中国科学院微生物研究所、中国微生物学会主办
文章信息
- 张浩, 邢志林, 汪军, 赵天涛
- Zhang Hao, Xing Zhilin, Wang Jun, Zhao Tiantao
- 异养同化降解氯代烃的研究现状、微生物代谢特性及展望
- Advances in microbial degradation of chlorinated hydrocarbons
- 生物工程学报, 2020, 36(6): 1083-1100
- Chinese Journal of Biotechnology, 2020, 36(6): 1083-1100
- 10.13345/j.cjb.190458
-
文章历史
- Received: October 12, 2019
- Accepted: April 9, 2020
- Published: May 15, 2020
2. 重庆市环境科学研究院,重庆 401147
2. Chongqing Academy of Environmental Sciences, Chongqing 401147, China
氯代烃(Chlorinated hydrocarbons,CAHs),包括氯代烷烃、氯代烯烃和氯代芳烃,其作为重要化工原材料和有机溶剂在工业生产和生活中被广泛使用[1]。由于CAHs存储和处置不当,造成了世界范围内工业园区、冶金矿业、医药企业等场地及地下水的严重污染[2-3]。美国环保局对39个小城镇地下水水源进行检测,在已处理和未经处理的地下水中都发现了11种CAHs[4],德国Bitterfeld地区经过近百年的化学工业发展,土壤和地下水受到了CAHs的严重污染,涉及的土壤面积高达25 km2,约有2亿m3的地下水遭受污染[5]。我国“水中优先控制污染物”中前9种均为CAHs,且CAHs具有潜在的“三致” (致癌、致畸、致突变)效应和遗传毒性效应,对人类健康和生态环境构成重大威胁[1]。尽管CAHs的污染控制已经持续研究了60多年,但当前治理现状依然不容乐观。因此,采取有效措施减少环境中的CAHs污染迫在眉睫。
生物法作为一种降解高效、环境友好和成本较低的修复方式,在CAHs的去除中一直备受关注[6]。厌氧还原脱氯、好氧共代谢和异养同化是CAHs生物降解的主要途径[7]。在严格厌氧条件下,CAHs作为电子受体,还原性物质作为电子供体,通过水解作用或亲核反应脱去氯原子的过程即为厌氧还原脱氯[8]。科研工作者针对厌氧脱氯开展了广泛系统的研究[9-11],包括厌氧脱氯相关微生物、脱氯机理、降解酶及影响因子等。厌氧脱氯主要发生在三氯乙烯(Trichloroethylene,TCE)、四氯乙烯(Perchloroethylene,PCE)、三氯苯(Trichlorobenzenes,TCBs)和六氯苯(Hexachlorobenzene,HCB)等高氯取代烃中,这些CAHs在还原脱卤酶的作用下转化为低氯取代物[12]。典型厌氧脱氯微生物有脱卤杆菌属Dehalobacter、脱卤拟球菌属Dehalococcoides、脱硫杆菌属Desulfifitobacterium、地杆菌属Geobacter[13],但只有Dehalococcoides可实现氯原子的全部脱除。好氧共代谢是CAHs生物降解的另一途径,微生物通过代谢生长基质诱导细胞产生单加氧酶或双加氧酶实现CAHs的降解[14],CAHs不作为碳源和能源也不能被微生物转化为生物质[15]。好氧共代谢研究持续多年,涉及共代谢降解微生物的筛选、生长基质的优化以及共代谢降解机理的解析[7, 16]。最典型共代谢微生物为甲烷氧化菌Methanotrophs[17]。这些研究为CAHs污染的修复提供了重要的理论基础,基于这些理论,研究者正开展CAHs的场地修复工作[7]。
异养同化作用是CAHs降解的第3种途径,与厌氧脱氯和共代谢降解相比,异养同化过程避免额外添加有机物,同时不会产生二次污染,在CAHs污染降解中具有重大应用潜力[18]。当前,已报道可通过微生物异养同化作用降解的CAHs种类十分有限,利用异养同化作用实现场地修复的研究还未有报道。开展CAHs异养同化过程的系统性认识,明晰CAHs异养同化规律,对于如何强化、应用异养同化过程,扩大CAHs的修复途径将具有重要的推动作用。国内外许多研究对厌氧脱氯和好氧共代谢过程已进行系统性总结[9-11, 15-16],对于如何优化调控厌氧脱氯和好氧共代谢过程有了一定的依据。而关于CAHs异养同化过程的总结还未见报道。基于此,本文将对异养同化降解的研究现状进行系统性总结,包括可发生异养同化CAHs的类型、降解特性,典型氯代烃异养同化的降解机理、相关微生物及其降解酶,并基于异养同化降解CAHs存在的问题提出未来重点关注的发展方向,将为CAHs异养同化的深入研究和场地修复实践提供重要的理论依据。
1 CAHs异养同化过程及优势CAHs三种代谢途径的特点如表 1所示。厌氧还原脱氯过程CAHs作为电子受体,还原性物质作为电子供体,通过水解作用或亲核反应脱去氯原子。所有氯代烃均可发生还原脱氯,但随着分子中氯原子数量的减少,还原脱氯的速率会迅速降低[19]。另外,厌氧脱氯发生环境要求苛刻,菌群数量和活性往往较低,且还需要不断添加电子供体。因此,该过程极易造成低氯取代物和其他副产物的大量积累[20-21]。好氧共代谢降解过程中CAHs被细胞代谢生长过程中产生的单加氧酶或双加氧酶降解,CAHs不为微生物提供能源和碳源,该过程需要添加大量生长基质,极易造成环境的二次污染;共代谢降解产生的环氧化合物和氯代醛类化合物也会造成微生物的活性降低;同时降解过程中生长基质和CAHs污染物会与关键酶竞争活性位点,产生竞争性抑制[22]。尽管已有厌氧脱氯和好氧共代谢过程用于场地修复的案例[23],但由于上述问题的限制,往往造成处理成本过高、修复效果差和修复时间长等问题。因此,面对日益严格环保需求,扩大CAHs的生物修复途径已十分必要。
| Reaction | Description | Reaction process | Characterization information | Toxic by-product | Half lives |
| Reductive dechlorination | Dechlorination of a compound where the compound is used as an electron acceptor, the bacteria may or may not gain energy by reduction of the compound. This reaction removes one chloride atom from the compound and replaces it with a proton |  |
Strictly anaerobic, requiring electron donors | Low chlorine substituted | Slow |
| Aerobic co-metabolism | Dechlorination of a compound where the compound is fortuitously degraded by an enzyme used in cellular metabolism-typically a monooxygenase enzyme |  |
Additional growth substrate | Epoxy compounds | Fast |
| Aerobic direct metabolism | Use of the chlorinated compound as an electron donor for aerobic metabolism |  |
No additional information is needed | None | Fast |
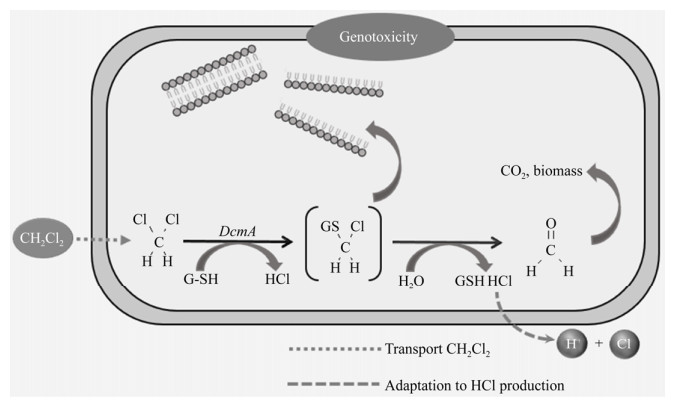
CAHs异养同化作为另一种途径为场地修复提供了新的选择,异养同化过程中微生物以CAHs为唯一碳源和能源进行生长,将其矿化为H2O、CO2和Cl–等小分子化合物并合成自身生物质[24]。CAHs在微生物细胞内的异养同化过程如图 1所示。CAHs通过主动运输进入微生物细胞内,在酶的作用下发生转化,为微生物生长提供必需的碳源和能源,同时中间产物通过丝氨酸途径转化为生物质,代谢生成的氯离子最终排出细胞外。与好氧共代谢和厌氧脱氯相比,异养同化优势体现在以下方面:(1)避免有毒中间产物的生成,特别是对降解酶和微生物有毒害作用的环氧化合物,同时异养同化避免产生低氯取代烃[21]。(2)无需添加额外生长基质,无二次污染,同时避免了在代谢过程中生长底物与CAHs竞争活性位点[22]以及生长底物对氧的消耗[21-22, 25]。如何利用CAHs异养同化过程也将是未来场地修复研究的热点。
