中国科学院微生物研究所、中国微生物学会主办
文章信息
- 肖娟, 马梦琪, 梁明星, 贺如阳, 陈华波
- Xiao Juan, Ma Mengqi, Liang Mingxing, He Ruyang, Chen Huabo
- 部分扩增与双片段连接相结合制作长基因定点突变
- Site-directed mutagenesis of long gene by partial amplification combining with double fragments ligation
- 生物工程学报, 2020, 36(6): 1232-1240
- Chinese Journal of Biotechnology, 2020, 36(6): 1232-1240
- 10.13345/j.cjb.190476
-
文章历史
- Received: October 22, 2019
- Accepted: December 5, 2019
基因定点突变是常用的分子生物学技术,是蛋白质结构与功能研究的基础手段。目前基因定点突变主要采用基于聚合酶链式反应(Polymerase chain reaction,PCR)的突变引入办法。最早开发出来的重叠延伸PCR (Overlap extension PCR,OE-PCR)是基因突变的主要方法[1],近年来还发展出大引物法与滚环扩增法等定点突变方法[2-3]。这些方法各有优劣,不同实验室在选择基因定点突变方法时有所偏好。
OE-PCR包括两轮扩增,第一轮PCR分别扩增基因上、下游片段,第二轮PCR通过上、下游片段的末端重叠延伸产物作为模板扩增全长基因[4]。很多科研工作者都有类似的经验,即上、下游片段较小时,第二轮PCR容易成功;随着上、下游基因片段长度增加,第二轮PCR成功率逐渐降低;当某一片段超过2 kb时,第二轮PCR不易获得目标产物。一般认为是较长的基因片段干扰了上、下游片段的末端互补所致。另一方面,PCR体外扩增DNA时总是存在随机突变的可能,而且随着目标产物长度增加,随机突变的概率也逐渐增加。由于以上两点,OE-PCR在制作长基因定点突变时往往并不顺利。因此,在遇到相应问题时,有科研工作者会转向滚环扩增法。滚环扩增法也是目前市场上大多数突变试剂盒所采用的基本原理,如NEB Q5、Stratagene QuikChange及碧云天QuickMutation基因定点突变试剂盒等[5-7]。然而,滚环扩增法也存在固有缺陷:其一,PCR扩增7–12 kb质粒依赖于高保真、快速且稳定的DNA聚合酶,否则无法得到目标产物或容易引入随机突变;其二,若DpnⅠ酶未能对模板质粒做彻底消化,则会严重干扰目标突变质粒的筛选。尽管所有试剂盒厂商都声称其所含上述两酶都满足要求,但随着试剂盒使用和存储时间增加,不可避免的酶活性降低就会极大地影响基因突变的效率。其三,滚环扩增法还会使目标质粒载体序列部分出现突变的风险,因为这部分序列经过了PCR扩增,但一般不会测序确认。
我们发现一般较长的基因内往往含有较多常见的限制性酶切位点。对于突变位点两侧有合适酶切位点的基因突变,仍可采取OE-PCR法扩增其两侧酶切位点之间的部分序列,再将其连入适当载体即可获得重组质粒。若这两个酶切位点在质粒序列上并不单一,则可以采用双片段连接法,利用恰当的片段组合,亦可得到完整的目标质粒。
视网膜母细胞瘤基因(Retinoblastoma gene 1,rb1)是典型的抑癌基因[8],rb1基因缺失与突变是众多肿瘤发生的重要诱因[9]。rb1基因转录终产物约4.7 kb,其开放阅读框全长2 787 bp,编码928个氨基酸。在细胞周期的G1期,Rb1蛋白相继被cyclin D1/CDK4与cyclin E/CDK2磷酸化,释放与之结合的E2F,从而启动G1/S转换[10-12]。Rb1蛋白C端有数个CDK潜在磷酸化位点,其中大多受cyclin E/CDK2修饰,唯有780Ser是较为可信的CDK4磷酸化位点[13-14],将Rb1蛋白780Ser(S)突变为780Glu(E)可模拟其磷酸化修饰效果,进而研究其调节机制[15]。由于rb1基因编码区较长,常规OE-PCR制作rb1S780E突变体时遇到困难。采用部分扩增与双片段连接法相结合,成功克隆了rb1S780E突变体并将其连入质粒载体,为进一步研究该基因的调节机制做好了基础准备,同时也提供了一种有效的长基因定点突变解决方案。
1 材料与方法 1.1 菌种与质粒大肠杆菌DH5α感受态购自天根生化科技(北京)有限公司;pEGFP_C3-rb1质粒由北京大学生命科学学院张传茂教授馈赠。
