中国科学院微生物研究所、中国微生物学会主办
文章信息
- 惠希武, 曹卫荣, 张迪, 盖文丽, 李书莉, 李银贵
- Hui Xiwu, Cao Weirong, Zhang Di, Ge Wenli, Li Shuli, Li Yingui
- 微生物来源的谷氨酰胺转氨酶(mTG)介导的单一定点修饰的PEG IFN-α2a
- Site-specific monoPEGylated interferon alpha2a mediated by microbial transglutaminase
- 生物工程学报, 2020, 36(4): 750-762
- Chinese Journal of Biotechnology, 2020, 36(4): 750-762
- 10.13345/j.cjb.190343
-
文章历史
- Received: August 1, 2019
- Accepted: November 1, 2019
- Published: November 27, 2019
Though we got tremendous advances in recombinant protein drugs in recent years, there're also some shortcomings that may limit their usefulness including susceptibility to degradation by proteases, short circulating half-life in vivo, low solubility, rapid kidney clearance and propensity to generate neutralizing antibodies[1-3]. Therefore, intensive research has been devoted to overcome these problems inherent to protein drugs. Various strategies have been explored to improve the clinical properties of protein drugs; among these efforts and techniques, the conjugation of PEG chains, termed PEGylation, is regard as the most useful approach for protein drug development[4-8]. There are several advantages of PEGylated protein drugs: increased stability and water solubility, increased resistance to proteolytic inactivation, low toxicity, improved pharmacokinetic profiles, and reduced renal clearance and immunogenicity[9-10]. PEGylation plays an important role in drug delivery, enhancing the potential of peptides and proteins as therapeutic agents due to these favorable properties.
PEGylation was first described in the 1970s by Davies and Abuchowski on albumin and catalase modification[11-12]. At present, chemical method is the most widely used modification method for protein PEGylation involves the covalent conjugation of activated monomethoxy-PEG (mPEG) at the level of N-terminal and/or ε-amino group of lysine residues. But there are also some limitations of this strategy, because of the potential multiple sites of conjugation and the consequent heterogeneity of the PEGylated proteins. Mixtures of multi-PEGylated isomers were generally obtained, resulting in significant losses of homogeneity and protein activity[13-15]. Thereafter improvements in PEGylation were achieved as shown by products like PEG-Intron® and PEGASYS®: mixtures of mono-PEGylated positional isomers. But it is heterogeneous, none site-specific, and there're still some problems in quality control[14-15]. In order to obtain site-specific PEGylation, other chemical approaches were developed, such as the selective PEGylation at the cysteines group or at the N-terminal amino group of a polypeptide chain by controlling the pH value or mutant the nonnative amino acid, which may influence the structure and activity of protein[16-17]. More recently, a very promising enzymatic method has been explored for devising novel and mild strategies of site-directed PEGylation of pharmaceutical proteins that use transglutaminase for the covalent linkage of PEG moieties at the c-carboxamide groups of Gln of proteins[18-20]. For this purpose, a PEG derivative bearing a primary amino group (PEG-NH2) is used; this becomes covalently linked to the protein at glutamines through a transglutamination reaction catalyzed by the enzyme according to the following scheme:
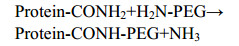
|
Where CO-NH2 is a carboxamide group[21].
The most commonly used transglutaminase in PEGylation is mTG, which derived from Streptomyces mobaraense, because the amino acid sequence of its 331-residue polypeptide chain and 3D structure are available[21-22]. mTG is calcium-independent, which exhibits no significant homology to mammalian transglutaminases, and has a molecular weight of 37.9 kDa and widely used to improve the physical and textural properties of protein-rich foods[23-24]. In the bio-pharmaceutical field, mTG has also been used in the preparation of peptides, recombinant proteins, antibody-drug conjugate (ADC) and other biological products, especially in conjugation[25-32]. Numerous studies have been conducted on the substrate specificity of transglutaminases using synthetic peptides, as well as proteins, with the aim of unravelling the pattern of amino acid sequences around the reactive Gln residues[33-35]. It was proposed that surface accessibility of a reactive Gln could dictate the transglutaminase-catalysed modification[36]. However, it was also recognized that surface exposure is not the only influence factor as it was often observed that several exposed Gln residues in native globular proteins of known 3D structure do not react with transglutaminase. Neighboring residues are unlikely to be PEGylated on the same molecule, because of steric hindrance; and the presence of a proline close to the putative site of PEGylation is a limiting factor that hampers the reaction[37].
Spolaore B et al obtained site-specific PEG-IFN mediated by mTG via linking PEG-20000 to IFN, which is the first reported PEGylation of a pharmaceutical protein at the level of Gln residues using a 20 kDa PEG. The PEGylated IFN derivatives maintained a high specific activity and a desired prolonged pharmacokinetic profile. From the perspective of druggability, However, the harsh reaction condition for protein, the low yield and complicated preparation process may limit its application, that do not meet the Pharmaceutical concept of low cost, simple process, and controllable quality[38].
