中国科学院微生物研究所、中国微生物学会主办
文章信息
- 陈雅维
- Chen Yawei
- ATP调控策略及其在微生物代谢产物合成中的应用
- ATP regulation strategy and its application in the synthesis of microbial metabolites
- 生物工程学报, 2020, 36(8): 1515-1527
- Chinese Journal of Biotechnology, 2020, 36(8): 1515-1527
- 10.13345/j.cjb.200163
-
文章历史
- Received: March 25, 2020
- Accepted: May 20, 2020
- Published: July 24, 2020
为提高微生物细胞工厂的效率,最有效的手段是加强目标化合物的代谢通量。越来越多的研究结果表明,利用传统代谢工程手段,如引入外源基因,过表达、弱化或者敲除代谢途径中的一个或多个基因,往往很难获得高浓度和高产率的目标代谢物,原因之一是忽视了辅因子在代谢途径中的重要调控作用[1-3]。
辅因子是一类可以和蛋白质结合并使蛋白质行使正常催化作用的非蛋白质类物质[4]。辅因子工程是代谢工程的一个新兴分支领域,主要包括ATP/ADP、NADH/NAD+、NADPH/NADP+、乙酰辅酶A、维生素等[5-7]辅因子的调控研究。与传统代谢工程针对酶的改造不同,辅因子工程通过直接调控细胞内关键酶的辅因子浓度和形式来实现代谢流的最大化,推进碳物质流导向目标代谢物[8-10]。最著名的一个实例就是关于如何提高糖酵解途径效率的研究。学者们发现无论是在真核生物还是原核生物中,单独或共同表达多个糖酵解途径中限速酶的基因并不能显著提高酵解途径的代谢速率[11-12]。随后发现ATP水平对于糖酵解代谢速率起着关键作用。有学者通过氧化磷酸化途径的调控降低了胞内ATP浓度,从而有效提高了糖酵解速率[11, 13-14]。与前人通过表达多个糖酵解途径中的关键酶来调节糖酵解通量相比,这种基于辅因子ATP水平的调控策略取得了更为显著的成效[3]。此外,ATP调控甚至可以驱动热力学不利反应向前推进。例如,在蓝藻Synechococcus elongatus中通过引入外源ATP依赖的酶重构代谢途径,首次直接利用CO2光催化合成1-丁醇[15]。这些无疑显示了ATP进行胞内复杂代谢网络调节的巨大潜力。
ATP作为微生物细胞内重要的辅因子参与了大量的酶催化反应,将物质代谢途径串联或并联成复杂的网络体系,最终使得物质代谢流的分配受到辅因子形式和浓度的牵制[3, 16]。ATP主要由原核细胞的细胞质基质、真核细胞线粒体或者叶绿体的ATP合酶产生[17-18]。在酶催化反应中ATP作为基团转移或者直接在各种生化过程中起到辅因子作用,参与RNA和DNA的合成,激活代谢中一些不易催化的反应,如醇、羧酸反应,酯化以及酸酐的形成[19]。此外,ATP还作为信号分子在转录后修饰中起到作用[20-21]。目前KEGG数据库(http://www.kegg.jp)显示ATP参与细胞内700余个代谢反应,涉及到工业菌株大部分目标代谢产物。因此ATP调控策略有望成为微生物菌株改造的有利工具,用于提高目标代谢物的浓度和产率[22-23]、增强微生物对环境的耐受度[24-25]以及促进底物利用[26]等。本文将从常用的ATP调控策略以及ATP调控对细胞代谢和产物合成的影响来论述,以期为实现微生物细胞工厂的高效构建提供参考。
1 胞内ATP调控策略基于目前对物质代谢和能量代谢有了更多的理解,再加上合成生物学的飞速发展,学者们在宿主微生物中引入了各种各样的ATP调控策略(图 1),用以满足微生物细胞工厂中不同目标代谢物产量提高或者环境耐受等需求[27-29]。表 1中列举了部分ATP调控策略及应用。
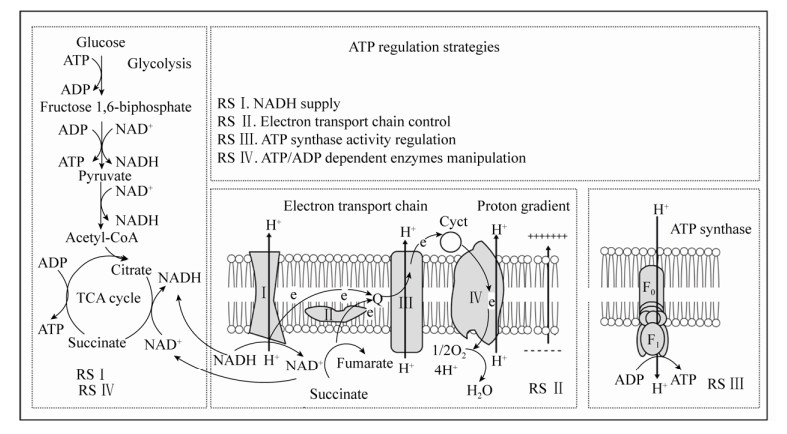
|
| 图 1 胞内ATP合成途径及ATP调控策略示意图[16, 30] Fig. 1 Schematic diagram of intracellular ATP synthesis pathway and ATP regulation strategy[16, 30]. Complex Ⅰ: NADH: ubiquinone oxidoreductase; Complex Ⅱ: succinate:ubiquinone oxidoreductase; Complex Ⅲ: ubiquinol:ferricytochrome c oxidoreductase; Complex Ⅳ: ferricytochrome c:oxygen oxidoreductase; RS: regulation strategy. |
| |
| Cofactor regulation strategies | Organism | Product | Titer or yield | Reference | |
| Regulation of NADH level by metabolic engineering | Deletion of adh1 and overexpression of ald3 |
Saccharomyces cerevisiae CENPK2 |
Glycerol | 0.46 g/g glucose | [32] |
| Overexpression of mitochondrial alternative oxidase gene aox | Aspergillus niger CGMCC 10142 |
Citric acid | 169.1 g/L | [33] | |
| Disruption of por1 gene | Candida utilis CCTCC M 209298 |
Co-production of S-adenosylmethionine and glutathione | 524.3 mg/L | [34] | |
| Control of NADH supply | Addition of 50 mmol/L citrate | Candida glabrata CCTCC M202019 |
Pyruvic acid | 68 g/L | [35] |
| Pulsed-feeding of citric acid | Bacillus licheniformis ATCC 9945A | Poly-γ-glutamic acid | 35 g/L | [36] | |
| Co-feeding of formate | Penicillium chrysogenum DS17690 |
Penicillin G | 0.5 g/g glucose | [26] | |
| Addition of 6 g/L sodium citrate | Saccharomyces cerevisiae CGMCC 2842 | S-adenosylmethionine | 1.85 g/L | [37] | |
| Addition of 62 mmol/L citrate | Lactobacillus panis PM1 |
Succinate, lactate | 40.9 mmol/L, 88.7 mmol/L | [38] | |
| Regulation of the activity of electron transport chain | Addition of rotenone, antimycin or oligomycin |
Torulopsis glabrata CCTCC M202019 |
Pyruvate | 49.8 g/L | [39] |
| Regulation of proton gradient |
Control of pH at 4.2 | Streptomyces albulus NBRC14147 | ε-Poly-L-Lysine | 1.5 g/L | [40] |
| Control of pH at 3.8 | Aureobasidium pullulans CCTCC M 2012259 | Pullulan | 26.8 g/L | [41] | |
| Control of oxygen supply | Optimizing the DO supply through changes of the agitation rate |
Candida utilis CCTCC M 209298 |
Co-production of S-adenosylmethionine and glutathione | 593.9 mg/L | [42] |
| Addition of n-heptane at the final concentrantions of 1% | Pichia pastoris GS115 | S-adenosylmethionine | 1.25 g/L | [43] | |
| Chromosomal integration of the Vitreoscilla hemoglobin gene | Saccharopolyspora spinosa SP06081 |
Spinosad | 466.6 mg/L | [44] | |