1.2 试剂2×EasyTaq PCR SuperMix购自北京全式金生物技术有限公司;Pyrobest DNA Polymerase、DNA Ligation Kit Ver.2.1购自TaKaRa;EcoRⅠ、MluⅠ、NheⅠ、SalⅠ购自NEB;DNA回收试剂盒、质粒小量抽提试剂盒购自杭州博日科技有限公司。
1.3 引物设计与合成引物采用Primer Premier 5. 0软件设计,由北京天润奥科生物科技有限公司合成,经PAGE纯化。引物序列见表 1。
1.4 基因克隆扩增含突变位点的目标基因采用高保真Pyrobest DNA Polymerase。方案如下:总体积20 µL,含14.3 µL ddH2O,2 µL 10×缓冲液,3 µL dNTPs,0.2 µL上游引物/0.2 µL下游引物,0.2 µL酶,0.1 µL (10 ng)质粒;运行程序:94 ℃ 5 min;94 ℃ 30 s,54 ℃ 30 s,72 ℃ 3 min,30个循环;72 ℃ 10 min。第二轮PCR方案:总体积20 µL,含13.4 µL ddH2O,2 µL 10×缓冲液,3 µL dNTPs,0.2 µL上游引物/0.2 µL下游引物,0.2 µL酶,0.5 µL上游片段/0.5 µL下游片段;运行程序:94 ℃ 5 min;94 ℃ 30 s,54 ℃ 30 s,72 ℃ 3 min,30个循环;72 ℃ 10 min。PCR产物经电泳检测后回收至20 μL ddH2O中。酶切方案:取PCR回收物17 µL,加2 µL 10×缓冲液,0.5 µL NheⅠ/0.5 µL SalⅠ;另取pEGFP_C3-rb1质粒3 µg稀释到17 µL ddH2O中,加2 µL 10×缓冲液、0.5 µL NheⅠ/0.5 µL EcoRⅠ;取pEGFP_C3-rb1质粒1 µg稀释到17 µL ddH2O中,加2 µL 10×缓冲液,0.5 EcoRⅠ/ 0.5 µL SalⅠ;37 ℃ 2 h。电泳检测后将目标片段回收至20 µL ddH2O中,用DNA Ligation Kit Ver.2.1连接:载体0.5 µL,两个片段各3.5 µL,solutionⅠ7.5 µL混合,16 ℃ 30 min。连接产物直接转化感受态大肠杆菌,然后涂布在含卡那霉素的LB固体培养基平板上,37 ℃培养过夜。
1.5 菌落检测与序列分析在过夜培养的平板上随机挑取数个成型菌落,置于4 mL含卡那霉素的LB培养基中继续培养6 h,以菌液为模板进行PCR检测。方案:5 µL 2×SuperMix,0.1 µL上游引物/0.1 µL下游引物,0.8 µL菌液,补水至10 μL;扩增条件为:94 ℃ 5 min;94 ℃ 30 s,54 ℃ 30 s,72 ℃ 1 min,30个循环。另对部分克隆取3 mL菌液小量提取质粒进行酶切检测,方案为:12.5 µL ddH2O,2 µL 10×缓冲液,0.5 µL内切酶,5 µL小提质粒;37 ℃ 2 h。PCR产物及酶切产物皆经1%琼脂糖凝胶电泳分析。取2个经验证的阳性质粒进行序列测定,质粒测序由北京天润奥科生物科技有限公司完成。
| Primer name | Primer sequence (5′–3′) | Size (bp) |
| S0 | AAAGGATCCATGCCGCCCAAAACCCCCCGAAA | 32 |
| S1 | TATGAATTCTCTTGGACTTGTAAC | 24 |
| S2 | GCCATTGAAATCTACCTCTCT | 21 |
| 780E_S | ACCTTGGAACCAATACCTCA | 20 |
| 780E_A | ATTGGTTCCAAGGTAGGGGG | 20 |
| pEGFP_C3′ | TATGGCTGATTATGATCAGT | 20 |
| Underline indicates that the corresponding nucleotide sequence is a restriction site. | ||
欲制作rb1S780E定点突变,需将rb1基因编码区2 338–2 339位“TC”突变为“GA”。考虑到该基因较长,采用常规OE-PCR法可能不易得到理想的第二轮PCR产物。对rb1作限制性酶切分析,发现目标突变位点S780E上游470 bp处有一个NheⅠ位点,另上游1 438 bp处有一个EcoRⅠ位点(图 1A)。因此,可以仅对rb1_F2 (900–2 787)或rb1_F3 (1 968–2 787)部分做OE-PCR扩增,再利用NheⅠ或EcoRⅠ酶切位点将该片段连入载体构建完整的突变体质粒。为寻找最佳解决方案,依上述思路按OE-PCR定点突变法进行对比操作。

|