2 CAHs异养同化降解研究概述 2.1 可发生异养同化降解的CAHs种类目前,研究报道可通过异养同化途径降解的CAHs有一氯甲烷(Chloromethane,CM)[28]、二氯甲烷(Dichloromethane,DCM)[29-30]、三氯甲烷(Chloroform,CF)、四氯化碳(Carbon tetrachloride,CT)[31]等氯代烷烃;氯乙烯(Vinyl chloride,VC)[32-33]、顺式二氯乙烯(cis-1, 2-dichloroethene,c-1, 2-DCE)[21, 34]、反式二氯乙烯(trans-1, 2- dichloroethylene,t-1, 2-DCE)[18]、TCE等氯代烯烃;氯苯(Chlorobenzene,CB)[35-36]、1, 2-二氯苯(1, 2-dichlorobenzene,1, 2-DCB)[37]、1, 4-二氯苯(1, 4-dichlorobenzene,1, 4-DCB)[38]、1, 2, 3-三氯苯(1, 2, 3-trichlorobenzene,1, 2, 3-TCB)、1, 3, 5-三氯苯(1, 3, 5-trichlorobenzene,1, 3, 5-TCB)[39]等氯代芳烃。典型CAHs的Cl/C比结果如图 2所示。随氯取代程度的增大,氯代烷烃异养同化降解性急剧下降,除CT外,其他全氯代烃无法通过异养同化途径降解。所有氯代甲烷都可作为微生物碳源和能源被降解[28-29, 31],表明氯代烷烃可以广泛地被微生物同化。除四氯乙烯外,其他氯代乙烯均可发生异养同化降解,发生异养同化的氯代烯烃Cl/C的比值低于2。氯代芳烃中,氯取代程度小于3的氯代芳烃可通过异养同化降解是广泛报道的。CAHs中氯取代程度越高,化合物电负性越强,在有氧条件下难以发生异养同化[40]。综上所述,能通过异养同化途径进行降解的CAHs以代氯取代烃(1–3个氯原子)为主,准确认识场地CAHs污染特性也是开展场地修复的重要前提。

|
| 图 2 典型氯代烃Cl/C比 Fig. 2 Cl/C ratio of typical CAHs |
| |
已报道CAHs异养同化微生物主要来自污染场地土壤、地下水、化工废水处理厂的活性污泥以及人工驯化的专性降解菌[31, 38],这些微生物包括纯菌株和混合菌,混合菌相对于纯菌对污染物具有更好的降解效率,且对环境具备更好的适应性[20]。已分离的典型CAHs异养同化降解菌属主要有生丝微菌属Hyphomicrobium、假单胞菌属Pseudomonas、分支杆菌属Mycobacterium、不动杆菌属Acinetobacter、类诺卡氏菌属Nocardioides、芽孢杆菌属Bacillus、寡养单胞菌属Stenotrophomonas等,大多数的降解菌只对特定的CAHs产生作用,但也有少部分降解菌如铜绿假单胞菌Pseudomonas aeruginosa strain S1-2[18]、肠杆菌Enterobacter sp. SA-2、Pseudomonas sp. SA-6[39]、红球菌Rhodococcus phenolicus G2PT[41]等可降解两种及以上的CAHs。对已分离筛选具有CAHs异养同化作用的微生物进行了系统总结,结果如表 2所示。
| Types of CAHs | Microorganisms | Concentration (mmol/L) | Cell growth rates (h–1) | Degradation rates | Experiment conditions | References |
| CH3Cl | Hyphomicrobium sp. MC1 | 0.35 | 0.09 | – | pH 7.2 30 ℃ | [53] |
| Hyphomicrobium sp. | 0.71 | 0.09–0.17 | – | pH 7.0 30 ℃ | [42] | |
| Aminobacter sp. | 0.15 | |||||
| Nocardioides sp. | 0.18 | |||||
| Leisingera methylohalidivorans MB2T | 0.37 | 0.05 | – | pH 7.7 27 ℃ 200 r/min | [28] | |
| CH2Cl2 | Pseudomonas sp. DM1 | 2–5 | 0.11 | – | pH 6.9 30 ℃ | [54] |
| Hyphomicrobium DM2 | 10.00 | 0.07 | 100% (60 h) | pH 7.2 30 ℃ | [52] | |
| Methylobacterium rhodesianum H13 | 5.00 | 2.49 | 100% (23 h) | pH 7.0 30 ℃ 160 r/min | [29] | |
| Lysinibacillus sphaericus wh22 | 32.00 | – | 93.8% (72 h) | pH 6.5 30 ℃ 200 r/min | [30] | |
| Hyphomicrobium sp. CM2 | 0.31 | 0.08 | – | pH 7.2 30 ℃ 140 r/min | [55] | |
| Methylobacterium sp. CM4 | – | |||||
| Paracoccus methylutens sp. nov. DM12 | 10.00 | – | – | pH 7.2 29 ℃ 180 r/min | [43] | |
| CHCl3 | Bacillus (GBB416) | 0.12 | 4.63±0.51 | – | pH 7.0 26 ℃ 160 r/min | [31] |
| Pseudomonas (GBB417) | 2.87±0.06 | |||||
| CCl4 | Bacillus (GBB416) | 0.10 | 6.69±0.31 | – | pH 7.0 26 ℃ 160 r/min | [31] |
| Pseudomonas (GBB417) | 2.41±0.04 | |||||
| VC | Pseudomonas aeruginosa MF1 | 6.00 | 0.002 | 100% (840 h) | pH 7.1 23 ℃ | [44] |
| Mixed culture | 0.25 | – | – | pH 7.0 30 ℃ | [20] | |
| Mycobacterium strains (JS60 JS61 JS616 JS617) | 0.15 | 0.007–0.010 | 99.0% (72 h) | pH 7.2 30 ℃ 150–165 r/min | [56] | |
| Nocardioides sp. JS614 | 0.03 | |||||
| Mycobacterium strains (JS622, JS623, JS624 and JS625) |
0.80–1.00 | – | – | pH 7.0 30 ℃ 200 r/min | [32] | |
| Ochrobactrum sp. TD | 0.29 | – | – | pH 7.2 30 ℃ | [57] | |
| Mycobacterium strain JS623 | 0.77 | – | – | pH 7.2 30 ℃ 200 r/min | [33] | |
| Mycobacterium rhodesiae JS60 | 0.69 | 0.017 | – | pH 6.5 30 ℃ 200 r/min | [58] | |
| Ralstonia sp. TRW-1 | 0.20 | 0.19 | 94.8% (132 h) | pH 7 30 ℃ | [45] | |
| c-1, 2-DCE | Polaromonas sp. JS666 | 0.04 | – | 100% (108 h) | pH 7.0 20 ℃ 150 r/min | [59] |
| Mixed culture | 0.80 | – | 100% (408 h) | pH 7.0 23 ℃ | [21] | |
| Corynebacterium sp. | 10.00 | 15.72±0.10 | – | pH 7.5 35 ℃ 150 r/min | [46] | |
| Bacillus sp. | 14.23±0.26 | |||||
| Burkholderia sp. | 9.77±0.09 | |||||
| Micrococcus sp. | 12.91±0.36 | |||||
| Pseudomonas sp. | 12.05±0.44 | |||||
| Jacksonville stream Bed sediments microorganism | 0.05 | – | – | pH 7.2 | [47] | |
| c-1, 2-DCE/ t-1, 2-DCE | Acinetobacter species | 1/1.5 | 0.014–0.023 0.019–0.028 |
70%–75%
(168 h) 60%–72% (168 h) |
pH 7.0 30 ℃ | [18] |
| Achromobacter xylosoxidans strain 2002-55549 |
||||||
| Klebsiella sp. HL1 | ||||||
| Bacillus species | ||||||
| Bacillus cereus strain ATCC BAA-1005 | ||||||
| Pseudomonas aeruginosa strain S 1-2 | ||||||
| TCE | Bacillus sp. 2479 | 1.00 | – | – | pH 6.9 37 ℃ 90 r/min | [49] |
| Stenotrophomonas maltophilia PM102 | 0.33 | – | 90.0% (2 h) | pH 7.0 37 ℃ | [48] | |
| CB | Delftia tsuruhatensis LW26 | 0.14 | 0.420 | 99.7% (16 h) | pH 7.2 30 ℃ 160 r/min | [36] |