In this study, the market-proven 40 kDa PEG was selected to directly couple with native IFNα2a mediated by mTG with high yield in mild reaction condition and with simple process. We choose IFN α2a as substrate to test, which is widely used in HBV and HCV by controlling replication of the virus. It is a challenging case, because it contains 12 potential PEGylation sites. The chemical PEGylation has been successfully applied to produce the PEGylated IFN, marketed under the brand name PEGASYS®. As mentioned above, this mixture still has many shortcomings. For this reason, the site-specific PEGylation of IFN α2a is essential to design of new and more effective therapeutic proteins. Firstly, we used computational approach to analyze the sequence and structure of IFNα2a and to predict a single potential PEGylation site in the IFN α2a, which was subsequently validated by PEGylation experiments and characteristics analysis of PEGylated IFN α2a by kinds of methods by SDS-PAGE, RP-HPLC, CD and MALDI-TOF. All of the data obtained from these experiments confirmed our computational prediction on the identification of a single IFN α2a residue-Gln101 that is the target of PEGylation modification by mTG. Moreover, in vitro and in vivo studies were carried out to evaluate the bioactivities and pharmacokinetic (PK) profiles of the new bioconjugate, respectively.
1 Materials and methods 1.1 MaterialsRecombinant human IFN α2a expressed by Escherichia coli was obtained by fermentation and purified to clinical grade. Microbial tranglutaminase (mTG) from Streptomyces mobaraensis was purchased from Yiming Biological (Jiangsu, China). Amino terminating Y shaped 40 kDa PEG (Y-PEG40k-NH2) was provided by JenKem (Beijing, China). N-α-carbobenzoxy-L-glutaminyl-glycine, L-glutamic acid γ-monohydroxamate were supplied by Sigma (Saint Louis, USA). HEK293 cells were purchased from ATCC (CRL-1573). All other chemical reagents were of analytical grade.
1.2 Sequence, secondary structure and solvent accessibilities analysisThe Sequence and structure information of IFN α2a were retrieved from NCBI and the Protein Data Bank (PDB code: 1ITF)[39]. Amino acid solvent- accessible surface area was calculated using the NetSurfP and the Scit web server[40].
1.3 Determination of mTG activityThe activity of mTG was determined by using the specific substrate, CBZ-L-Glutaminyl-glycine (Z-Gln-Gly), as described elsewhere[41]. Briefly, 0.2 mL of 200 mmol/L Tris-buffer pH 6.0 containing 36 mmol/L carbobenzoxy-glutaminyl-glycine, 100 mmol/L hydroxylamine and 10 mmol/L reduced glutathione were incubated for 10 min at 37 ℃ with 30 μL of test sample. The enzymatic reaction was stopped by adding 0.5 mL of 12% trichloroacetic acid. After that, 0.5 mL of 5% ferric chloride exahydrate solutions were added and the resulting red color was measured at 525 nm and the enzymatic activity was calculated.
1.4 PEGylation of IFN α2a with Y-PEG40k-NH2IFN α2a solutions (2 mg/mL) were mixed with Y-PEG40k-NH2 at the molar ratio of 1:8 in 20 mmol/L Tris-Cl buffer (pH 8.0). Then the mTG solution was added to the IFN-α2a/Y-PEG40k-NH2 solution to obtain the final mTG concentrations of 0.5 U/mL. The conjugation reaction mixtures were stirred for 5 h at room temperature and then stopped by 3 mol/L ammonium acetate. The pH of the solution was adjusted and then purified using a SPHP column previously equilibrated with 20 mmol/L sodium acetate, pH 5.0. The column was washed with the equilibration buffer, and the PEGylated protein was eluted with NaCl gradient from 0 to 70 mmol/L in sodium acetate buffer, pH 5.0. The PEGylated protein pools were collected, and then characterized by SDS-PAGE, SP-HPLC, CD, and MALDI-TOF mass spectrometry.
1.5 Identification of PEGylation siteNative IFN α2a and PEG-Gln101-IFN α2a were digested with endoproteinase Trypsin and Glu-C after treated with UA buffer (8 mol/L Urea, 150 mmol/L Tris-HCl, pH 8.0) and DTT buffer. After that, samples were purified by RP-HPLC on Agilent ZORBAX 300SB C8 column (4.6 mm× 150 mm, 5 μm), and collect the peak appeared in PEG-Gln101-IFN α2a hydrolysate separation, which is not found in the IFN α2a hydrolysates, then lyophilized for subsequent N-terminal sequencing by automated Protein Sequencer PPSQ-33A (SHIMADZU corporation) according to Edman degradation and molecular weight (Mw) by MALDI-TOF/TOF (4800 Plus MALDI TOF/TOF, AB SCIEX).