| 图 1 OE-PCR扩增含突变位点的基因片段 Fig. 1 Amplification of the gene fragments containing target mutation by OE-PCR. (A) rb1 gene map and primers position. (B) Agarose gel electrophoresis of the first round of PCR. The upstream primers for lane 'a' to 'c' were S0, S1, S2 respectively and all of their downstream primer was 780E_A. The primers for lane 'd' were 780E_S/pEGFP_C3′. (C) Agarose gel electrophoresis of the second round of PCR. The template for #1 to #3 were mixture of product a & d, b & d, c & d, respectively. The template for '+' was pEGFP_C3-rb1 plasmid. The upstream primers for group 1 to 3 were S0, S1, S2 respectively and all of their downstream primer was pEGFP_C3′. M stands for DNA marker. |
| |
以原始质粒pEGFP_C3-rb1为模板,分别以S0、S1、S2搭配780E_A为引物扩增上游片段,以780E_S/pEGFP_C3′为引物扩增下游片段。PCR产物经电泳检测,得到4个预期大小的DNA片段(图 1B)。在第二轮PCR时,发现上游片段a与下游片段d组合未能扩增出全长rb1,上游片段b与下游片段d组合扩增出的产物主带不明,且杂带较多;而上游片段c与下游片段d组合扩增出预期的目标产物片段。经多次调整PCR反应条件,仍未能得到理想的全长rb1产物及F23片段。以上结果说明OE-PCR时较长片段不易重叠延伸而扩增出目标产物,与经验相符。
2.2 目标质粒的双片段连接理论上,将上述#3产物以NheⅠ/SalⅠ双酶切即可得到rb1_F3片段,再连接到切除对应片段的载体即可得到完整的突变体质粒。然而,限制性酶切分析发现原始质粒多克隆位点上游还有另一个NheⅠ位点,因此F3片段无法直接与载体连接得到目标质粒。尽管如此,可采用双片段连接法解决上述问题。即以NheⅠ/EcoRⅠ双酶切原始质粒获得rb1_F2 (900–1968)片段,以NheⅠ/SalⅠ双酶切#3 PCR产物获得F3片段,将上述两个片段一起与经EcoRⅠ/SalⅠ酶切的质粒载体部分连接,从而得到完整目标质粒(图 2)。

|
| 图 2 双片段连接示意图 Fig. 2 The diagram of double fragments ligation. F2 (yellow line) and F3 (red line) are ligated with vector, which contain F1 (purple line), to obtain the complete plasmid. Color dots represent different restriction sites and black dot represents the target mutation site. |
| |
上述#3 PCR产物双酶切简单易行(图 3 a泳道)。将原始质粒以NheⅠ/EcoRⅠ双酶切,得到1 068 bp、1 662 bp、4 773 bp三个片段,其中最小的片段即所需的F2片段,它与1 622 bp片段相距较远,容易分离回收(图 3 b 泳道)。将原始质粒以EcoRⅠ/SalⅠ双酶切可得到1 888 bp、5 615 bp两个片段,其中大片段即所需的载体部分(图 3 c泳道),它含有rb1_F1 (1–900)部分。
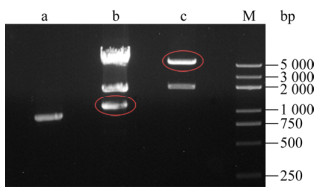
|
| 图 3 基因片段及载体的限制性酶切 Fig. 3 Restriction enzymatic digestion of gene fragments and vector. Agarose gel electrophoresis of the OE-PCR product #3 digested by NheⅠ/SalⅠ (lane a), and pEGFP_C3-rb1 plasmid digested by NheⅠ/EcoRⅠ (lane b) or EcoRⅠ/SalⅠ (lane c). M stands for DNA marker. DNA fragments that need to be retrieve from gel were encircled by an ellipse. |