| Acinetobacter sp. CB001 | 0.44 | 0.002 | 98.2% (120 h) | pH 7.2 25 ℃ 160 r/min | [35] | |
| 1, 2-DCB | Acidovorax avenae | 0.17 | 0.004 | – | pH 7.0 30 ℃ 100 r/min | [37] |
| Pseudomonas sp. JS100 | 0.002–0.004 | – | – | pH 7.0 30 ℃ 200 r/min | [37] | |
| 1, 4-DCB | Flavobacterium sp. DEB-1 | 0.68 | 0.05 | 94.5% (24 h) | pH 7.8 30 ℃ 130 r/min | [38] |
| Xanthobacter flavus 14p1 | 0.27 | 0.035 | – | pH 7.5 30 ℃ | [60] | |
| CB 1, 4-DCB | Rhodococcus phenolicus G2PT | – | – | – | pH 7.0 30 ℃ | [41] |
| 1, 4-DCB 1, 2, 3-DCB 1, 3, 5-TCB |
Enterobacter sp. SA-2 | 0.43–0.9 | 0.02–0.04 | 80.0%–90.0% (200 h) |
pH 7.0 25 ℃ | [39] |
已报道分离的氯代烷烃异养同化降解菌属主要包括Hyphomicrobium、Pseudomonas、Nocardioides、氨基杆菌属Aminobacter、甲基杆菌属Methylobacterium、副球菌属Paracoccus、Bacillus等。氯代烷烃类污染物分布广泛,多种氯代烷烃均可作为微生物底物,研究人员从多种环境分离了这些菌株。McAnulla等[42]从陆地、河口、海洋中分离出6株属于Hyphomicrobium sp.、Aminobacter sp.和Nocardioides sp.的CM降解菌。Jeffra等[28]从海水中分离的菌株Leisingera methylohalidivorans MB2T不仅能以CM为唯一碳源进行生长还能同化溴甲烷和碘甲烷。Doronina等[43]从污染地下水中分离出一种新型的革兰氏阴性菌株Paracoccus methylutens sp. nov. DM12,该菌株可以DCM为唯一碳源。而Wu等[30]从制药污水中分离出的DCM降解微生物芽孢杆菌Lysinibacillus sphaericus wh22,还能以1, 2-二氯乙烷(1, 2-dichloroethane,1, 2-DCA)、氯溴甲烷(Chlorobromomethane,CBM)、1, 1, 1-三氯乙烷(1, 1, 1-trichloroethane,1, 1, 1-TCA)和TCE为唯一碳源进行生长。CF和CT等高氯代烷烃,难以作为微生物生长的唯一碳源,Olaniran等[31]分离出两株能以底物浓度为0.1% (W/V)的CF和CT为唯一碳源和能源的降解菌Bacillus (GBB416)和Pseudomonas (GBB417),这是唯一一篇报道的对CT具有异养同化降解功能的菌株,实现了高氯代烷烃异养同化降解研究的突破。氯代烷烃类污染物分布广泛,多种氯代烷烃均可作为微生物底物,在氯代烷烃污染场地,异养同化降解是十分有效的修复途径。
2.2.2 氯代烯烃异养同化微生物氯代烯烃在环境中的检出率较高,毒性更大,研究人员在污染场地中分离出许多氯代烯烃异养同化降解菌,主要包括Pseudomonas、Mycobacterium、Nocardioides、Stenotrophomonas和苍白杆菌属Ochrobactrum等。大多数氯代烯烃异养同化微生物是以特定的基质为生长底物、通过长期驯化衍生的特异性菌株。Verce等[44]报道了第一株以VC为唯一碳源的Pseudomonas aeruginosa MF1,该菌株以VC为唯一基质长期驯化筛选获得,菌株MF1不仅具有长时间的耐饥饿能力,还具有很强的VC耐受能力,当VC浓度增加到7.3 mmol/L时对生长也无抑制作用。除纯菌外,Singh等[20]通过长期驯化从污染场地中筛选出一组由Mycobacteria和Rhodococcus构成的混合菌,当VC浓度为50–250 μmol/L时可被混合菌迅速降解。相同功能的不同菌株降解能力差异明显,Elango发现从高浓度VC环境分离出的罗尔斯通菌Ralstonia sp. TRW-1对VC的降解能力是其他分枝杆菌的3–10倍[45]。
除VC外,c-1, 2-DCE也能作为微生物生长的唯一碳源。Schmidt等研究了污染地下水中c-1, 2-DCE的异养同化过程,在多种条件下观察到了c-1, 2-DCE生物降解过程的稳定性同位素分馏,通过蛋白质的形成证明了c-1, 2-DCE可作为微生物生长的唯一碳源和能源[21]。随后,Olaniran等利用富集培养技术从尼日利亚和南非的污染场地分离出5株c-1, 2-DCE的异养同化降解菌,分别属于棒状杆菌属Corynebacterium sp.、Bacillus sp.、伯克氏菌属Burkholderia sp.、微球菌属Micrococcus sp.和Pseudomonas sp.,这些菌均可以c-1, 2-DCE为唯一碳源和能源,降解率为59%–86%[46]。此外,Bradley等[47]从黑臭水体中分离出一组c-1, 2-DCE降解混合菌,在浓度为50 μmol/L的条件下可以实现c-1, 2-DCE的完全降解并检测出大量CO2的生成,表明该混合菌具有高效的c-1, 2-DCE同化能力。尽管VC和DCE容易作为微生物碳源被异养同化降解,但随着氯取代程度的增大,异养同化降解难度急剧增大。迄今为止,只发现嗜麦芽窄食单胞菌Stenotrophomonas maltophilia PM102[48]和Bacillus sp. 2479[49]具有TCE的异养同化降解能力,还未有直接证据证明PCE可通过异养同化途径实现降解。因此,在多氯取代烯烃的污染场地,采用异养同化与厌氧还原脱氯结合的修复措施可实现污染物的有效去除。
2.2.3 氯代芳烃异养同化微生物氯代芳烃作为持久性有机污染物,其异养同化微生物的筛选分离研究已开展多年,研究主要集中在氯取代数小于3的氯苯类有机物。研究人员以CB为唯一碳源,分离筛选的不动杆菌Acinetobacter sp. CB001[35]和代尔夫特菌Delftia tsuruhatensis LW26[36]除了能以CB为唯一碳源外,也能以二氯苯(Dichlorobenzenes,DCBs)为唯一碳源进行生长。另外,Haigler等[50]从活性污泥中分离出一株以1, 2-DCB为唯一碳源和能源生长的菌株Pseudomonas sp. JS100,该菌株也能以CB为唯一碳源和能源进行生长。此外,Monferrán等[37]以1, 2-DCB为唯一碳源筛选的燕麦食酸菌Acidovorax avenae降解污染物范围更广,能够以CB、1, 3-DCB和1, 4-DCB为碳源和能源生长。TCBs异养同化降解菌也存在类似的情况,Adebusoye等[39]分离的菌株Enterobacter sp. SA-2和Pseudomonas sp. SA-6不仅可以异养同化1, 2, 3-TCB和1, 3, 5-TCB,也能以1, 4-DCB为唯一碳源和能源生长。以上结果表明,氯代芳烃异养同化微生物之间可能存在较高的同源性。
2.2.4 异养同化微生物的生长代谢特性氯代烷烃异养同化微生物生长pH值范围为6.5–7.7,温度为25–30 ℃。当CAHs浓度为0.1–10 mmol/L时,最大比生长速率范围为0.05–6.63 h–1,氯代烷烃的平均降解率均在92%以上。在异养同化过程中,生长基质浓度对微生物生长和代谢速率有较大的影响。CAHs浓度过高,产生毒性抑制,微生物生长缓慢;CAHs浓度过低,微生物底物匮乏,限制微生物生长。菌株Hyphomicrobium sp. MC1在气相浓度为1%的MC中最大比生长速率为0.09 h–1,当气相浓度增到10%时最大比生长速率降为0.04 h–1[51]。此外,不同微生物对CAHs利用效率也具有很大差别,当DCM浓度相同时,菌株Hyphomicrobium DM2和Pseudomonas sp. DM1的最大比生长速率分别为0.07 h–1和0.11 h–1[52]。为进一步解析代谢机理,Wu等[30]从DCM异养同化菌株Lysinibacillus sphaericus wh22提取质粒pRC11,并转移到大肠杆菌中,该重组大肠杆菌菌株能在5–16 mmol/L DCM的条件下正常生长,表明该基因可能是DCM代谢的关键基因。
氯代烯烃作为微生物生长的唯一碳源被广泛报道[44],这些氯代烯烃同化微生物的最适生长温度为20–37 ℃,pH为6.5–7.5。当氯代烯烃浓度为0.15–10.00 mmol/L时,微生物最大比生长速率范围为0.002–15.720 h–1,降解率在60%–100%。菌株Pseudomonas aeruginosa MF1以VC为唯一碳源生长时,6 mmol/L的VC在35 d内可完全降解[44]。氯代烯烃异构体同样影响微生物代谢特性,Olaniran从非洲污染场地中分离出7株DCE异养同化微生物,在1 mmol/L的c-1, 2-DCE和1.5 mmol/L的t-1, 2-DCE为唯一碳源进行生长时,比生长速率分别为0.014–0.023 h–1和0.019–0.028 h–1,平均降解率为70%–75%和60%–72%,其中t-1, 2-DCE培养基中的细胞密度要高于c-1, 2-DCE,可能是因为c-1, 2-DCE氧化生成环氧化合物具有更高的毒性[18]。与纯菌相比,混合菌降解能力更高,Sing分离的混合菌可迅速降解浓度为50–250 μmol/L的VC[20]。研究表明含氧量和pH可影响氯代烯烃的异养同化过程,VC降解菌株Pseudomonas aeruginosa MF1在缺氧条件下培养2.5 d,并不能完全恢复在VC上的生长能力,且降解过程中有VC环氧化合物的生成[44]。同样的,VC降解混合菌在缺氧条件下也有VC环氧化合物生成[20],表明缺氧状态使得VC的降解机理可能发生了改变。TCE异养同化降解菌Stenotrophomonas maltophilia PM102在pH为中性时降解率可达到90%,而pH降为5时降解率下降到77%[48]。