| Overexpression of Vitreoscilla hemoglobin | Aureobasidium melanogenum P16 |
Pullulan | 102 g/L | [45] | |
| Regulation of F0F1-ATP synthase activity | Heterogenous expression of ATP6 gene |
Candida utilis CCTCC M 209298 |
Co-production of S-adenosylmethionine and glutathione | 455.6 mg/L | [46] |
| Deletion of cg1360 | Corynebacterium glutamicum ATCC 13032 | L-valine | 9.2 g/L | [47] | |
| Manipulation of ATP-dependent key enzymes | Introduction of phosphoenolpyruvate carboxykinase, disruption of the genes involved in the glucose phosphotransferase system | Enterobacter aerogenes AJ110637 | Succinate | 0.727 g/g glucose | [48] |
| Overexpression of phosphoenolpyruvate carboxykinase | Escherichia coli BW25113 |
Succinate | 5.97 g/L | [49] | |
| Overexpression of the ATP-related key enzymes, such as phosphoglycerate kinase, pyruvate kinase and adenylosuccinate lyase |
Bacillus amyloliquefaciens |
Putrescine | 5.51 g/L | [2] | |
| Knock-down the genes of proB, glnA and argB in ATP consumption pathway through synthetic sRNA approach | Escherichia coli BL21 (DE3) | S-adenosylmethionine | 1.21 mg/L | [50] | |
| Knock-down the ATP-related genes of metK and proB through CRISPR interference system | Escherichia coli BL21 (DE3) | Pinocembrin | 110.44 mg/L | [51] | |
| Re-construction of the ATP-driven malonyl-CoA synthesis pathway | Synechococcus elongates PCC 7942 |
1-butanol | 29.9 mg∕L | [15] | |
| Compound regulation strategy | Deletion of cydB and cydC, introduction of VHb; overexpression of the genes purB and adK in ATP-biosynthetic pathway | Bacillus licheniformis WX-02 | Poly-γ-glutamic acid | 43.81 g/L | [29] |
微生物中ATP水平调控方式主要有两种:一是ATP合成或者消耗途径相关酶的代谢调控,主要涉及ADP依赖酶的再生过程,例如磷酸甘油酸激酶、丙酮酸激酶和/或聚磷酸激酶等[19]。另一种方式是氧化磷酸化水平的调控。在有氧条件下,ATP主要通过氧化磷酸化途径合成,并且氧化磷酸化比底物水平磷酸化的产能效率更高。因此通过氧化磷酸化途径来调控胞内ATP水平更为有效[19, 31]。其中NADH水平、电子传递链、ATP合酶活性等是胞内ATP水平调节的主要位点。
1.1 胞内NADH供给的调控细胞内NADH主要来源于糖酵解、脂肪酸氧化以及三羧酸循环。在有氧条件下,NADH通过电子传递链氧化,以氧气为终端电子受体产生ATP[3]。在厌氧条件下,NADH经由发酵途径的乙醛脱氢酶或乳酸脱氢酶等作用氧化[52-53],此时底物水平磷酸化是ATP的主要来源[3]。因此,可以通过代谢工程或者能量底物加入等方式来操控NADH水平,最终实现ATP水平的调控[54-55]。
1.1.1 基于代谢工程的NADH调控根据胞内NADH代谢途径,利用基因重组技术外源引入、过表达、敲除或者弱化NADH (NAD+)依赖的关键酶,从而通过操控NADH水平来调控胞内ATP水平[54]。例如敲除酿酒酵母Saccharomyces cerevisiae中消耗NADH的乙醇脱氢酶,并过表达生成NADH的乙醛脱氢酶,有效提高了胞内NADH的水平,也因此提高了NADH依赖的产物甘油的产量[32]。
为避免NADH调控过程中对于胞内其他物质代谢的影响,可选择亚磷酸脱氢酶和NADH氧化酶等,直接作用于胞内NADH水平的调控。施氏假单胞菌Pseudomonas stutzeri来源的亚磷酸脱氢酶PTDH几乎不可逆转地将亚磷酸氧化成磷酸,同时NAD+还原为NADH[56-57]。除此之外,NADH也可由氧化酶直接氧化生成水[58]。其中形成水的NADH氧化酶定位于细胞质中,直接将细胞质中NADH氧化成水。例如在酿酒酵母中表达来源于乳酸乳球菌Lactococcus lactis中形成水的NADH氧化酶NoxE,胞内NADH浓度降低5倍,NADH/NAD+比率降低6倍[59]。而交替氧化酶Aox是线粒体中交替呼吸途径的末端氧化酶,其氧化效率更高。在线粒体中过表达Aox,使得NADH的氧化从电子传递链转向交替氧化酶途径,从而降低ATP水平及NADH/NAD+比值[60]。Li等在黑曲霉Aspergillus niger中过表达交替氧化酶基因aox,重组菌细胞生长速率和产物柠檬酸得率都有所增加[33]。
与线粒体NADH不同,胞质NADH必须穿过线粒体外膜中的孔道,才能将电子传递至NADH脱氢酶[61]。因此,有学者将产朊假丝酵母Candida utilis CCTCCM 209298中编码线粒体孔道蛋白的基因por1敲除,在分批式发酵中重组菌胞内NADH和ATP浓度均有所增加,S-腺苷蛋氨酸和谷胱甘肽联产浓度增加34.9%[34]。
1.1.2 NADH能量底物的利用直接在培养基中添加依赖NAD+脱氢酶的相关底物,可以有效提高胞内ATP的水平。柠檬酸(盐)和甲酸等辅助能量底物通过NAD+依赖脱氢酶的反应,产生额外的NADH,增强了NADH对电子的贡献。NADH通过电子传递链产生质子梯度,再经由膜定位的F0F1-ATP合成酶促进ATP合成[26, 62]。例如通过脉冲流加柠檬酸,地衣芽孢杆菌Bacillus licheniformis中聚γ-谷氨酸浓度可达35 g/L[36]。在S-腺苷蛋氨酸和谷胱甘肽联产的研究中添加柠檬酸钠作为能量底物,有效提高了目标化合物的产量[37]。此外,在以葡萄糖为主要底物的培养基中添加甲酸作为能量底物,产黄青霉菌Penicillium chrysogenum中青霉素的产率也有所提高[24]。通过加入柠檬酸钠,光滑假丝酵母Candida glabrata胞内ATP水平增加,不仅提高了丙酮酸的产率,而且促进了细胞对于酸性环境的耐受性[35]。由于丙酮酸的增加,下游乙酸和乳酸的产量也得到提升[38]。
基于NADH调控胞内ATP的策略较为容易操作,而且效率高,适用于上调或者下调胞内ATP水平。但是NADH的变化会影响胞内氧化还原状态,同时也会作用于细胞生长代谢及产物合成。因此,NADH水平改变并不是ATP调控的直接方法,需要综合考虑细胞的氧化还原状态是否利于目标代谢物的合成。
1.2 电子传递链的调控电子传递链是由复合物Ⅰ、Ⅱ、Ⅲ和Ⅳ组成的复杂体系,主要通过氧化还原反应将电子从电子供体转至终端电子受体氧气,过程产生质子梯度用于ATP的合成[63]。电子传递链对于ATP合成至关重要,但由于涉及基因众多,难以通过单个或多个相关基因的操控来提高ATP水平。然而破坏电子传递链中的任一环节,都可能会影响ATP的合成[3]。
1.2.1 调控电子传递链的活性目前电子传递链活性调控主要用于下调胞内ATP水平,其调控策略主要包括添加电子传递链抑制剂和代谢工程手段。抑制剂可以和电子传递链的某个部分相结合,不可逆转地阻止电子进一步传递,从而阻碍ATP合成。例如异戊巴比妥和鱼藤酮可以破坏复合物Ⅰ的电子传递,叠氮化物和氰化物则会抑制复合物Ⅳ的活性[39]。其次可以通过代谢工程方式破坏电子传递链的功能,如在电子传递链中引入交替氧化酶Aox,从而减少ATP生成[60]。最后还可以通过突变方式选育缺失电子传递链关键部分的菌株[3]。Liu等在基质中添加10 mg/L的鱼藤酮或者制霉素A来分别抑制复合物Ⅰ和复合物Ⅱ,使得光滑球拟酵母Torulopsis glabrata胞内ATP水平分别降低43%和27.7%,葡萄糖的消耗速率和丙酮酸的生产速率显著增加[39]。