| |
连接产物转化感受态大肠杆菌后经卡那霉素选择性平板培养基筛选,得到数十个菌落(图 4A),明显少于本实验室单片段连接时的菌落形成数[16]。从中任意挑取8个菌落,经适当培养后进行检测、鉴定。以两种不同的引物组合进行PCR检测,方案一选用S2/780E_A引物组合;阳性结果表示质粒含有rb1基因后半部分(图 4B)。检测得到#1、#4、#5三个阳性克隆,其产物大小约500 bp,与预期(514 bp)相符(图 4C)。方案二选用780E_S/pEGFP_C3′引物组合;阳性结果表示质粒为pEGFP_C3载体,且含有rb1后半部分。该方案检测结果与方案一一致,其产物大小约400 bp,与预期(421 bp)相符(图 4D)。为进一步确认检测结果的可靠性,对#1–5号克隆提取质粒作酶切检测。分别以MluⅠ与NheⅠ作单酶切检测,结果显示#1、#4、#5这3个质粒能切出预期片段,而#2、#3号质粒无预期片段(图 4E,F)。酶切检测结果与PCR检测结果一致。

|
| 图 4 阳性克隆的筛选 Fig. 4 Screening positive colonies. (A) Bacterial colonies on plate. (B) The position of the primers and restriction enzymes which were used for detection of positive colonies. Agarose gel electrophoresis showed the results of PCR with S2/780E_A primers (C) or 780E_S/pEGFP_C3′ primers (D) and the restriction enzymatic digestion results with Mlu I (E) or Nhe I (F). –/+ stand for negative/positive control, #1–8 stand for the selected colony and M stands for DNA marker. Expected fragment sizes were showed at the bottom of each figure. |
| |
为鉴定目标质粒序列准确性,对其中#1、#4阳性质粒进行序列测定。由于经PCR扩增的F3片段位于基因下游,且目标突变位点S780E距下游SalⅠ位点仅490 bp,故选择pEGFP_C3′引物对该质粒反义链进行序列测定。结果显示两者目标位点为TC碱基,且F3部分序列内未引入其他非预期突变点(图 5)。表明其正义链对应位置为GA碱基,符合预期的rb1S780E突变体序列特征。由于目标质粒其他部分未经PCR扩增,故未作序列测定。综合分析,#1、#4克隆皆是符合要求的pEGFP_C3-rb1S780E目标突变体。

|
| 图 5 阳性质粒的关键序列 Fig. 5 Sequence of the positive plasmids. (A) Amino acid and nucleotide sequence of target mutant. Peak trace of antisense of plasmid #1 (B) and #4 (C). Critical sequences of mutant position were encircled by square frame. |
| |
长片段干扰重叠延伸效率是制约OE-PCR应用于长基因定点突变的重要因素;另一方面,制作长基因突变体时,往往得到了预期定点突变,却引入了新的非预期突变(图 6A)。滚环扩增法在一定程度上可以解决长基因定点突变的问题,但其也存在其他固有缺陷。若仍以OE-PCR制作长基因定点突变,则仅扩增含突变位点的部分基因序列成为克服上述困难的良好解决方案。一则OE-PCR扩增较短的基因片段成功率较高,二则扩增短基因片段时引入非预期突变的几率相对较小(图 6B)。

|