氯代芳烃异养同化微生物最适pH值为7.0–7.8,最适温度为25–30 ℃,当氯代芳烃浓度为0.002–1.000 mmol/L时,微生物最大比生长速率为0.002–0.420 h–1,降解效率范围77%–99%,表明氯代芳烃异养同化微生物的活性差异较大。李明堂等[35]分离的菌株Acinetobacter sp. CB001,在0.44 mmol/L的CB条件下,120 h降解率可达98.2%。该菌株还可同化DCBs,异养同化能力顺序为1, 3-DCB > 1, 2-DCB > 1, 4-DCB。Monferrán等[37]以1, 2-DCB唯一碳源筛选的菌株Acidovorax avenae在2 d内可将0.136 mmol/L的1, 2-DCB完全降解,释放全部氯原子。同样地,菌株Enterobacter sp. SA-2和Pseudomonas sp. SA-6以1, 2, 3-TCB (0.44 mmol/L)和1, 3, 5-TCB (0.43 mmol/L)为唯一碳源和能源生长时,约80%–90%的TCBs可以在200 h内降解,且细胞浓度增加3个数量级[39]。与氯代烷烃和氯代烯烃相比,氯代芳烃降异养同化微生物特异性较低,对多种氯代芳烃均具有降解能力,在复合污染环境中适用性更强。基于当前对CAHs异养同化降解菌筛选分离和生长特性的研究,绝大多数降解菌具有较强的耐受性和降解能力,能较好地适用于氯代烃类污染物场地的修复工作。
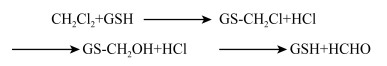
3 典型氯代烃的异养同化降解酶及机理 3.1 氯代烷烃的异养同化MC和DCM是异养同化降解研究最多的氯代烷烃,现已充分揭示了两种氯代烷烃的代谢机理。基于文献报道总结的MC和DCM的降解机理如图 3所示。MC和DCM具有相似的代谢途径,MC首先发生脱氯反应,生成的甲基在CmuA和CmuB甲基转移酶的作用下转移到四氢叶酸(Tetrahydrofolate,H4folate)上形成甲基-H4folate,四氢叶酸连接的中间体在亚甲基-H4folate还原酶、亚甲基-H4folate脱氢酶、甲基-H4folate环水解酶、甲酰-H4folate水解酶的相继作用下进一步氧化生成甲酸,甲酸在甲酸脱氢酶的作用下生成CO2。此外,中间体亚甲基-H4folate可以通过丝氨酸途径被微生物同化为自身的组成物质。与MC不同,DCM首先在还原型谷胱甘肽(Glutathione,GSH)的作用下失去一个氯原子并与GSH结合生成中间产物S-氯甲基谷胱甘肽,中间会自发生成S-羟甲基谷胱甘肽并最终生成甲醛(HCHO)和谷胱甘肽(图 4)。形成的HCHO分别依赖于H4folate和四氢甲蝶呤(Tetrahydromethanopterin,H4MPT)途径进行代谢,H4MPT降解途径普遍存在于大多数甲基营养菌中[61]。HCHO在甲醛活化酶的作用下与四氢甲蝶呤缩合成亚甲基-H4MPT,而亚甲基-H4MPT是丝氨酸循环和大多数微生物同化必需的[62-63]。亚甲基-H4MPT通过亚甲基-H4MPT脱氢酶、甲基-H4MPT环水解酶、甲酰甲基呋喃转移酶和甲酸脱氢酶的相继作用最终生成CO2。
3.2 氯代烯烃的异养同化氯代烯烃异养同化降解中,只有VC异养同化代谢机理被系统解析,降解途径如图 5所示。VC首先在烯烃单加氧酶(Alkene monooxygenase,AkMO)作用下转化为环氧氯乙烯[22],由辅酶M转移酶(Coenzyme M transferase,EaCoMT)介导的辅酶M (Coenzyme M,CoM)进一步与环氧乙烯结合形成2-氯-羟乙基-CoM,2-氯-羟乙基-CoM自发地消除氯原子生成2-酮乙基-CoM,随后,在CoM-还原酶/羧化酶作用下生成丙二酸半醛和还原性的CoM。丙二酸半醛通过双官能醛/醇脱氢酶的氧化生成丙二酸,在脱羧酶、辅酶A (Coenzyme A,CoA)合成酶和CoA转移酶的循环中转化为丙二酰-CoA,最终进入三羧酸循环(Tricarboxylic acid cycle,TCA)[22]。研究者针对氯代烯烃降解酶展开了深入研究,AkMO和EaCoMT是VC异养同化微生物的重要酶系[33],Mycobacterium sp. strain E20降解乙烯的研究中首次发现AkMO[22],研究还发现其他酶也参与VC的异养同化过程,Danko等[57]研究证明Ochrobactrum sp. TD菌株中的脱羧酶参与VC的降解过程,对同化过程酶生成的编码基因进行了深入分析,发现2–3个ORFs编码参与Mycobacterium JS60和Nocardioides JS614菌株的CoA合成酶和CoA转移酶的合成。Chuang等[65]通过蛋白质组学和RT-PCR确认了VC代谢基因etnA、etnC、etnE和comA与双功能醇/醛脱氢酶、FAD/NAD(P)H依赖性还原酶/羧化酶和CoA转移酶的表达过程受到了VC的诱导。这些为VC异养同化过程的有效调控提供了重要的理论依据。
3.3 氯代芳烃的异养同化CB、DCBs和TCBs等典型氯代芳烃的异养同化过程具有相似的代谢途径,同化过程普遍遵循“先开环再脱氯”的降解机制[36],降解过程如图 6所示。氯代芳烃在双加氧酶的作用下发生邻位或间位裂环脱氯,CB被依次氧化生成邻氯苯酚、3-氯邻苯二酚,然后在邻苯二酚1, 2-双加氧酶的作用下,中间先后经历邻位开环、脱氯、氧化等过程,最终将其矿化为CO2和转化为生物质;1, 2-DCB和1, 4-DCB分别在1, 2-二氯苯双加氧酶和1, 4-二氯苯双加氧酶等酶的作用下,先后经历开环、脱氯、氧化后生成脱氯产物5-氯马来酸和2-氯马来酸[50]。研究表明,参与氯代芳烃异养同化过程的邻苯二酚双加氧酶种类很多,叶杰旭等[36]在Delftia tsuruhatensis同化CB过程中发现3-氯邻苯二酚在邻苯二酚1, 2-双加氧酶的作用下邻位开环生成2-氯-粘康酸。在有氧条件下黄色杆菌Xanthobacter flavus 14p1对1, 4-DCB的降解过程中,邻苯二酚1, 2-双加氧酶将3, 6-二氯邻苯二酚转化为2, 5-二氯-粘康酸[60]。菌株Enterobacter sp. SA-2和Pseudomonas sp. SA-6在降解1, 3, 5-TCB的过程中,降解3, 6-二氯邻苯二酚的关键酶为邻苯二酚2, 3-双加氧酶,且菌株SA-2比菌株SA-6酶活性略高[39]。综上表明,邻苯二酚双加氧酶是氯代芳烃异养同化过程中的关键酶。
不同CAHs异养同化过程功能基因差异很大,不同CAHs具有不同的特征基因,对特征基因的检测分析可明晰微生物或环境样品的异养同化降解功能。典型的CAHs异养同化过程特征基因的引物如表 3所示。氯甲烷同化过程的特征基因为cmuA和cmuB[66],其中cmuA作为分子探针而广泛应用。Miller等[67]设计了cmuA的引物cmuA802和cmuA1609r用于检测卤甲烷降解物。随后,Nadalig开发了两个新的反向引物cmuA1802r和MF2,建立了cmuA的qPCR方法并用于研究绿藻球中的氯甲烷降解物[68]。DCM异养同化特征基因为dcmA,dcmA基因具有高度保守性,开发的引物CFOR/CREV[69]和DMfor/DMrev[70]可准确定量环境中DCM异养同化微生物[71]。VC异养同化的特征基因为etnC和etnE,当前开发的引物可以准确定量功能微生物和地下水环境中特征基因含量[71-72]。氯代芳烃异养同化的特征基因为adhA1,Monferrán等[37]利用引物17F和1406R证实了菌株Acidovorax avenae对1, 2-DCB的代谢途径。CAHs特征基因的明确,对明晰CAHs的降解产物、同化途径和实际污染场地功能微生物的定量工作具有重要的指导意义。
| Gene | CAHs | Primer name | Primer sequence (5′–3′) | Size (bp) | References |
| cmuA | CH3Cl | cmuA802 | TTCAACGGCGAYATGTATCCYGG | 807 | [67] |
| cmuA1609r | TCTCGATGAACTGCTCRGGCT | 984 | |||
| cmuA1802r | TTVGCRTCRAGVCCGTA | 167 | [68] | ||
| MF2 | CCRCCRTTRTAVCCVACYTC | ||||
| dcmA | CH2Cl2 | Cfor | ATSATCYKGCRTCMCAGC | 441–450 | [69] |
| Crev | TMAGCMAGTAWTYCTA | ||||
| Dmfor | AAAAAAAACATCTAGAGAATGACAACCGTGCGC | 1 225 | [70] | ||