The digestion mixtures were also analyzed by LC-MS/MS. Briefly, mixtures was separated by RP-HPLC on Agilent Zorbax SB C18 column (2.1 mm×50 mm, 3.5 μm), the chromatographic peaks of native and PEGylated IFN-α2a were collected, lyophilized, and analyzed by LC-MS/MS (TripleTOF 5600+, AB SCIEX).
1.6 Bioactivity studiesThe procedure is comprised of three simple steps. In brief, HEK293 cells were transiently transfected with pISRE-Luciferase DNA (Beyotime Institute of Biotechnology, Shanghai, China). 96-well Costar plates were seeded with 0.04×106 cells in the presence of IFNα2a, PEG-Gln101-IFN α2a or controls in a total volume of 100 μL/well. The plate was incubated at 37 ℃ with 5% CO2 for 21 hours. The culture supernatants were then removed by aspiration and the cells washed once gently with PBS, followed by addition of 70 μL PromegaGlo Lysis buffer. The plates were subsequently shaken for 5 min on a titer-plate shaker, followed by the addition of 70 μL of Promega Bright Glo-Luciferase Assay reagent. Luciferase activity was then determined by reading on a Luminoscan Ascent plate reader and data analyzed using SoftMax pro.
1.7 Pharmacokinetic studiesTwenty-four male SD rats were randomly divided into 3 groups of 6 animals each and subcutaneously injected with normal saline (NS) containing IFN α2a, PEG-Gln101-IFN α2a and PEGASYS®. Blood samples were withdrawn from the tail vein at scheduled times after administration 15, 30, 60 min and 2, 4, 8 and 24 h for IFNα2a- treated rats, and 1, 2, 4, 8, 24, 32, 48, 72, 96, 120, 144 and 168 h for the PEG-Gln101-IFN α2a and PEGASYS® treated rats. The blood samples were centrifuged, and the IFN α2a concentration in the serum was determined by RGA. The data were elaborated to determine the mean concentration value, standard deviation (±SD) and the following main PK parameters: half life (t1/2), maximal concentration (Cmax), time of maximal concentration (Tmax) and area under the curve (AUC). The pharmacokinetic data elaboration was performed by GraphPad Prism software.
2 Results and discussion 2.1 IFN-α2a sequence and structure analysisHuman IFN α2a is a single chain protein of 165 amino acid residues displaying several biological activities, including anti-virus, anti-tumor and activation of immuoregulatory effects. IFNs are nowadays extensively used to treat HBV and HCV. The structure of IFN α2a has been determined by multidimensional heteronuclear NMR spectroscopy. The IFN α2a primary structure includes 12 glutamines that, in principle, are candidates for transglutamination by mTG (Fig. 1)[39].

|
| Fig. 1 The sequence and structure of IFN-α2a. (A) Model of the three-dimensional structure of IFN-α2a. The location of the helical segments (A to E) is shown; the position of the site-specific PEGylation at the Gln101 residue is indicated by an arrow. (B) Amino acid sequence of human IFN-α2a. Red boxes show the position of helices A to E. The Gln residues, potential sites for mTG- PEGylation, are written in bold and the Gln101 residue is underlined. |
| |
Our aim was to identify the IFN α2a reactive glutamine(s) involved in the transglutamination process, under the assumptions that they are exposed to the solvent, highly flexible, and in a region that can undergo favorable interactions with the enzyme active site. As a first step, we evaluated the accessible surface area of each of these 12 glutamines by NetSurfP. Glutamines were considered to be buried when < 25% of their total area was exposed to solvent: there are nine glutamines satisfying this condition (Table 1)[42]. The substrate specificity of mTG is rather broad[43-44]. In general, broad specificity requires flexibility of the substrate, which is expected to be able to adapt to the enzyme conformation. It follows that the site of PEGylation should not be part of regular secondary structure elements[20, 36], and this latter requirement reduced our candidate list to six glutamines: Gln5, Gln40, Gln46, Gln48, Gln101 and Gln158. Neighboring residues are unlikely to be PEGylated on the same molecule, because of steric hindrance; and the presence of a proline close to the putative site of PEGylation is a limiting factor that hampers the reaction[37], and then Gln5, Gln40, Gln46, Gln48 were reduced because of Pro, Phe, Asn or Arg on either side of them. Overall, sequence, structure and dynamic analysis of IFN α2a molecule indicate that Gln101 is the most likely transglutamination site.