1.2.2 调节质子梯度质子梯度是在电子传递链氧化NADH,驱动F0F1-ATP合酶生成ATP时产生。二甲苯和细菌素等都可以破坏细胞膜或者线粒体膜的透性,破坏质子梯度从而影响ATP的合成[3, 16]。而控制pH在酸性条件下可以显著增强微生物胞内ATP的供应。这是因为较低的外界pH有利于质子梯度的产生,从而利于驱动呼吸链中的F0F1-ATP合酶。例如控制酸性pH条件可促进白色链霉菌Streptomyces albulus细胞内ATP的供应,激发ATP依赖的ε-聚-l-赖氨酸合成酶的活性,大大提高目标化合物ε-聚-l-赖氨酸的合成[40]。在有氧、酸性pH条件下,出芽短梗霉Aureobasidium pullulans细胞内ATP/ADP比值在pH 3.5–4.5范围内与外界酸度成比例增加。通过外界pH调控增加了胞内ATP的水平,提高了普鲁兰多糖的产量,并促进了多糖的排出和细胞的耐酸性[41]。
1.2.3 调控氧气供给在有氧发酵中,氧气是电子传递链的终端受体。因此发酵体系中的氧气浓度是ATP合成的重要条件[64]。特别是对于高密度、高粘度和高能量需求的发酵体系的控制过程更要保证氧气的供应[65]。研究表明调控发酵罐的搅拌速度、在发酵过程通入纯氧或者加入正庚烷、正己烷等携氧载体均可以促进胞内氧气的供给[3, 42-43]。Li等在培养基中加入4% (V/V)正己烷,提高了酿酒酵母BZ66胞内ATP水平,2.5 L发酵罐中S-腺苷蛋氨酸产量提高23.37% (2.27 g/L)[43]。此外,在细胞内引入透明颤菌Vitreoscilla来源的血红蛋白(VHB),已成功用于多种ATP依赖的目标代谢物的增产[45, 65-66]。在低溶氧条件下,微生物胞内的血红蛋白与氧气结合,提高胞内溶氧,从而促进ATP合成[27, 67]。Luo等报道在刺糖多胞菌Saccharopolyspora spinosa中表达血红蛋白后,在正常和有限的氧气条件下天然杀虫剂的产量均有所增加[44]。还有学者研究发现,在大肠杆菌的限氧发酵过程中,血红蛋白的引入使得细胞生物量增加11%左右[68]。
虽然很难通过直接操作某个或多个相关基因来调控电子传递链活性,但是通过提高电子传递链最终受体氧气供给的方式可以有效地提高ATP合成能力。此外还可以通过外界pH的控制来调节质子梯度。但是要注意在微生物细胞工厂中ATP生成和消耗之间的平衡可能与最佳pH条件不同。因此需要平衡微生物细胞中目标化合物的生产能力和pH耐受性之间的关系。
1.3 F0F1-ATP合酶活性的调节F0F1-ATP合酶是氧化磷酸化的最终反应部分,也是ATP合成中最关键的部分。学者们认为过表达F0F1-ATP合酶是提高ATP水平最直接高效的方法,可惜迄今为止并未取得显著成效[3]。Zhang等将来源于拟南芥Arabidopsis thaliana的线粒体ATP6基因在酿酒酵母中表达,结果表明F0F1-ATP合酶活性和ATP合成都有所提升,重组菌株对各种胁迫因素的耐受性也显著提高[69]。后有人将拟南芥来源的线粒体ATP6基因整合至产朊假丝酵母CCTCC M 209298中,胞内谷胱甘肽和S-腺苷蛋氨酸产量分别提高46.6%和28.7%[46]。
目前主要有3种方法可用于下调F0F1-ATP合酶活性:在基质中添加ATP合酶抑制剂,代谢工程手段改造ATP合酶以及诱变育种筛选ATP合酶突变株[70-71]。目前已知的天然或者合成的抑制剂超过250种[30]。如一种抗结核药物二芳基喹啉(Diarylquinoline,TMC207),通过靶向结合细菌ATP合酶的c亚基来阻断ATP的合成[30]。Liu等通过亚硝基胍诱变筛选得到的光滑球拟酵母突变菌株,F0F1-ATP合酶活性降低65%,胞内ATP浓度降低24%[31]。将枯草芽孢杆菌中ATP合酶的cg1360基因敲除,胞内ATP浓度降低72%,NADH和NADPH浓度有所增加[47]。下调F0F1-ATP合酶活性,导致经由呼吸链合成的ATP减少,从而将碳代谢流导向糖酵解和三羧酸循环,利于丙酮酸及下游代谢物的合成。
目前学者们仍致力于ATP合酶三维结构与功能的解析,这也为ATP合酶作用机理的揭示和ATP合酶的分子改造奠定了基础[72-73]。
1.4 ATP (ADP)依赖酶的代谢调控根据胞内ATP合成或者消耗的相关代谢途径(图 2),通过基因重组技术外源引入、过表达、敲除或者弱化关键酶,从而直接调控ATP水平。最常用的就是利用ADP依赖的酶催化过程进行ATP再生,例如过表达或者外源引入磷酸甘油酸激酶、丙酮酸激酶和/或聚磷酸激酶等提高胞内ATP水平。将来自琥珀酸放线杆菌Actinobacillus succinogenes的磷酸烯醇丙酮酸羧激酶(PCK)在大肠杆菌中表达,有效地促进了细胞生长和琥珀酸的产生[49]。此外,通过相似的ATP调控策略,在产气肠杆菌Enterobacter aerogenes中异源表达PCK,并敲除葡萄糖磷酸转移酶系统来增加ATP的生成,提高了琥珀酸的产量[48]。Kozak等将外源的非ATP依赖途径替换酿酒酵母中天然依赖ATP的途径,从而实现乙醇到乙酰辅酶A等平台化合物的代谢途径重构[74]。
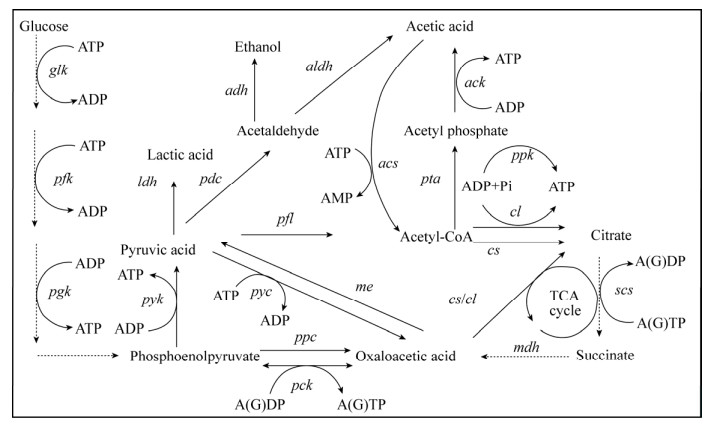
|
| 图 2 胞内ATP合成及消耗途径示意图 Fig. 2 Schematic diagram of intracellular ATP synthesis and consumption pathway. glk: glucokinase; pfk: 6-phosphofructokinase; pgk: phosphoglycerate kinase; pyk: pyruvate kinase; ldh: lactate dehydrogenase; adh: alcohol dehydrogenase; pdc: pyruvate decarboxylase; aldh: aldehyde dehydrogenase; pta: phosphate acetyltransferase; ack: acetate kinase; pyc: pyruvate carboxylase; pck: phosphoenolpyruvate carboxy kinase; ppc: phosphoenolpyruvate carboxylase; pfl: pyruvate-formate lyase; cs: citrate synthase; cl: citrate lyase; scs: succinyl-CoA synthase; mdh: malate dehydrogenase; me: malic enzyme; acs: acetyl-CoA synthetase; ppk: polyphosphate kinase. |
| |
聚磷酸激酶(ppk2编码)是近年来新发现的可用于ATP再生的酶。它可以将廉价的无机聚磷酸催化为ATP而不涉及胞内其他代谢物。但是该反应效率较低,反应条件严苛,在微生物细胞中应用较少,常用于体外酶催化体系中ATP再生系统的构建[75]。大多数聚磷酸激酶只在具有10多个磷酸盐残基的长聚磷酸存在下才具有活性。最近从谷氨酸棒杆菌Corynebacterium glutamate中发现的聚磷酸激酶可以利用三聚或四聚磷酸盐作为磷酸盐供体再生ATP。廉价底物的使用大大拓宽了聚磷酸激酶的应用范围[76-77]。例如鼠李树胶糖激酶和磷酸激酶偶联用于D-果糖的生产[78]。
除了强化ATP再生途径,此外还可以利用高效的基因操作方法来减少非目标合成途径对于ATP的消耗。例如利用小分子RNA技术[50]、CRISPR或者CRISPR干扰(CRISPRi)技术[51]敲除、弱化ATP消耗途径的基因,从而使得目标代谢物合成的ATP供应增加。
2 ATP调控对细胞代谢的影响在细胞的生长代谢活动中,ATP主要参与细胞内物质的转运和代谢。例如ATP的水平直接影响有机酸等小分子代谢物的分泌[79-80]。Hara等通过在酿酒酵母中引入消耗ATP的ADP1基因从而使得胞内的谷胱甘肽得以运输到胞外,进而提高了谷胱甘肽的产量[81]。细胞内的能量状态也会影响细胞生长和代谢反应的速率[82-83],因此ATP对于蛋白质、脂类、核苷酸和氨基酸等物质的合成代谢均有重要影响。例如胞内ATP供应的增加可以促进聚氨基酸和多糖等目标代谢物的合成[84-85]。而胞内ATP浓度的减少会致使中心代谢途径通量的增加,从而促进丙酮酸、谷氨酸等目标代谢物的合成[31, 86]。此外微生物在目标代谢物合成过程中对于ATP需求的程度不同,例如,在重组的酿酒酵母生产乳酸时,较低的ATP水平有利于从葡萄糖合成乳酸,而乳酸分泌则需要较高的ATP能量供应[79]。
此外许多研究表明,ATP调控可以影响基因的转录和表达。例如随着酵母细胞内ATP水平的改变,胞内中心碳代谢中糖酵解和三羧酸循环的众多基因转录水平均发生上调或者下调[22, 60]。ATP调控也会影响全局转录调控因子和信号转导系统。研究表明许多全局转录因子例如ArcA、Fnr、CRP和IHF通常都与NADH和ATP水平有关,其他的转录因子也与ATP或者NADH水平有关[87-88]。可以通过操作这些全局转录调控因子来改变胞内NADH或ATP的水平。例如,在敲除arc基因的大肠杆菌中,三羧酸循环通量增加了4.4倍,ATP/ADP比值增加了2倍[89]。这些结果表明,全局转录调控因子可以影响许多新的内源性靶点,以有效地调节细胞内的能量状态。