| Dmrev | AAAAAAAAAAGGATCCGGTCATCGAAGGAATGC | ||||
| etnC | VC | NVC105 | CAGGAGTCSCTKGACCGTCA | 360 | [73] |
| NVC106 | CARACCGCCGTAKGACTTTGT | ||||
| RTC_F | ACCCTGGTCGGTGTKSTYTC | 360 | [74] | ||
| RTC_R | TCATGTAMGAGCCGACGAAGTC | 106 | |||
| MRTC_F | ACACTCGTCGGCGTTGTTTC (+7 others) | 106 | [72] | ||
| MRTC_R | TCATGTACGAGCCGACGAAGTC (+5 others) | ||||
| etnE | VC | CoM-F1L | AACTACCCSAAYCCSCGCTGGTACGAC | 891 | [58] |
| CoM-R2 | TCGTCGGCAGTTTCGGTGATCGTGCTCTT | ||||
| RTE_F | CAGAAYGGCTGYGACATYATCCA | 151 | [74] | ||
| RTE_R | CSGGYGTRCCCGAGTAGTTWCC | ||||
| MRTE_F | CAGAATGGCTGTGACATTATCCA (+5 others) | 151 | [72] | ||
| MRTE_R | CTGGTGTGCCGGAGTAGTTTCC (+8 others) | ||||
| adhA1 | 1, 2-DCB | 17F | GAGTTTGATCCTGGCTCAG | 730 | [37] |
| 1406R | ACGGGCGGTGTGTA/GC |
异养同化作为CAHs的生物降解方式之一为污染场地生物修复提供了可选的修复方案,与其他生物代谢过程相比具有显著优势。本文对可发生异养同化的CAHs进行了系统性的总结,对典型CAHs的相关微生物、异养同化降解途径及其降解酶进行了系统阐述。目前,报道的CAHs异养同化微生物和可降解CAHs的种类较少,已分离的可进行CAHs异养同化微生物仅有十几个属,可作为碳源和能源被微生物利用的CAHs仅限于低氯取代烃,如VC、DCE、DCM和CM,且只有一篇文章报道了TCE的异养同化降解[44];总结发现,大多数微生物生长条件(pH 6.5–7.5,25–35 ℃)均比较温和,对于极端条件下的微生物研究甚少;另外,CAHs的异养同化研究主要集中在分离的纯菌或群落结构简单的混合菌中,对于实际CHAs的污染场地中,异养同化降解菌的丰度、酶的种类、异养同化代谢强度、环境因子氧气的影响特性等的信息报道还十分有限;此外,污染场地中氯代烃组成复杂,有些氯代烃无法作为异养同化微生物的碳源和能源,共存氯代烃还会抑制其他氯代烃的同化降解[75],同化降解过程氯代烃间的相互作用影响还未得到有效解决。这些问题限制了异养同化降解微生物在CAHs污染处理中的应用。因此,面对日益严峻的CAHs污染,未来研究中关于CAHs的异养同化要更加关注以下几个方面:(1)扩大菌株筛选范围,获得具有抗逆性的多种CAHs降解能力的菌株;通过基因工程和酶工程开发工程菌,如已有利用基因工程手段获得具有CAHs异养同化降解功能的大肠杆菌[30]。(2)充分利用异养同化降解特点及快速、无二次污染的优势,开发厌氧脱氯-异养同化,共代谢-异养同化,厌氧脱氯-共代谢-异养同化,物化处理(铁还原)-异养同化等联合CAHs处理工艺,已有研究发现以VC为底物可共代谢降解DCE[20]。(3)利用宏基因组和转录组测序等多组学技术,明晰污染场地异养同化微生物基因组信息,解析不同种属微生物对CAHs的代谢机理[76]。(4)逐步开展异养同化降解的场地应用,明晰场地环境中异养同化降解的影响因素,建立有效的异养同化实施、监控和模拟方案。
附:缩略词索引
CAHs:氯代烃,Chlorinated hydrocarbons
TCE:三氯乙烯,Trichloroethylene
PCE:四氯乙烯,Perchloroethylene
TCBs:三氯苯,Trichlorobenzenes
HCB:六氯苯,Hexachlorobenzene
CM:一氯甲烷,Chloromethane
DCM:二氯甲烷,Dichloromethane
CF:三氯甲烷,Chloroform
CT:四氯化碳,Carbon tetrachloride
VC:氯乙烯,Vinyl chloride
c-1, 2-DCE:顺式二氯乙烯,cis-1, 2-dichloroethene
t-1, 2-DCE:反式二氯乙烯,trans-1, 2-dichloroethylene
CB:氯苯,Chlorobenzene
1, 2-DCB:1, 2-二氯苯,1, 2-dichlorobenzene
1, 4-DCB:1, 4-二氯苯,1, 4-dichlorobenzene
1, 2, 3-TCB:1, 2, 3-三氯苯,1, 2, 3-trichlorobenzene
1, 3, 5-TCB:1, 3, 5-三氯苯,1, 3, 5-trichlorobenzene
1, 2-DCA:1, 2-二氯乙烷,1, 2-dichloroethane
CBM:氯溴甲烷,Chlorobromomethane
1, 1, 1-TCA:1, 1, 1-三氯乙烷,1, 1, 1-trichloroethane
DCBs:二氯苯,Dichlorobenzenes
H4folate:四氢叶酸,Tetrahydrofolate
GSH:谷胱甘肽,Glutathione
H4MPT:四氢甲蝶呤,Tetrahydromethanopterin
AkMO:烯烃单加氧酶,Alkene monooxygenase
EaCoMT:辅酶M转移酶,Coenzyme M transferase
CoM:辅酶M,Coenzyme M
CoA:辅酶A,Coenzyme A
TCA:三羧酸循环,1, 1, 2-trichloroethane
| [1] |
Lei C, Sun YQ, Khan E, et al. Removal of chlorinated organic solvents from hydraulic fracturing wastewater by bare and entrapped nanoscale zero-valent iron. Chemosphere, 2018, 196: 9-17. |
| [2] |
Chen HM, Wu MT. Residential exposure to chlorinated hydrocarbons from groundwater contamination and the impairment of renal function-An ecological study. Sci Rep, 2017, 7: 40283. |
| [3] |
Liu S, Xing ZL, Li C, et al. The biotransformation mechanism of chloroform in landfill cover. China Environ Sci, 2018, 38(12): 4581-4590 (in Chinese). 刘帅, 邢志林, 李宸, 等. 典型污染物包覆层中氯仿的沿程生物转化机制. 中国环境科学, 2018, 38(12): 4581-4590. DOI:10.3969/j.issn.1000-6923.2018.12.024 |
| [4] |
Lu Q, Li H, Lin KF, et al. Investigation of chlorinated hydrocarbons in groundwater from a typical contaminated site in Pudong District, Shanghai. Acta Sci Circum, 2016, 36(5): 1730-1737 (in Chinese). 陆强, 李辉, 林匡飞, 等. 上海浦东某氯代烃场地地下水污染现状调查. 环境科学学报, 2016, 36(5): 1730-1737. |
| [5] |
Heidrich S, Weiß H, Kaschl A. Attenuation reactions in a multiple contaminated aquifer in Bitterfeld (Germany). Environ Pollut, 2004, 129(2): 277-288. |
| [6] |
Xing ZL, Zhang LJ, Zhao TT. Advances in degradation of chlorinated hydrocarbons by obligate and facultative methanotrophs. Chin J Biotech, 2014, 30(4): 531-544 (in Chinese). 邢志林, 张丽杰, 赵天涛. 专一营养与兼性甲烷氧化菌降解氯代烃的研究现状、动力学分析及展望. 生物工程学报, 2014, 30(4): 531-544. |
| [7] |
Liu S, Zhao TT, Xing ZL, et al. Advances in biotic and abiotic mutual promoting mechanism for chlorinated aliphatic hydrocarbons degradation. Chin J Biotech, 2018, 34(4): 510-524 (in Chinese). 刘帅, 赵天涛, 邢志林, 等. 氯代脂肪烃生物与非生物共促降解机制研究进展. 生物工程学报, 2018, 34(4): 510-524. |