| Gln | Buried or exposed | Surface accessibility | Secondary structure |
| 5 (PQ5) | E | 0.613 | N-terminal |
| 20 | E | 0.611 | α-helix A |
| 40 (PQ40) | E | 0.437 | In the loop between α-helix A and B |
| 46 (NQFQK) | E | 0.474 | In the loop between α-helix A and B |
| 48 (NQFQK) | E | 0.478 | In the loop between α-helix A and B |
| 61 | B | 0.267 | α-helix B |
| 62 | B | 0.164 | α-helix B |
| 90 | E | 0.413 | α-helix C |
| 91 | B | 0.115 | α-helix C |
| 101 | E | 0.587 | In the loop between α-helix C and D |
| 124 | E | 0.441 | α-helix D |
| 158 | B | 0.203 | C-terminal |
In order to optimize the conditions for the enzymatic PEGylation of IFN α2a with mTG, PEGylation studies were carried out under different operative conditions: Y-PEG40k-NH2: IFN α2a ratios, mTG concentrations, reaction pH and times, etc. Consequently, the reaction was carried out at 5 h, PEG:protein ratio of 8:1 and pH 8.0. Under the optimized condition, the monoPEGylated conjugate PEG-Gln101-FN-α2a was obtained. The analysis of the reaction mixture after 5 h by SP-HPLC showed the following peak percentages: 61.8% of PEG-Gln101-FN-α2a (TR=8.4 min), 22.5% of native FN-α2a (TR=17.5 min), and 15.7% of by-product (TR=14.7 min) (Fig. 2).
|
| Fig. 2 The HPLC and SDS-PAGE profiles of PEGylated IFN-α2a and native IFN-α2a. (A) HPLC profiles of IFN-α2a enzymatic PEGylation reaction after 5 h (dot line) and 0 h (solid line) by SP-stat column. The peaks with retention time of 8.4 min and 17.5 min correspond to PEG-Gln101-IFN-α2a and native IFN-α2a, respectively. (B) SDS-PAGE analysis of reduced (lane 3) and non-reduced (lane 2) samples of PEG-Gln101-IFN-α2a stained by Coomassie Brilliant Blue. Lane 1, protein markers. |
| |
The purification of PEG-Gln101-IFN α2a was performed by a single column chromatography on SPHP, a high performance strong cation exchange resin used to separate the PEGylated proteins from the unmodified protein and other impurities. Since the conjugated PEG molecules mask the exposed amino acid residues, PEG-Gln101-IFN α2a is more rapidly eluted from the column than the unreacted IFN α2a that tightly binds to the resin. Moreover, we also found that mTG had higher affinity for the resin than the PEGylated IFN α2a while other impurities had lower affinity: the unreacted PEG passed through the column without binding. Therefore, the different elution time of PEG-Gln101-IFN α2a and the by-products, namely unreacted IFN α2a, mTG, IFN α2a aggregates and Y-PEG40k-NH2, allowed for the extensive purification of the bioconjugate. The data reported in Table 2 show that the chromatographic process yields high purified PEG-Gln101-IFN α2a recovery.
| Process step | Protein amount (mg) | Yields (%) | Cumulative yields (%) |
| IFN α2a | 100.0 | − | − |
| PEGylation reaction | 61.8 | 61.8 | 61.8 |
| SPHP | 57.7 | 93.4 | 57.7 |
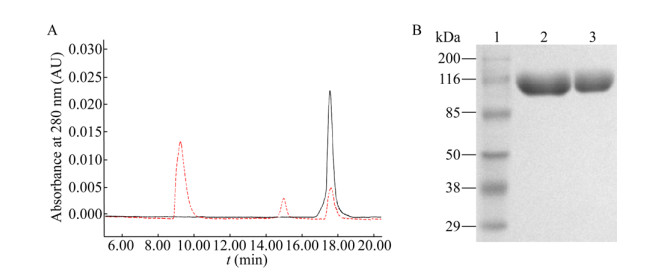
The molecular weight of purified PEG- Gln101-IFN α2a was determined by MALDI-TOF mass spectrometry and SDS-PAGE. The mass spectrum reported in Fig. 3A shows the major signal with average Mw of 58 495.6 Da, which is consistent with the theoretical molecular weight of PEG-Gln101-IFN α2a calculated by summing the mass of IFNα2a (19 269 Da) and the nominal mass of 40 kDa Y-PEG-NH2. The other signals with average mass of 29 260 Da correspond to the doubly-charged PEG-Gln101-IFN α2a. The SDS-PAGE analysis showed in Fig. 2B shows that PEG-Gln101-IFN α2a migrates with an apparent molecular weight of about 100 kDa. In this regard, it should be noted that the hydrated PEG, which coordinates 2-3 water molecules per ethoxy unit, bestows the hydrodynamic radius on the bioconjugate. Therefore, PEGylated proteins migrate with a higher apparent molecular mass in the electrophoresis gel, which usually corresponds to the molecular weight of the non-PEGylated protein plus about 2-fold the molecular weight of the PEG moiety[44-46].