3 总结与展望以辅因子调控为核心的辅因子工程成为代谢工程和合成生物学的一个重要研究方向。其中以ATP为辅因子的调控研究更是成为提高微生物细胞工厂效率的重要技术手段。虽然ATP调控策略在许多代谢物合成中得到广泛的应用,但仍然存在一些困难和问题。其一,目前对于呼吸链和ATP合酶的直接代谢工程调控研究较少。主要原因是它们结构太复杂,涉及基因数量众多。随着越来越多的呼吸链及ATP合酶结构被解析,对于ATP合成机理的理解更加深入,结合基因操作技术的迅速发展,将来有望通过代谢工程方式直接改造呼吸链和ATP合酶,从而实现ATP的高效调控。其二,如何平衡ATP调控在目标代谢物和副产物之间的分配。为此我们需要探究ATP扰动对碳代谢及其有效生成目标化合物的代谢流分配机制。例如通过代谢组、转录组等组学方式来研究ATP调控在胞内能量和物质代谢中的分配。也可以借助COBRA等工具通过代谢流平衡分析(Flux balance analysis,FBA)来辅助研究。其三,如何满足代谢物合成过程中对于ATP的不同需求。如果胞内ATP浓度过高,还可能会抑制微生物细胞生长。这就要求我们开发更为精确的ATP调控策略,例如ATP的动态调控。目前已有利用ATP响应的核糖开关ydaO模块来动态调控胞内ATP水平,从而利于S-腺苷蛋氨酸和谷胱甘肽等小分子物质合成的研究。未来通过计算机模拟结合定点突变技术改变天然ydaO模块序列,从而改变该核糖开关对于ATP响应的浓度范围,有望将该方法进一步推广,以适应不同ATP依赖代谢物的生产。
此外,ATP还参与活细胞的各种病理过程,因此迫切需要能在活细胞、组织和环境样品中灵敏、有选择性地检测ATP浓度。因此,实时检测ATP浓度的荧光传感器的开发也引起了学者们的高度关注。将来有望利用ATP荧光传感器来解析胞内ATP浓度对于代谢物合成的机理研究。
| [1] |
Wang M, Chen BQ, Fang YM, et al. Cofactor engineering for more efficient production of chemicals and biofuels. Biotechnol Adv, 2017, 35(8): 1032-1039. DOI:10.1016/j.biotechadv.2017.09.008 |
| [2] |
Li L, Zou D, Ji AY, et al. Multilevel metabolic engineering of Bacillus amyloliquefaciens for production of the platform chemical putrescine from sustainable biomass hydrolysateS. ACS Sustainable Chem Eng, 2020, 8(5): 2147-2157. DOI:10.1021/acssuschemeng.9b05484 |
| [3] |
Chen YW. Cofactor-based regulation for S-adenosylmethionine production[D]. Beijing: Beijing University of Chemical Technology, 2016 (in Chinese). 陈雅维.基于辅因子调控的S-腺苷蛋氨酸合成研究[D].北京: 北京化工大学, 2016. |
| [4] |
Akhtar MK, Jones PR. Cofactor engineering for enhancing the flux of metabolic pathways. Front Bioeng Biotechnol, 2014, 2: 30. |
| [5] |
Chen XL, Li SB, Liu LM. Engineering redox balance through cofactor systerms. Trends Biotechnol, 2014, 32(6): 337-343. DOI:10.1016/j.tibtech.2014.04.003 |
| [6] |
Tan ZT, Zhu CJ, Fu JW, et al. Regulating cofactor balance in vivo with a synthetic flavin analogue. Angewand Chem Int Ed, 2018, 57(50): 16464-16468. DOI:10.1002/anie.201810881 |
| [7] |
Wu JJ, Zhang X, Zhou P, et al. Improving metabolic efficiency of the reverse beta-oxidation cycle by balancing redox cofactor requirement. Metabol Eng, 2017, 44: 313-324. DOI:10.1016/j.ymben.2017.11.001 |
| [8] |
Lee JY, Kang CD, Lee SH, et al. Engineering cellular redox balance in Saccharomyces cerevisiae for improved production of L-lactic acid. Biotechnol Bioeng, 2015, 112(4): 751-758. DOI:10.1002/bit.25488 |
| [9] |
Kim S, Hahn JS. Efficient production of 2, 3-butanediol in Saccharomyces cerevisiae by eliminating ethanol and glycerol production and redox rebalancing. Metabol Eng, 2015, 31: 94-101. DOI:10.1016/j.ymben.2015.07.006 |
| [10] |
Chen YW, Xu DB, Fan LH, et al. Manipulating multi-system of NADPH regulation in Escherichia coli for enhanced S-adenosylmethionine production. RSC Adv, 2015, 5(51): 41103-41111. DOI:10.1039/C5RA02937F |
| [11] |
Larsson C, Pahlman IL, Gustafsson L. The importance of ATP as a regulator of glycolytic flux in Saccharomyces cerevisiae. Yeast, 2000, 16(9): 797-809. DOI:10.1002/1097-0061(20000630)16:9<797::AID-YEA553>3.0.CO;2-5 |
| [12] |
Shimizu K, Matsuoka Y. Regulation of glycolytic flux and overflow metabolism depending on the source of energy generation for energy demand. Biotechnol Adv, 2019, 37(2): 284-305. DOI:10.1016/j.biotechadv.2018.12.007 |
| [13] |
Koebmann BJ, Westerhoff HV, Snoep JL, et al. The glycolytic flux in Escherichia coli is controlled by the demand for ATP. J Bacteriol, 2002, 184(14): 3909-3916. DOI:10.1128/JB.184.14.3909-3916.2002 |
| [14] |
Sekine H, Shimada T, Hayashi C, et al. H+-ATPase defect in Corynebacterium glutamicum abolishes glutamic acid production with enhancement of glucose consumption rate. Appl Microbiol Biotechnol, 2001, 57(4): 534-540. DOI:10.1007/s002530100778 |
| [15] |
Lan EI, Liao JC. ATP drives direct photosynthetic production of 1-butanol in cyanobacteria. Proc Natl Acad Sci USA, 2012, 109(16): 6018-6023. DOI:10.1073/pnas.1200074109 |
| [16] |
Zhou JW, Liu LM, Shi ZP, et al. ATP in current biotechnology: regulation, applications and perspectives. Biotechnol Adv, 2009, 27(1): 94-101. DOI:10.1016/j.biotechadv.2008.10.005 |