| [8] |
Aulenta F, Majone M, Tandoi V. Enhanced anaerobic bioremediation of chlorinated solvents: Environmental factors influencing microbial activity and their relevance under field conditions. J Chem Technol Biotechnol, 2006, 81(9): 1463-1474. DOI:10.1002/jctb.1567 |
| [9] |
Wang Q, Liu H, Jiang L, et al. Effect of the coexistence of chlorobenzene homologue on anaerobic degradation of hexachlorobenzene. Environ Sci, 2014, 35(4): 1358-1365 (in Chinese). 王琪, 刘辉, 姜林, 等. 共存氯苯类同系物对六氯苯厌氧降解活性的影响. 环境科学, 2014, 35(4): 1358-1365. |
| [10] |
McLean JE, Ervin J, Zhou J, et al. Biostimulation and bioaugmentation to enhance reductive dechlorination of TCE in a long-term flow through column study. Ground Water Monit Remed, 2015, 35(3): 76-88. |
| [11] |
Nijenhuis I, Kuntze K. Anaerobic microbial dehalogenation of organohalides — state of the art and remediation strategies. Curr Opin Biotechnol, 2016, 38: 33-38. |
| [12] |
Tiehm A, Schmidt KR. Sequential anaerobic/aerobic biodegradation of chloroethenes-aspects of field application. Curr Opin Biotechnol, 2011, 22(3): 415-421. |
| [13] |
Richardson RE. Genomic insights into organohalide respiration. Curr Opin Biotechnol, 2013, 24(3): 498-505. |
| [14] |
Jesus J, Frascari D, Pozdniakova T, et al. Kinetics of aerobic cometabolic biodegradation of chlorinated and brominated aliphatic hydrocarbons: A review. J Hazard Mater, 2016, 309: 37-52. DOI:10.1016/j.jhazmat.2016.01.065 |
| [15] |
Teng F, Yang XL, Li FM, et al. Microbial co-metabolism of persistent organic pollutants in environment. J Microbiol, 2016, 36(3): 80-85 (in Chinese). 滕菲, 杨雪莲, 李凤梅, 等. 微生物对环境中难降解有机污染物共代谢作用. 微生物学杂志, 2016, 36(3): 80-85. DOI:10.3969/j.issn.1005-7021.2016.03.015 |
| [16] |
Tobajas M, Monsalvo VM, Mohedano AF, et al. Enhancement of cometabolic biodegradation of 4-chlorophenol induced with phenol and glucose as carbon sources by Comamonas testosteroni. J Environ Manage, 2012, 95(2): S116-S121. |
| [17] |
Yang X, Xing ZL, Zhang LJ. Advances in transformation and regulation biodegradation of chlorinated hydrocarbons in landfill. Acta Microbiol Sin, 2017, 57(4): 468-479 (in Chinese). 杨旭, 邢志林, 张丽杰. 填埋场氯代烃生物降解过程的机制转化与调控研究及展望. 微生物学报, 2017, 57(4): 468-479. |
| [18] |
Olaniran AO, Pillay D, Pillay B. Aerobic biodegradation of dichloroethenes by indigenous bacteria isolated from contaminated sites in Africa. Chemosphere, 2008, 73(1): 24-29. |
| [19] |
Hauke S, Willem M DV. Anaerobic microbial dehalogenation. Annu Rev Microbiol, 2004, 58(1): 43-73. DOI:10.1146/annurev.micro.58.030603.123600 |
| [20] |
Sing H, Löffler FE, Fathepure BZ. Aerobic biodegradation of vinyl chloride by a highly enriched mixed culture. Biodegradation, 2004, 15(3): 197-204. DOI:10.1023/B:BIOD.0000026539.55941.73 |
| [21] |
Schmidt KR, Augenstein T, Heidinger M, et al. Aerobic biodegradation of cis-1, 2-dichloroethene as sole carbon source: Stable carbon isotope fractionation and growth characteristics. Chemosphere, 2010, 78(5): 527-532. DOI:10.1016/j.chemosphere.2009.11.033 |
| [22] |
Mattes TE, Alexander AK, Coleman NV. Aerobic biodegradation of the chloroethenes: pathways, enzymes, ecology, and evolution. FEMS Microbiol Rev, 2010, 34(4): 445-475. DOI:10.1111/j.1574-6976.2010.00210.x |
| [23] |
Frascari D, Zanaroli G, Danko AS. In situ aerobic cometabolism of chlorinated solvents: a review. J Hazard Mater, 2015, 283: 382-399. DOI:10.1016/j.jhazmat.2014.09.041 |
| [24] |
Taylor AE, Dolan ME, Bottomley PJ, et al. Utilization of fluoroethene as a surrogate for aerobic vinyl chloride transformation. Environ Sci Technol, 2007, 41(18): 6378-6383. DOI:10.1021/es0701255 |
| [25] |
Giddings CGS, Liu F, Gossett JM. Microcosm assessment of Polaromonas sp. JS666 as a bioaugmentation agent for degradation of cis-1, 2-dichloroethene in aerobic, subsurface environments. Ground Water Monit Remediat, 2010, 30(2): 106-113. DOI:10.1111/j.1745-6592.2010.01283.x |
| [26] |
Vangelas K, Truex MJ, Newell CJ, et al. Scenarios Evaluation Tool for Chlorinated Solvent MNA. United States: Washington Savannah River Company, 2007: 18-46.