|
| Fig. 3 Molecular weight and CD spectrum of PEG-Gln101-IFN-α2a. (A) MALDI-TOF spectra of PEG-Gln101-IFN-α2a. PEG-Gln101-IFN-α2a with the single charge peak (58 295 Da) and the double charge peak (29 260 Da). (B) Circular dichroism profiles of PEG-Gln101-IFN-α2a and IFN-α2a. |
| |
To develop products for human use, PEGylated protein drugs have to be subjected to secondary structure determination after conjugation. Spectrometric studies were carried out to evaluate the influence of the enzymatic PEGylation on the structural conformation of IFNα2a. Overlapped CD profiles were obtained with IFNα2a and PEG-Gln101-IFN α2a in the near and far UV. Native IFNα2a has a predominantly α-helical structure in physiological conditions with two characteristic, strong bands at 222 and 208 nm, as revealed by the far UV-CD spectrum shown in Fig. 3B. It was found that the two protein forms had similar molar ellipticities and the calculated distribution of the secondary structure elements were the same for both proteins. Taken together, the spectroscopy data suggest that both secondary and tertiary structure of IFNα2a was not influenced by PEGylation. These results are consistent with previous studies showing that the protein's secondary structure is not influenced by PEGylation.
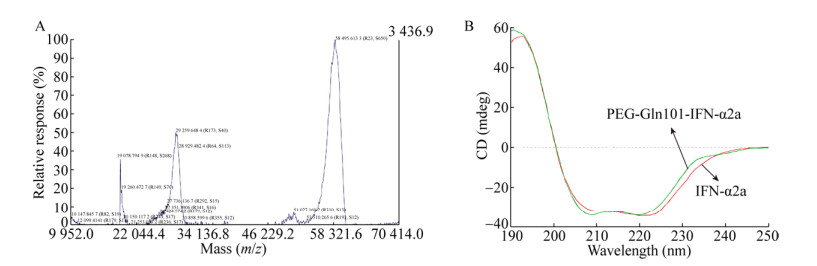
2.4 Identification of the sites of PEG-Gln101- IFN α2aThe computational study of sequence, structure and dynamic analysis of IFN α2a allowed for the identification of Gln101 as glutamine target for enzymatic PEGylation. The peptide mapping was concomitantly performed by IFN α2a and PEG-Gln101-IFN α2a digestion with endoproteinase trypsin and Glu-C, which takes advantage of the large number of glutamic acid residues in IFNα2a sequence as well as of specific cleavage on the COOH-terminal glutamic acids[47]. Fig. 4A shows that there is one obvious peak appeared in 48-49 min in PEG-Gln101-IFN α2a hydrolysate separation, which is not found in native IFNα2a hydrolysates. And the LC-MS/MS profile shows the same result (Fig. 4B). The peak was collected, and then lyophilized for subsequent N-terminal sequencing and Mw by MALDI-TOF. The results reported that the molecular weight mainly around 40 kDa (Fig. 4C), consistent with theoretical molecular weight peptides to PEG, and the N-terminal sequence is ACVIQGVGVTETPLMK-(Fig. 4D).
|
| Fig. 4 Identification of the Sites of PEG-IFN. (A) RP-HPLC chromatographic and (B) LC-MS/MS profiles of trypsin and Glu-C digests of PEG-Gln101-IFN-α2a and IFN-α2a. There is one obvious peak appeared in 48-49 min in PEG-IFN hydrolysate separation, which is not found in the IFN hydrolysates. (C) MALDI-TOF spectra of PEGylated peptide. (D) N-terminal sequence of PEGylated peptide detected by automated Protein Sequencer. |
| |
The peptide fragments were also identified by LC-MS/MS. Table 3 reports the amino acid sequence of each fragment obtained by IFNα2a digestion along with corresponding elution times and molecular masses. The data reported in Table 3 show that the peptide mapping of IFNα2a covers 92% of the primary protein sequence. The peptide mapping of PEG-Gln101-IFN α2a produced the same sequence pattern of IFNα2a except for the absence of the peptide: ACVIQGVGVTETPLMK- (Table 3), which putatively contains the PEGylation sites. In order to verify the conjugation site of IFNα2a, a LC-MS/MS investigation was undertaken by endoproteinase trypsin and Glu-C digestion. The resulting amino acid sequence ACVIQGVGVTETPLMK- (where the symbol means a missing signal) spans from Asp 97 to Arg 111 of IFNα2a primary sequence was found in IFNα2a, but not PEG-Gln101-IFN α2a, which support the Gln101 as site-specific target for enzymatic PEGylation catalyzed by mTG. Although the IFNα2a primary structure includes 12 glutamines, the results obtained in the present work show that a monoPEGylated IFNα2a derivative is obtained providing for direct experimental evidence that only one single glutamine residue, Gln101, is the target of enzymatic PEGylation.