| [17] |
Watt IN, Montgomery MG, Runswick MJ, et al. Bioenergetic cost of making an adenosine triphosphate molecule in animal mitochondria. Proc Natl Acad Scie USA, 2010, 107(39): 16823-16827. DOI:10.1073/pnas.1011099107 |
| [18] |
Andexer JN, Richter M. Emerging enzymes for atp regeneration in biocatalytic processes. Chem Bio Chem, 2015, 16(3): 380-386. DOI:10.1002/cbic.201402550 |
| [19] |
Hara KY, Kondo A. ATP regulation in bioproduction. Microb Cell Fact, 2015, 14: 198. DOI:10.1186/s12934-015-0390-6 |
| [20] |
Sivaramakrishnan V, Fountain SJ. Evidence for extracellular ATP as a stress signal in a single-celled organism. Eukaryot Cell, 2015, 14(8): 775-782. DOI:10.1128/EC.00066-15 |
| [21] |
Hill S, Van Remmen H. Mitochondrial stress signaling in longevity: a new role for mitochondrial function in aging. Redox Biol, 2014, 2: 936-944. DOI:10.1016/j.redox.2014.07.005 |
| [22] |
Chen YW, Tan TW. Enhanced S-Adenosylmethionine Production by increasing atp levels in baker's yeast (Saccharomyces cerevisiae). J Agric Food Chem, 2018, 66(20): 5200-5209. DOI:10.1021/acs.jafc.8b00819 |
| [23] |
Causey TB, Shanmugam KT, Yomano LP, et al. Engineering Escherichia coli for efficient conversion of glucose to pyruvate. Proc Natl Acad Sci USA, 2004, 101(8): 2235-2240. DOI:10.1073/pnas.0308171100 |
| [24] |
Shima J, Ando A, Takagi H. Possible roles of vacuolar H+-ATPase and mitochondrial function in tolerance to air-drying stress revealed by genome-wide screening of Saccharomyces cerevisiae deletion strains. Yeast, 2008, 25(3): 179-190. DOI:10.1002/yea.1577 |
| [25] |
Sheng JY, Marquis RE. Enhanced acid resistance of oral streptococci at lethal pH values associated with acid-tolerant catabolism and with ATP synthase activity. FEMS Microbiol Lett, 2006, 262(1): 93-98. DOI:10.1111/j.1574-6968.2006.00374.x |
| [26] |
Harris DM, Van Der Krogt ZA, Van Gulik WM, et al. Formate as an auxiliary substrate for glucose-limited cultivation of Penicillium chrysogenum: impact on penicillin g production and biomass yield. Appl Environ Microbiol, 2007, 73(15): 5020-5025. DOI:10.1128/AEM.00093-07 |
| [27] |
Zhang H, Kang X, Xiao N, et al. Intracellular expression of Vitreoscilla haemoglobin improves lipid production in Yarrowia lipolytica. Lett Appl Microbiol, 2019, 68(3): 248-257. DOI:10.1111/lam.13111 |
| [28] |
Chen YW, Zhou HY, Wang M, et al. Control of ATP concentration in Escherichia coli using an ATP-sensing riboswitch for enhanced S-adenosylmethionine production. RSC Adv, 2017, 7(36): 22409-22414. DOI:10.1039/C7RA02538F |
| [29] |
Cai DB, Chen YZ, He PH, et al. Enhanced production of poly-γ-glutamic acid by improving ATP supply in metabolically engineered Bacillus licheniformis. Biotechnol Bioeng, 2018, 115(10): 2541-2553. DOI:10.1002/bit.26774 |
| [30] |
Neupane P, Bhuju S, Thapa N, et al. ATP Synthase: structure, function and inhibition. Biomol Concepts, 2019, 10(1): 1-10. |
| [31] |
Liu LM, Li Y, Du GC, et al. Increasing glycolytic flux in Torulopsis glabrata by redirecting ATP production from oxidative phosphorylation to substrate-level phosphorylation. J Appl Microbiol, 2006, 100(5): 1043-1053. DOI:10.1111/j.1365-2672.2006.02871.x |
| [32] |
Cordier H, Mendes F, Vasconcelos I, et al. A metabolic and genomic study of engineered Saccharomyces cerevisiae strains for high glycerol production. Metabol Eng, 2007, 9(4): 364-378. DOI:10.1016/j.ymben.2007.03.002 |
| [33] |
Hou L, Liu L, Zhang HF, et al. Functional analysis of the mitochondrial alternative oxidase gene (aox1) from Aspergillus niger CGMCC 10142 and its effects on citric acid production. Appl Microbiol Biotechnol, 2018, 102(18): 7981-7995. DOI:10.1007/s00253-018-9197-9 |
| [34] |
Wang DH, Li DC, Zhang GC, et al. Disruption of por1 gene in Candida utilis improves co-production of S-adenosylmethionine and glutathione. J Biotechnol, 2019, 290: 16-23. DOI:10.1016/j.jbiotec.2018.12.005 |
| [35] |
Zhou J, Liu L, Chen J. Improved ATP supply enhances acid tolerance of Candida glabrata during pyruvic acid production. J Appl Microbiol, 2011, 110(1): 44-53. |