|
| [27] |
Trotsenko YA, Torgonskaya ML. The aerobic degradation of dichloromethane: Structural-functional aspects (a review). Appl Biochem Micro, 2009, 45(3): 233-247. |
| [28] |
Schaefer JK, Goodwin KD, Mcdonald IR, et al. Leisingera methylohalidivorans gen. nov., sp. nov., a marine methylotroph that grows on methyl bromide. Int J Syst Evol Micr, 2002, 52(3): 851-859. |
| [29] |
Liu HX, Zhu RY, Ouyang DJ, et al. Isolation and degradation characteristics of dichloromethane- degradation bacterial strain by Methylobacterium rhodesianum H13. Environ Sci, 2013, 34(9): 3613-3619 (in Chinese). 刘洪霞, 朱润晔, 欧阳杜娟, 等. 二氯甲烷降解菌Methylobacterium rhodesianum H13的分离鉴定及降解特性研究. 环境科学, 2013, 34(9): 3613-3619. |
| [30] |
Wu SJ, Hu ZH, Zhang LL, et al. A novel dichloromethane-degrading Lysinibacillus sphaericus strain wh22 and its degradative plasmid. Appl Microbiol Biot, 2009, 82(4): 731-740. |
| [31] |
Olaniran AO, Okoh AI, Ajisebutu S, et al. The aerobic dechlorination activities of two bacterial species isolated from a refuse dumpsite in Nigeria. Int Microbiol, 2002, 5(1): 21-24. |
| [32] |
Jin YO, Mattes TE. Adaptation of aerobic, ethene-assimilating Mycobacterium strains to vinyl chloride as a growth substrate. Environ Sci Technol, 2008, 42(13): 4784-4789. DOI:10.1021/es8000536 |
| [33] |
Jin YO, Cheung S, Coleman NV, et al. Association of missense mutations in epoxyalkane coenzyme M transferase with adaptation of Mycobacterium sp. strain JS623 to growth on vinyl chloride. Appl Environ Microb, 2010, 76(11): 3413-3419. DOI:10.1128/AEM.01320-09 |
| [34] |
Mundle SOC, Spain JC, Lacrampe-Couloume G, et al. Branched pathways in the degradation of cDCE by cytochrome P450 in Polaromonas sp. JS666. Sci Total Environ, 2017, 605-606: 99-105. DOI:10.1016/j.scitotenv.2017.06.166 |
| [35] |
Li MT, Hao LL, Cui JT, et al. Identification and characterization of an aerobic bacterium degrading chlorobenzene. Acta Microbiol Sin, 2010, 50(5): 586-592 (in Chinese). 李明堂, 郝林琳, 崔俊涛, 等. 好氧氯苯降解菌的分离鉴定. 微生物学报, 2010, 50(5): 586-592. |
| [36] |
Ye JX, Lin TH, Luo YH, et al. Isolation and identification of a chlorobenzene-degrading bacterium and its degradation characteristics. Environ Sci, 2017, 38(2): 802-808 (in Chinese). 叶杰旭, 林彤晖, 骆煜昊, 等. 1株氯苯高效降解菌的分离鉴定及降解特性. 环境科学, 2017, 38(2): 802-808. |
| [37] |
Monferrán MV, Echenique JR, Wunderlin DA. Degradation of chlorobenzenes by a strain of Acidovorax avenae isolated from a polluted aquifer. Chemosphere, 2005, 61(1): 98-106. DOI:10.1016/j.chemosphere.2005.03.003 |
| [38] |
Dai QH, Cao XD, Sun XW. Study on isolation and characterization of a dichlorobenzene-degrading bacterial strain. Chin J Environ Eng, 2009, 3(12): 2219-2222 (in Chinese). 戴青华, 曹晓丹, 孙向武. 1, 4-二氯苯降解菌的分离及其降解特性研究. 环境工程学报, 2009, 3(12): 2219-2222. |
| [39] |
Adebusoye SA, Picardal FW, Ilori MO, et al. Aerobic degradation of di- and trichlorobenzenes by two bacteria isolated from polluted tropical soils. Chemosphere, 2007, 66(10): 1939-1946. DOI:10.1016/j.chemosphere.2006.07.074 |
| [40] |
Liu H, Jiang L, Wang Q, et al. Progress on microbial degradation of hexachlorobenzene. Environ Pollut Control, 2013, 35(1): 86-92 (in Chinese). 刘辉, 姜林, 王琪, 等. 六氯苯微生物降解研究进展. 环境污染与防治, 2013, 35(1): 86-92. DOI:10.3969/j.issn.1001-3865.2013.01.019 |
| [41] |
Rehfuss M, Urban J. Rhodococcus phenolicus sp. nov., a novel bioprocessor isolated actinomycete with the ability to degrade chlorobenzene, dichlorobenzene and phenol as sole carbon sources. Syst Appl Microbiol, 2005, 28(8): 695-701. DOI:10.1016/j.syapm.2005.05.011 |
| [42] |
McAnulla C, McDonald IR, Murrell JC. Methyl chloride utilising bacteria are ubiquitous in the natural environment. FEMS Microbiol Lett, 2001, 201(2): 151-155. DOI:10.1111/j.1574-6968.2001.tb10749.x |
| [43] |
Szymanik M, Welc-Faleciak R, Bartosik D, et al. Replication system of plasmid pMTH4 of Paracoccus methylutens DM12 contains an enhancer. Pol J Microbiol, 2006, 55(4): 261-270. |
| [44] |
Verce MF, Ulrich RL, Freedman DL. Characterization of an isolate that uses vinyl chloride as a growth substrate under aerobic conditions. Appl Environ Microb, 2000, 66(8): 3535-3542. DOI:10.1128/AEM.66.8.3535-3542.2000 |
| [45] |
Elango VK, Liggenstoffer AS, Fathepure BZ. Biodegradation of vinyl chloride and cis-dichloroethene by a Ralstonia sp. strain TRW-1. Appl Microbiol Biot, 2006, 72(6): 1270-1275. DOI:10.1007/s00253-006-0424-4 |
| [46] |
Olaniran AO, Pillay D, Pillay B. Haloalkane and haloacid dehalogenases from aerobic bacterial isolates indigenous to contaminated sites in Africa demonstrate diverse substrate specificities. Chemosphere, 2004, 55(1): 27-33. DOI:10.1016/j.chemosphere.2003.10.067 |
| [47] |
Bradley PM, Chapelle FH. Aerobic microbial mineralization of dichloroethene as sole carbon substrate. Environ Sci Technol, 2000, 34(1): 221-223. |
| [48] |
Mukherjee P, Roy P. Purification and identification of trichloroethylene induced proteins from Stenotrophomonas maltophilia PM102 by immuno-affinity-chromatography and MALDI-TOF Mass spectrometry. SpringerPlus, 2013, 2: 207. DOI:10.1186/2193-1801-2-207 |
| [49] |
Dey K, Roy P. Degradation of trichloroethylene by Bacillus sp.: isolation strategy, strain characteristics, and cell immobilization. Curr Microbiol, 2009, 59(3): 256-260. DOI:10.1007/s00284-009-9427-6 |
| [50] |
Haigler BE, Nishino SF, Spain JC. Degradation of 1, 2-dichlorobenzene by a Pseudomonas sp. Appl Environ Microb, 1988, 54(2): 294-301. DOI:10.1128/AEM.54.2.294-301.1988 |
| [51] |
Fetzner S. Aerobic degradation of halogenated aliphatics//Timmis KN, Ed. Handbook of Hydrocarbon and Lipid Microbiology. Berlin, Heidelberg: Springer, 2010: 865-885
|
| [52] |