| Sequence | Length | Charge | m/z | Retention time | Score | Intensity |
| ISLFSCLK | 8 | 2 | 484.267 7 | 35.893 | 50.040 | 97 537 000 |
| SFSLSTNLQESLR | 13 | 2 | 741.383 4 | 33.887 | 50.088 | 8 283 300 |
| ISLFSCLKDR | 10 | 2 | 619.831 7 | 32.748 | 55.314 | 26 022 000 |
| CDLPQTHSLGSR | 12 | 2 | 706.833 0 | 23.303 | 62.732 | 1.2E+08 |
| SFSLSTNLQE | 10 | 2 | 563.274 8 | 31.247 | 67.704 | 8.79E+08 |
| MIQQIFNLFSTK | 12 | 2 | 735.394 7 | 46.322 | 80.690 | 1.19E+08 |
| DSSAAWDETLLDKFYTE | 17 | 2 | 995.949 5 | 43.699 | 81.346 | 8.87E+08 |
| ACVIQGVGVTETPLMK | 16 | 2 | 851.947 0 | 34.407 | 83.540 | 7.38E+08 |
| RTLMLLAQMR | 10 | 2 | 616.851 9 | 34.074 | 101.380 | 1.52E+08 |
| EDSILAVR | 8 | 2 | 451.750 7 | 24.519 | 101.600 | 1.79E+09 |
| TLMLLAQMR | 9 | 2 | 538.801 4 | 37.188 | 109.710 | 7.09E+08 |
| FGNQFQK | 7 | 2 | 434.719 2 | 17.582 | 122.980 | 6.13E+09 |
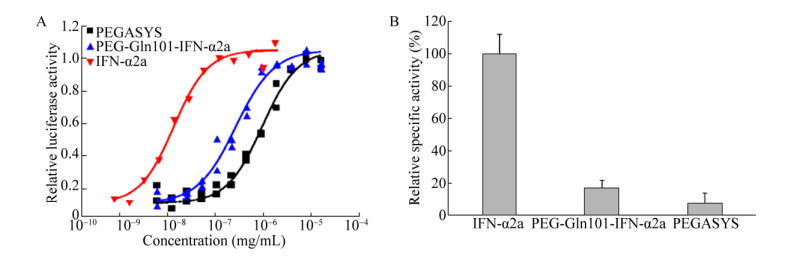
After binding to interferon-specific receptors on cell surface, interferon triggers the signaling cascade, resulting in translocation of interferon-stimulated gene factors (ISGFs) into the cell nucleus to interact with interferon stimulated response elements (ISRE). This activates the transcription of interferon- stimulated genes, producing antiviral, anti- proliferative and immunomodulatory effects[48-49]. By detecting ISRE-driven luciferase reporter gene expression, the biological activity of interferon was measured. Table 4 and Fig. 5 show the bioactivities of IFN-α2a and PEGylated IFN-α2a in vitro. The result from three independent experiments showed that the EC50 of IFN-α2a, PEG-Gln101-IFNα2a and PEGASYS were 2.07×108 U/mg, 3.74×107 U/mg and 1.63×107 U/mg, respectively. The activity of PEG-Gln101-IFNα2a and PEGASYS were decreased to 18% and 8% of IFN-α2a, respectively, which is ascribed to the hindering effect of the polymer on the protein surface that can partially mask the recognition site of the cytokine to impair its interaction with the cell surface receptor. Moreover, PEG-Gln101-IFNα2a was about 2.3-fold activity of PEGASYS, which underlines the relevance of the PEGylation site for the protein activity. The different anchoring group on the protein surface and the homogeneity may affect cytokine accessibility to cellular receptor. Therefore, PEG conjugation to Gln101, which is located in a flexible loop, seems to have lower interfering effect on the cytokine-cellular receptor interaction as compared to polymer conjugation to the terminal amino group.
| Sigmoidal dose-response (Variable slope) | PEG-Gln101-IFNα2a | PEGASYS | IFNα2a |
| Best-fit values | |||
| BOTTOM | 0 | 0 | 0 |
| TOP | 100 | 100 | 100 |
| LOGEC50 | -7.19 | -7.719 | -8.969 |
| HILLSLOPE | 1.223 | 1.128 | 1.275 |
| EC50 | 3.74E-07 | 1.63E-07 | 2.07E-08 |
| Std.Error | |||
| LOGEC50 | 0.028 16 | 0.036 91 | 0.052 19 |
| HILLSLOPE | 0.086 16 | 0.095 65 | 0.173 2 |
| 95%Confidence Intervals | |||
| LOGEC50 | -7.348 to -7.732 | -7.196 to -7.443 | -8.085 to -8.453 |
| HILLSLOPE | 1.044 to 1.401 | 0.929 8 to 1.327 | 0.889 2 to 1.661 |
| EC50 | 3.644 to 4.386e-007 | 1.399 to 1.875e-007 | 1.819 to 2.404e-008 |
| Goodness of Fit | |||
| Degrees of Freedom | 22 | 22 | 10 |
| R2 | 0.982 9 | 0.974 3 | 0.971 9 |
| Absolute Sum of Squares | 543.1 | 861.4 | 442.3 |
|
| Fig. 5 Bioactivity of IFN-α2a, PEG-Gln101-IFN-α2a and PEGASYS in vitro by RGA. The X-axis is presented as a logarithmic scale. Values are x±s (n=3). Statistical differences between groups were compared by the Student's t-test (P < 0.01). The activity was showed by specific activity, which is calculated from the sample concentration (mg/mL) and EC50 (U/mL) from the graphic analysis. Specific activity (U/mg)= EC50 (U/mL)/sample concentration (mg/mL). |
| |
Because hepatitis B virus can only infect human beings and chimpanzees, there is no good animal model to choose to directly reflect the antiviral activity of IFNs in vivo. Therefore, anti-tumor experiments were usually chose to detect the activity of IFNs in vivo. We detected the anti-tumor activities of IFNs in Nude mice xenograft models, as well as PEGASYS. And the result showed that PEG-Gln101-IFNα2a was more effective in anti-tumor effect compared with IFNα-2a, and equivalent to PEGASYS (data not show).