| [36] |
Yoon SH, Do JH, Lee SY, et al. Production of poly-γ-glutamic acid by fed-batch culture of Bacillus licheniformis. Biotechnol Lett, 2000, 22(7): 585-588. DOI:10.1023/A:1005625026623 |
| [37] |
Chen HL, Wang Z, Wang ZL, et al. Improving methionine and ATP availability by MET6 and SAM2 co-expression combined with sodium citrate feeding enhanced SAM accumulation in Saccharomyces cerevisiae. World J Microbiol Biotechnol, 2016, 32: 56. DOI:10.1007/s11274-016-2010-y |
| [38] |
Kang TS, Korber DR, Tanaka T. Contributions of citrate in redox potential maintenance and ATP production: metabolic pathways and their regulation in Lactobacillus panis PM1. Appl Microbiol Biotechnol, 2013, 97(19): 8693-8703. DOI:10.1007/s00253-013-5108-2 |
| [39] |
Liu LM, Li Y, Li HZ, et al. Significant increase of glycolytic flux in Torulopsis glabrata by inhibition of oxidative phosphorylation. FEMS Yeast Res, 2006, 6(8): 1117-1129. DOI:10.1111/j.1567-1364.2006.00153.x |
| [40] |
Yamanaka K, Kito N, Imokawa Y, et al. Mechanism of ε-poly-L-lysine production and accumulation revealed by identification and analysis of an ε-poly-L-lysine-degrading enzyme. Appl Environ Microbiol, 2010, 76(17): 5669-5675. DOI:10.1128/AEM.00853-10 |
| [41] |
Wang DH, Yu XL, Wei GY. Pullulan production and physiological characteristics of Aureobasidium pullulans under acid stress. Appl Microbiol Biotechnol, 2013, 97(18): 8069-8077. DOI:10.1007/s00253-013-5094-4 |
| [42] |
Li DC, Wang DH, Wei GY. Efficient co-production of S-adenosylmethionine and glutathione by Candida utilis: effect of dissolved oxygen on enzyme activity and energy supply. J Chem Technol Biotechnol, 2017, 92(8): 2150-2158. DOI:10.1002/jctb.5226 |
| [43] |
Li MH, Meng XM, Diao EJ, et al. Productivity enhancement of S-adenosylmethionine in Saccharomyces cerevisiae using n-hexadecane as oxygen vector. J Chem Technol Biotechnol, 2012, 87(10): 1379-1384. DOI:10.1002/jctb.3752 |
| [44] |
Luo YS, Kou XX, Ding XZ, et al. Promotion of spinosad biosynthesis by chromosomal integration of the Vitreoscilla hemoglobin gene in Saccharopolyspora spinosa. Sci China Life Sci, 2012, 55(2): 172-180. DOI:10.1007/s11427-012-4276-0 |
| [45] |
Xue SJ, Jiang H, Chen L, et al. Over-expression of Vitreoscilla hemoglobin (VHb) and flavohemoglobin (FHb) genes greatly enhances pullulan production. Int J Biol Macromol, 2019, 132: 701-709. DOI:10.1016/j.ijbiomac.2019.04.007 |
| [46] |
Xu RY, Wang DH, Wang CL, et al. Improved S-adenosylmethionine and glutathione biosynthesis by heterologous expression of an ATP6 gene in Candida utilis. J Basic Microbiol, 2018, 58(10): 875-882. DOI:10.1002/jobm.201800151 |
| [47] |
Wang XC, Yang HY, Zhou W, et al. Deletion of cg1360 Affects ATP synthase function and enhances production of L-Valine in Corynebacterium glutamicum. J Microbiol Biotechnol, 2019, 29(8): 1288-1298. DOI:10.4014/jmb.1904.04019 |
| [48] |
Tajima Y, Yamamoto Y, Fukui K, et al. Impact of an energy-conserving strategy on succinate production under weak acidic and anaerobic conditions in Enterobacter aerogenes. Microb Cell Fact, 2015, 14: 80. DOI:10.1186/s12934-015-0269-6 |
| [49] |
Singh A, Soh KC, Hatzimanikatis V, et al. Manipulating redox and ATP balancing for improved production of succinate in E. coli. Metabol Eng, 2011, 13(1): 76-81. DOI:10.1016/j.ymben.2010.10.006 |
| [50] |
Chen YW, Lou SY, Fan LH, et al. Control of ATP concentration in Escherichia coliusing synthetic small regulatory RNAs for enhanced S-adenosylmethionine production. FEMS Microbiol Lett, 2015, 362(15): fnv115. DOI:10.1093/femsle/fnv115 |
| [51] |
Tao S, Qian Y, Wang X, et al. Regulation of ATP levels in Escherichia coli using CRISPR interference for enhanced pinocembrin production. Microb Cell Fact, 2018, 17: 147. DOI:10.1186/s12934-018-0995-7 |
| [52] |
Zhang YP, Li Y, Du CY, et al. Inactivation of aldehyde dehydrogenase: A key factor for engineering 1, 3-propanediol production by Klebsiella pneumoniae. Metabol Eng, 2006, 8(6): 578-586. DOI:10.1016/j.ymben.2006.05.008 |
