12): 1956-1965. DOI:10.1039/C8EW00663F
|
| [92] |
Vanwonterghem I, Jensen PD, Ho DP, et al. Linking microbial community structure, interactions and function in anaerobic digesters using new molecular techniques. Curr Opin Biotechnol, 2014, 27: 55-64. DOI:10.1016/j.copbio.2013.11.004
|
| [93] |
Kakuk B, Wirth R, Maróti G, et al. Early response of methanogenic archaea to H2 as evaluated by metagenomics and metatranscriptomics. Microb Cell Fact, 2021, 20(1): 127. DOI:10.1186/s12934-021-01618-y
|
| [94] |
王丹蕊, 沈文丽, 魏子艳, 等. 单细胞测序技术在微生物生态领域中的应用. 生物技术通报, 2020, 36(10): 237-246. Wang DR, Shen WL, Wei ZY, et al. Applications of single-cell sequencing technology in microbial ecology. Biotechnol Bull, 2020, 36(10): 237-246 (in Chinese). |
| [95] |
Lewis WH, Tahon G, Geesink P, et al. Innovations to culturing the uncultured microbial majority. Nat Rev Microbiol, 2021, 19(4): 225-240. DOI:10.1038/s41579-020-00458-8
|
| [96] |
Harmsen HJ, Kengen HM, Akkermans AD, et al. Detection and localization of syntrophic propionate-oxidizing bacteria in granular sludge by in situ hybridization using 16S rRNA-based oligonucleotide probes. Appl Environ Microbiol, 1996, 62(5): 1656-1663. DOI:10.1128/aem.62.5.1656-1663.1996
|
| [97] |
Godon JJ, Zumstein E, Dabert P, et al. Molecular microbial diversity of an anaerobic digestor as determined by small-subunit rDNA sequence analysis. Appl Environ Microbiol, 1997, 63(7): 2802-2813. DOI:10.1128/aem.63.7.2802-2813.1997
|
| [98] |
LaPara TM, Nakatsu CH, Pantea L, et al. Phylogenetic analysis of bacterial communities in mesophilic and thermophilic bioreactors treating pharmaceutical wastewater. Appl Environ Microbiol, 2000, 66(9): 3951-3959. DOI:10.1128/AEM.66.9.3951-3959.2000
|
| [99] |
Collins G, Woods A, McHugh S, et al. Microbial community structure and methanogenic activity during start-up of psychrophilic anaerobic digesters treating synthetic industrial wastewaters. FEMS Microbiol Ecol, 2003, 46(2): 159-170. DOI:10.1016/S0168-6496(03)00217-4
|
| [100] |
Abram F, Enright AM, O'Reilly J, et al. A metaproteomic approach gives functional insights into anaerobic digestion. J Appl Microbiol, 2011, 110(6): 1550-1560. DOI:10.1111/j.1365-2672.2011.05011.x
|
| [101] |
Zakrzewski M, Goesmann A, Jaenicke S, et al. Profiling of the metabolically active community from a production-scale biogas plant by means of high-throughput metatranscriptome sequencing. J Biotechnol, 2012, 158(4): 248-258. DOI:10.1016/j.jbiotec.2012.01.020
|
| [102] |
Yang D, Fan X, Shi X, et al. Metabolomics reveals stage-specific metabolic pathways of microbial communities in two-stage anaerobic fermentation of corn-stalk. Biotechnol Lett, 2014, 36(7): 1461-1468. DOI:10.1007/s10529-014-1508-3
|
| [103] |
Deng Y, Jiang YH, Yang YF, et al. Molecular ecological network analyses. BMC Bioinformatics, 2012, 13: 113. DOI:10.1186/1471-2105-13-113
|
 2021, Vol. 37
2021, Vol. 37