Liu L. Cloning and characterization of dichloromethane dehalogenase for dichloromethane degradation[D]. Hangzhou: Zhejiang University of Technology, 2015 (in Chinese). 刘靓. DCM脱卤酶基因的克隆表达及特性的研究[D].杭州: 浙江工业大学, 2015. http://cdmd.cnki.com.cn/Article/CDMD-10337-1015434451.htm |
| [53] |
Nadalig T, Greule M, Bringel F, et al. Hydrogen and carbon isotope fractionation during degradation of chloromethane by methylotrophic bacteria. Microbiology Open, 2013, 2(6): 893-900. DOI:10.1002/mbo3.124 |
| [54] |
Krausova VI, Robb FT, González JM. Bacterial degradation of dichloromethane in cultures and natural environments. J Microbiol Meth, 2003, 54(3): 419-422. DOI:10.1016/S0167-7012(03)00062-9 |
| [55] |
Studer A, Stupperich E, Vuilleumier S, et al. Chloromethane: tetrahydrofolate methyl transfer by two proteins from Methylobacterium chloromethanicum strain CM4. Eur J Biochem, 2001, 268(10): 2931-2938. DOI:10.1046/j.1432-1327.2001.02182.x |
| [56] |
Coleman NV, Mattes TE, Gossett JM, et al. Phylogenetic and kinetic diversity of aerobic vinyl chloride-assimilating bacteria from contaminated sites. Appl Environ Microb, 2002, 68(12): 6162-6171. DOI:10.1128/AEM.68.12.6162-6171.2002 |
| [57] |
Danko AS, Freedman DL. Involvement of carbon dioxide in the aerobic biodegradation of ethylene oxide, ethene, and vinyl chloride. Process Biochem, 2008, 43(5): 517-521. DOI:10.1016/j.procbio.2008.01.008 |
| [58] |
Fung HKH, Gadd MS, Drury TA, et al. Biochemical and biophysical characterisation of haloalkane dehalogenases DmrA and DmrB in Mycobacterium strain JS60 and their role in growth on haloalkanes. Mol Microbiol, 2015, 97(3): 439-453. |
| [59] |
Coleman NV, Mattes TE, Gossett JM, et al. Biodegradation of cis-dichloroethene as the sole carbon source by a β-proteobacterium. Appl Environ Microb, 2002, 68(6): 2726-2730. DOI:10.1128/AEM.68.6.2726-2730.2002 |
| [60] |
Spiess E, Sommer C, Görisch H. Degradation of 1, 4-dichlorobenzene by Xanthobacter flavus 14p1. Appl Environ Microb, 1995, 61(11): 3884-3888. DOI:10.1128/AEM.61.11.3884-3888.1995 |
| [61] |
Studer A, McAnulla C, Büchele R, et al. Chloromethane-induced genes define a third C1 utilization pathway in Methylobacterium chloromethanicum CM4. J Bacteriol, 2002, 184(13): 3476-3484. DOI:10.1128/JB.184.13.3476-3484.2002 |
| [62] |
Kayser MF, Ucurum Z, Vuilleumier S. Dichloromethane metabolism and C1 utilization genes in Methylobacterium strains. Microbiology, 2002, 148(6): 1915-1922. DOI:10.1099/00221287-148-6-1915 |
| [63] |
Vorholt JA, Marx CJ, Lidstrom ME, et al. Novel formaldehyde-activating enzyme in Methylobacterium extorquens AM1 required for growth on methanol. J Bacteriol, 2000, 182(23): 6645-6650. DOI:10.1128/JB.182.23.6645-6650.2000 |
| [64] |
Trotsenko YA, Doronina NV. The biology of Methylobacteria capable of degrading halomethanes. Mikrobiologiia, 2003, 72(2): 149-160. |
| [65] |
Chuang AS, Mattes TE. Identification of polypeptides expressed in response to vinyl chloride, ethene, and epoxyethane in Nocardioides sp. strain JS614 by using peptide mass fingerprinting. Appl Environ Microb, 2007, 73(13): 4368-4372. DOI:10.1128/AEM.00086-07 |
| [66] |
Cox MJ, Schäfer H, Nightingale PD, et al. Diversity of methyl halide-degrading microorganisms in oceanic and coastal waters. FEMS Microbiol Lett, 2012, 334(2): 111-118. DOI:10.1111/j.1574-6968.2012.02624.x |
| [67] |
Miller LG, Warner KL, Baesman SM, et al. Degradation of methyl bromide and methyl chloride in soil microcosms: Use of stable C isotope fractionation and stable isotope probing to identify reactions and the responsible microorganisms. Geochim Cosmochim Ac, 2004, 68(15): 3271-3283. DOI:10.1016/j.gca.2003.11.028 |
| [68] |
Nadalig T, Ul Haque MF, Roselli S, et al. Detection and isolation of chloromethane-degrading bacteria from the Arabidopsis thaliana phyllosphere, and characterization of chloromethane utilization genes. FEMS Microbiol Ecol, 2011, 77(2): 438-448. DOI:10.1111/j.1574-6941.2011.01125.x |
| [69] |
Vuilleumier S, Ivos N, Dean M, et al. Sequence variation in dichloromethane dehalogenases/ glutathione S-transferases. Microbiology, 2001, 147(3): 611-619. DOI:10.1099/00221287-147-3-611 |
| [70] |
Firsova JE, Doronina NV, Trotsenko YA. Analysis of the key functional genes in new aerobic degraders of dichloromethane. Microbiology, 2010, 79(1): 66-72. DOI:10.1134/S0026261710010091 |
| [71] |
Coleman NV. Primers: functional genes for aerobic chlorinated hydrocarbon-degrading microbes// McGenity T, Timmis K, Nogales B, Eds. Hydrocarbon and Lipid Microbiology Protocols. Berlin, Heidelberg: Springer, 2015: 141-175.
|
| [72] |
Jin YO, Mattes TE. Assessment and modification of degenerate qPCR primers that amplify functional genes from etheneotrophs and vinyl chloride-assimilators. Lett Appl Microbiol, 2011, 53(5): 576-580. DOI:10.1111/j.1472-765X.2011.03144.x |
| [73] |
Coleman NV, Bui NB, Holmes AJ. Soluble di-iron monooxygenase gene diversity in soils, sediments and ethene enrichments. Environ Microbiol, 2006, 8(7): 1228-1239. DOI:10.1111/j.1462-2920.2006.01015.x |
| [74] |
Jin YO, Mattes TE. A quantitative PCR assay for aerobic, vinyl chloride- and ethene-assimilating microorganisms in groundwater. Environ Sci Technol, 2010, 44(23): 9036-9041. DOI:10.1021/es102232m |
| [75] |
Zhao HP, Schmidt KR, Tiehm AT. Inhibition of aerobic metabolic cis-1, 2-di-chloroethene biodegradation by other chloroethenes. Water Res, 2010, 44(7): 2276-2282. DOI:10.1016/j.watres.2009.12.023 |
| [76] |
Xing ZL, Zhao TT, Zhang LJ, et al. Effects of copper on expression of methane monooxygenases, trichloroethylene degradation, and community structure in methanotrophic consortia. Eng Life Sci, 2018, 18(4): 236-243. DOI:10.1002/elsc.201700153 |
 2020, Vol. 36
2020, Vol. 36