| [53] |
Zhu J, Shimizu K. The effect of pfl gene knockout on the metabolism for optically pure D-lactate production by Escherichia coli. Appl Microbiol Biotechnol, 2004, 64(3): 367-375. DOI:10.1007/s00253-003-1499-9 |
| [54] |
Hou J, Suo F, Wang CQ, et al. Fine-tuning of NADH oxidase decreases byproduct accumulation in respiration deficient xylose metabolic Saccharomyces cerevisiae. BMC Biotechnol, 2014, 14: 13. DOI:10.1186/1472-6750-14-13 |
| [55] |
Xu YL, Niu XY, Chen HL, et al. Switch-on fluorescence sensor for ascorbic acid detection based on MoS2 quantum dots-MnO2 nanosheets system and its application in fruit samples. Chin Chem Lett, 2017, 28(2): 338-344. DOI:10.1016/j.cclet.2016.10.003 |
| [56] |
Woodyer R, van der Donk WA, Zhao HM. Relaxing the nicotinamide cofactor specificity of phosphite dehydrogenase by rational design. Biochemistry, 2003, 42(40): 11604-11614. DOI:10.1021/bi035018b |
| [57] |
Vrtis JM, White AK, Metcalf WW, et al. Phosphite dehydrogenase: An unusual phosphoryl transfer reaction. J Am Chem Soc, 2001, 123(11): 2672-2673. DOI:10.1021/ja004301k |
| [58] |
Riebel BR, Gibbs PR, Wellborn WB, et al. Cofactor regeneration of NAD+ from NADH: novel water-forming NADH oxidases. Adv Synth Catal, 2002, 344(10): 1156-1168. DOI:10.1002/1615-4169(200212)344:10<1156::AID-ADSC1156>3.0.CO;2-# |
| [59] |
Heux S, Cachon R, Dequin S. Cofactor engineering in Saccharomyces cerevisiae: expression of a H2O-forming NADH oxidase and impact on redox metabolism. Metabol Eng, 2006, 8(4): 303-314. DOI:10.1016/j.ymben.2005.12.003 |
| [60] |
Vemuri GN, Eiteman MA, McEwen JE, et al. Increasing NADH oxidation reduces overflow metabolism in Saccharomyces cerevisiae. Proc Natl Acad Sci USA, 2007, 104(7): 2402-2407. DOI:10.1073/pnas.0607469104 |
| [61] |
Rigoulet M, Aguilaniu H, Avéret N, et al. Organization and regulation of the cytosolic NADH metabolism in the yeast Saccharomyces cerevisiae. Mol Cell Biochem, 2004, 256-257(1/2): 73-81. |
| [62] |
Sánchez C, Neves AR, Cavalheiro J, et al. Contribution of citrate metabolism to the growth of Lactococcus lactis CRL264 at low pH. Appl Environ Microbiol, 2008, 74(4): 1136-1144. DOI:10.1128/AEM.01061-07 |
| [63] |
Vasava AA, Mashiyava PH. Electron transport chain: role in reactive oxygen species production and aging. Scholars J Agric Vet Sci, 2016, 3(5): 378-388. |
| [64] |
Stark BC, Pagilla KR, Dikshit KL. Recent applications of Vitreoscilla hemoglobin technology in bioproduct synthesis and bioremediation. Appl Microbiol Biotechnol, 2015, 99(4): 1627-1636. DOI:10.1007/s00253-014-6350-y |
| [65] |
Kahraman H, Erenler SO. Rhamnolipid production by Pseudomonas aeruginosa engineered with the Vitreoscilla hemoglobin gene. Appl Biochem Microbiol, 2012, 48(2): 188-193. DOI:10.1134/S000368381202007X |
| [66] |
Zhang L, Li YJ, Wang ZN, et al. Recent developments and future prospects of Vitreoscilla hemoglobin application in metabolic engineering. Biotechnol Adv, 2007, 25(2): 123-136. |
| [67] |
Wang XT, Ding YT, Gao XY, et al. Promotion of the growth and plant biomass degrading enzymes production in solid-state cultures of Lentinula edodes expressing Vitreoscilla hemoglobin gene. J Biotechnol, 2019, 302: 42-47. DOI:10.1016/j.jbiotec.2019.06.301 |
| [68] |
Wang ZN, Xiao Y, Chen WS, et al. Functional expression of Vitreoscilla hemoglobin (VHb) in Arabidopsis relieves submergence, nitrosative, photo-oxidative stress and enhances antioxidants metabolism. Plant Sci, 2009, 176(1): 66-77. |
| [69] |
Zhang XX, Liu SK, Takano T. Overexpression of a mitochondrial ATP synthase small subunit gene (AtMtATP6) confers tolerance to several abiotic stresses in Saccharomyces cerevisiae and Arabidopsis thaliana. Biotechnol Lett, 2008, 30(7): 1289-1294. DOI:10.1007/s10529-008-9685-6 |
| [70] |
Johnson KM, Cleary J, Fierke CA, et al. Mechanistic basis for therapeutic targeting of the mitochondrial F1F0-ATPase. ACS Chem Biol, 2006, 1(5): 304-308. DOI:10.1021/cb600143j |
| [71] |