2.6 Pharmacokinetic studiesPharmacokinetic studies were carried out by s.c. injection of 8E+7 IU/kg of IFNα2a and 700 μg/kg of PEG-Gln101-IFN α2a in rats. For comparison, the pharmacokinetic profile of PEGASYS® was also investigated. The protein concentrations in blood at the experimental time points were assessed by RGA. As shown in Fig. 6 and Table 5, the native IFNα2a was rapidly absorbed and eliminated from the circulation. The Cmax (10 697 IU/mL) was achieved in 2 h, the plasma half-life (t1/2) was 3.4 h and the AUC was 35 415 h×IU/mL. As compared to the IFNα2a, PEG-Gln101-IFN α2a displayed delayed absorption, and long plasma half-life (26.5 h). And the results showed that the pharmacokinetic profile obtained with PEG-Gln101-IFN α2a was better than PEGASYS®. The prolonged permanence of PEG-Gln101-IFN α2a in the circulation resulted in higher AUC as compared to IFNα2a. The AUC of PEG-Gln101-IFN α2a and the AUC obtained with PEGASYS® was 21 917 880 h×IU/mL and 15 083 030 h×IU/mL, respectively. The pharmacokinetic data show that the polymer size and shape, which are known to affect the bioconjugate distribution through biomembranes and the glomerular ultrafiltration, are not the only parameters determining the bioconjugate fate in vivo. Indeed, PEG-Gln101-IFN α2a and PEGASYS® have substantially the same composition as both of them have been obtained by conjugation of 40 kDa branched PEG to IFNα2a. Therefore, the pharmacokinetic profile of PEGylated IFNα2a seems to be also affected by binding site of the protein and homogeneity of the bioconjugate. The PEGylation site, for example, may affect the proteolytic digestion of the cytokine as well as its interaction with IFN receptor that has been found to be involved in the cytokine clearance[8].

|
| Fig. 6 Pharmacokinetic profiles of IFN-α2a, PEG- Gln101-IFN-α2a and PEGASYS. The Y-axis is presented as a logarithmic scale. |
| |
| Sample | t1/2 (h) | Tmax (h) | Cmax (IU/mL) | AUC (h×IU/mL) | CL (h) |
| IFN-α2a | 3.36 | 2.00 | 10 697.0 | 35 415.0 | 228 |
| PEG-Gln101- IFN-α2a | 26.5 | 32.5 | 756 655.3 | 21 917 880.1 | 1.2E-05 |
| PEGASYS | 14.3 | 26.0 | 511 890.0 | 15 083 030.4 | 2.4E-05 |
The market approval of recombinant IFNs opened an exciting new era for the treatment of patients with hepatitis. Nevertheless, IFNs is rapidly cleared from blood circulation due to a combination of renal filtration and receptor mediate endocytosis. To overcome the limitations of repetitive injections, non-selective chemical PEGylated derivatives have been developed. The PEGylated IFN is marketed with the brand name of PEGASYS® and its clinical use was authorized by USA and European regulatory authorities in 2002 for the treatment of HBV and HCV. However, PEGASYS® is a mixture. There are about 8-10 different positional isomers in PEGASYS®, and these isomers have different bioactivity[15, 50-51]. These characteristics will lead to the corresponding problems that the quality is hard to control.
The results reported in this paper show that the Y shaped PEG40kDa-NH2 conjugation to IFNα2a at the presence of mTG enabled the production of a new IFNα2a derivative, which is site-specifically monoPEGylated on Gln101 residue. The optimization of the conjugation conditions resulted in high bioconjugation yield and prevented the protein degradation and denaturation. Furthermore, the clinical grade purity of the final PEG-Gln101- IFNα2a bulk preparation could be obtained by simple efficient ion exchange chromatography. According to the results obtained in the preclinical studies, PEG-Gln101-IFNα2a is a new chemical entity (NCE) fully comparable in terms of pharmacodynamic profile to PEGASYS®. Clinical trials will be conducted to evaluate the potential clinical differences between PEG-Gln101-IFNα2a and PEGASYS®, which are both PEGylated IFN-α2a, only differing in conjugation sites. To our knowledge, this is the first report that used an easy and mild method to obtain a site-specific PEGylated IFN α2a mediated by mTG, which has better bioactivity and PK profiles to PEGASYS.