| 图 6 常规OE-PCR与部分扩增法的比较 Fig. 6 Comparison of two protocols. (A) In conventional protocol, it is difficult to get target product by OE-PCR with long upstream and/or downstream fragments. (B) It is relatively easy to amplify a short DNA fragment containing mutation site. Red lines indicate that there may be unexpected mutations in DNA fragments which were amplified by PCR. |
| |
采取部分扩增结合酶切-连接法制作长基因定点突变要求突变位点两侧附近皆有合适的酶切位点,以便于将扩增的基因片段重新连入载体获得完整目标质粒。最理想的情况是其两侧附近各有一个单一酶切位点,此时按常规基因克隆方法即可重新连接得到完整目标质粒。然而往往遇到另外一种情况,即突变位点两侧虽有酶切位点,但它们在目标质粒上并不单一。如本例中目标位点S780E上游虽有一个NheⅠ位点,但载体序列本身还有另一个NheⅠ位点。若以NheⅠ/SalⅠ酶切原始质粒再连接扩增基因片段,则目标质粒将丢失rb1基因上游大部分序列。对于这种情况,双片段连接法是一个很好的解决方案:本例中两个片段同时与一个载体连接的办法成功避开了双NheⅠ位点对片段连接的限制(图 2)。该方案所获得的完整质粒中基因上游F1部分与中部F2皆源自原始质粒,一般认为序列忠实,不会出现新的突变(图 6B)。唯一存在随机突变可能的只有经PCR扩增的F3部分,只对这部分DNA进行序列测定即可判断目标质粒的准确性。
结合实际工作经验,我们发现相对常规单片段连接方案,双片段连接时得到的转化克隆数较少,且阳性率偏低;调整连接体系中两片段和载体的比例可适当提高转化克隆数。本例中平板上只形成数十个菌落,检测阳性率只有37.5%,都低于本实验室常规方案下的经验值。尽管如此,若常规OE-PCR难以解决某些长基因定点突变问题时,本方法不失为一种有效的解决方案。
| [1] |
Ho SN, Hunt HD, Horton RM, et al. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene, 1989, 77(1): 51-59. |
| [2] |
Kooi CWV. Megaprimer method for mutagenesis of DNA. Methods Enzymol, 2013, 529: 259-269. |
| [3] |
Zhao SZ, Shen XQ. A High efficient site-directed mutagenesis method by RCA and megaprimer PCR. Acta Agric Jiangxi, 2009, 21(8): 7-8, 11 (in Chinese). 赵松子, 沈向群. 用滚环扩增与大引物PCR法高效构建定点突变序列. 江西农业学报, 2009, 21(8): 7-8, 11. |
| [4] |
Yang L, Wang LY, Li HM, et al. Multi-site specific mutagenesis by multi-fragment overlap extension PCR. China Biotechnol, 2019, 39(8): 52-58 (in Chinese). 杨林, 王柳月, 李慧美, 等. 改进的多片段重叠延伸PCR制作基因多位点突变. 中国生物工程杂志, 2019, 39(8): 52-58. |
| [5] |
Qi RH, Otting G. Mutant T4 DNA polymerase for easy cloning and mutagenesis. PLoS ONE, 2019, 14(1): e211065. |
| [6] |
Zhang J, Liu Y, Zhu Z, et al. Role of microRNA508-3p in melanogenesis by targeting microphthalmia transcription factor in melanocytes of alpaca. Animal, 2017, 11(2): 236-243. |
| [7] |
Tang SW, Li H, Cui SY, et al. Construction of a novel plasmid system for epitope tagging and gene deletion in Saccharomyces cerevisiae. Acta Microbiol Sin, 2019, 59(5): 939-949 (in Chinese). 唐仕伟, 李辉, 崔时媛, 等. 新型酵母蛋白表位标记和基因敲除质粒系统的构建及可行性验证. 微生物学报, 2019, 59(5): 939-949. |