Zhang Y, Yang HH, Feng JL, et al. Overexpressing F0/F1 operon of ATPase in Rhodobacter sphaeroides enhanced its photo-fermentative hydrogen production. Int J Hydrog Energy, 2016, 41(16): 6743-6751. DOI:10.1016/j.ijhydene.2016.03.061 |
| [72] |
Srivastava AP, Luo M, Zhou WC, et al. High-resolution cryo-EM analysis of the yeast ATP synthase in a lipid membrane. Science, 2018, 360(6389): eaas9699. DOI:10.1126/science.aas9699 |
| [73] |
Hahn A, Vonck J, Mills DJ, et al. Structure, mechanism, and regulation of the chloroplast ATP synthase. Science, 2018, 360(6389): eaat4318. DOI:10.1126/science.aat4318 |
| [74] |
Kozak BU, Van Rossum HM, Niemeijer MS, et al. Replacement of the initial steps of ethanol metabolism in Saccharomyces cerevisiae by ATP-independent acetylating acetaldehyde dehydrogenase. FEMS Yeast Res, 2016, 16(2): fow006. DOI:10.1093/femsyr/fow006 |
| [75] |
Kameda A, Shiba T, Kawazoe Y, et al. A novel ATP regeneration system using polyphosphate-AMP phosphotransferase and polyphosphate kinase. J Biosci Bioeng, 2001, 91(6): 557-563. DOI:10.1016/S1389-1723(01)80173-0 |
| [76] |
Cao H, Li CC, Zhao J, et al. Enzymatic production of glutathione coupling with an ATP regeneration system based on polyphosphate kinase. Appl Biochem Biotechnol, 2018, 185(2): 385-395. DOI:10.1007/s12010-017-2664-4 |
| [77] |
Lindner SN, Vidaurre D, Willbold S, et al. NCgl2620 encodes a class Ⅱ polyphosphate kinase in Corynebacterium glutamicum. Appl Environ Microbiol, 2007, 73(15): 5026-5033. DOI:10.1128/AEM.00600-07 |
| [78] |
Xiao Q, Niu JR, Liu H, et al. High conversion of d-fructose into d-allulose by enzymes coupling with an atp regeneration system. Mol Biotechnol, 2019, 61(6): 432-441. DOI:10.1007/s12033-019-00174-6 |
| [79] |
van Maris AJA, Winkler AA, Porro D, et al. Homofermentative lactate production cannot sustain anaerobic growth of engineered Saccharomyces cerevisiae: possible consequence of energy-dependent lactate export. Appl Environ Microbiol, 2004, 70(5): 2898-2905. DOI:10.1128/AEM.70.5.2898-2905.2004 |
| [80] |
Hara KY, Kobayashi J, Yamada R, et al. Transporter engineering in biomass utilization by yeast. FEMS Yeast Res, 2017, 17(7): fox061. |
| [81] |
Kiriyama K, Hara KY, Kondo A. Extracellular glutathione fermentation using engineered Saccharomyces cerevisiae expressing a novel glutathione exporter. Appl Microbiol Biotechnol, 2012, 96(4): 1021-1027. DOI:10.1007/s00253-012-4075-3 |
| [82] |
Holm AK, Blank LM, Oldiges M, et al. Metabolic and transcriptional response to cofactor perturbations in Escherichia coli. J Biol Chem, 2010, 285(23): 17498-17506. DOI:10.1074/jbc.M109.095570 |
| [83] |
Na YA, Lee JY, Bang WJ, et al. Growth retardation of Escherichia coli by artificial increase of intracellular ATP. J Ind Microbiol Biotechnol, 2015, 42(6): 915-924. DOI:10.1007/s10295-015-1609-6 |
| [84] |
Candela T, Fouet A. Poly-gamma-glutamate in bacteria. Mol Microbiol, 2006, 60(5): 1091-1098. DOI:10.1111/j.1365-2958.2006.05179.x |
| [85] |
Blank LM, McLaughlin RL, Nielsen LK. Stable production of hyaluronic acid in Streptococcus zooepidemicus chemostats operated at high dilution rate. Biotechnol Bioeng, 2005, 90(6): 685-693. DOI:10.1002/bit.20466 |
| [86] |
Aoki R, Wada M, Takesue N, et al. Enhanced glutamic acid production by a H+-ATPase-defective mutant of Corynebacterium glutamicum. Biosci Biotechnol Biochem, 2005, 69(8): 1466-1472. DOI:10.1271/bbb.69.1466 |
| [87] |
Berthoumieux S, De Jong H, Baptist G, et al. Shared control of gene expression in bacteria by transcription factors and global physiology of the cell. Mol Syst Biol, 2013, 9: 634. DOI:10.1038/msb.2012.70 |
| [88] |
Hengge R. Bacterial Signal Transduction: Networks and Drug Targets. New York: Springer Press, 2008, 631: 40-53. |
| [89] |
Toya Y, Nakahigashi K, Tomita M, et al. Metabolic regulation analysis of wild-type and arcA mutant Escherichia coli under nitrate conditions using different levels of omics data. Mol Biosyst, 2012, 8(10): 2593-2604. DOI:10.1039/c2mb25069a |
 2020, Vol. 36
2020, Vol. 36