| [8] |
Liu SH, Wang SZ, Zhang H, et al. Research advances on RB1 gene. Hereditas (Beijing), 2010, 32(11): 1097-1104 (in Chinese). 刘双虎, 王守志, 张慧, 等. 视网膜母细胞瘤基因1(RB1)研究进展. 遗传, 2010, 32(11): 1097-1104. |
| [9] |
Du Q, Jiang YH, Galli BL. Low-penetrance retinoblastoma due to exons 24 and 25 deletions in the Rb1 gene. Chin J Med Genet, 2002, 19(5): 370-374 (in Chinese). 杜琴, 江悦华, Galli BL. Rb1基因第24和25外显子缺失导致低外显性视网膜母细胞瘤. 中华医学遗传学杂志, 2002, 19(5): 370-374. |
| [10] |
Yang Y, Ma B, Li L, et al. CDK2 and CDK4 play important roles in promoting the proliferation of SKOV3 ovarian carcinoma cells induced by tumor-associated macrophages. Oncol Rep, 2014, 31(6): 2759-2768. |
| [11] |
Taylor-Harding B, Aspuria PJ, Agadjanian H, et al. Cyclin E1 and RTK/RAS signaling drive CDK inhibitor resistance via activation of E2F and ETS. Oncotarget, 2015, 6(2): 696-714. |
| [12] |
Delfau-Larue MH, Klapper W, Berger F, et al. High-dose cytarabine does not overcome the adverse prognostic value of CDKN2A and TP53 deletions in mantle cell lymphoma. Blood, 2015, 126(5): 604-611. |
| [13] |
Liu JC, Granieri L, Shrestha M, et al. Identification of CDC25 as a common therapeutic target for triple-negative breast cancer. Cell Rep, 2018, 23(1): 112-126. |
| [14] |
Shi Y, Qian Z, Zhang S, et al. Cell Cycle protein expression in neuroendocrine tumors: association of CDK4/CDK6, CCND1, and phosphorylated retinoblastoma protein with proliferative index. Pancreas, 2017, 46(10): 1347-1353. |
| [15] |
Zhang XY, Chen Q, Feng J, et al. Sequential phosphorylation of Nedd1 by Cdk1 and Plk1 is required for targeting of the gammaTuRC to the centrosome. J Cell Sci, 2009, 122(13): 2240-2251. |
| [16] |
Wang LY, Li HM, Ma MQ, et al. Improve the site-directed mutagenesis efficiency of overlap extension PCR by outboard-primers. Biotechnol Bull, 2019, 35(12): 196-202 (in Chinese). 王柳月, 李慧美, 马梦琪, 等. 利用旁侧引物提高重叠延伸PCR定点突变效率. 生物技术通报, 2019, 35(12): 196-202. |
 2020, Vol. 36
2020, Vol. 36