| [1] |
Pavlou AK, Reichert JM. Recombinant protein therapeutics-success rates, market trends and values to 2010. Nat Biotechnol, 2004, 22(12): 1513-1519. DOI:10.1038/nbt1204-1513 |
| [2] |
Harris JM, Chess RB. Effect of pegylation on pharmaceuticals. Nat Rev Drug Discov, 2003, 2(3): 214-221. DOI:10.1038/nrd1033 |
| [3] |
Malik DK, Baboota S, Ahuja A, et al. Recent advances in protein and peptide drug delivery systems. Curr Drug Deliv, 2007, 4(2): 141-151. DOI:10.2174/156720107780362339 |
| [4] |
Harris JM, Martin NE, Modi M. Pegylation: a novel process for modifying pharmacokinetics. Clin Pharmacokinet, 2001, 40(7): 539-551. DOI:10.2165/00003088-200140070-00005 |
| [5] |
Greenwald RB, Choe YH, Mcguire J, et al. Effective drug delivery by pegylated drug conjugates. Adv Drug Deliv Rev, 2003, 55(2): 217-250. DOI:10.1016/S0169-409X(02)00180-1 |
| [6] |
Veronese FM, Pasut G. Pegylation: posttranslational bioengineering of protein biotherapeutics. Drug Discov Today Technol, 2008, 5(2/3): e57-e64. |
| [7] |
Pasut G, Guiotto A, Veronese FM. Protein, peptide and non-peptide drug pegylation for therapeutic application. Exp Opin Therapeut Pat, 2004, 14(6): 859-894. DOI:10.1517/13543776.14.6.859 |
| [8] |
Veronese FM, Pasut G. Pegylation, successful approach to drug delivery. Drug Discov Today, 2005, 10(21): 1451-1458. DOI:10.1016/S1359-6446(05)03575-0 |
| [9] |
Roberts MJ, Bentley MD, Harris JM. Chemistry for peptide and protein pegylation. Adv Drug Deliv Rev, 2012, 64(S1): 116-127. |
| [10] |
Wattendorf U, Merkle HP. Pegylation as a tool for the biomedical engineering of surface modified microparticles. J Pharm Sci, 2008, 97(11): 4655-4669. DOI:10.1002/jps.21350 |
| [11] |
Abuchowski A, van Es T, Palczuk NC, et al. Alteration of immunological properties of bovine serum albumin by covalent attachment of polyethylene glycol. J Biol Chem, 1977, 252(11): 3578-3581. |
| [12] |
Abuchowski A, McCoy JR, Palczuk NC, et al. Effect of covalent attachment of polyethylene glycol on immunogenicity and circulating life of bovine liver catalase. J Biol Chem, 1977, 252(11): 3582-3586. |
| [13] |
Fee CJ, Alstine JMV. Peg-proteins: reaction engineering and separation issues. Chem Eng Sci, 2006, 61(3): 924-939. DOI:10.1016/j.ces.2005.04.040 |
| [14] |
Wang YS, Youngster S, Grace M, et al. Structural and biological characterization of pegylated recombinant interferon alpha-2b and its therapeutic implications. Adv Drug Deliv Rev, 2002, 54(4): 547-570. DOI:10.1016/S0169-409X(02)00027-3 |
| [15] |
Monkarsh SP, Ma YM, Aglione A, et al. Positional isomers of monopegylated interferon α-2a: isolation, characterization, and biological activity. Anal Biochem, 1997, 247(2): 434-440. |
| [16] |
Kinstler O, Molineux G, Treuheit M, et al. Mono-n-terminal poly(ethylene glycol)-protein conjugates. Adv Drug Deliv Rev, 2002, 54(4): 477-485. DOI:10.1016/S0169-409X(02)00023-6 |
| [17] |
Chi YS, Zhang HB, Huang WL, et al. Microwave-assisted solid phase synthesis, pegylation, and biological activity studies of glucagon-like peptide-1(7-36) amide. Bioorg Med Chem, 2008, 16(16): 7607-7614. DOI:10.1016/j.bmc.2008.07.019 |
| [18] |
Sato H. Enzymatic procedure for site-specific pegylation of proteins. Adv Drug Deliv Rev, 2002, 54(4): 487-504. DOI:10.1016/S0169-409X(02)00024-8 |
| [19] |
Fontana A, Spolaore B, Mero A, et al. Site-specific modification and pegylation of pharmaceutical proteins mediated by transglutaminase. Adv Drug Deliv Rev, 2008, 60(1): 13-28. DOI:10.1016/j.addr.2007.06.015 |
| [20] |
Mero A, Schiavon M, Veronese FM, et al. A new method to increase selectivity of transglutaminase mediated pegylation of salmon calcitonin and human growth hormone. J Controlled Release, 2011, 154(1): 27-34. DOI:10.1016/j.jconrel.2011.04.024 |
| [21] |
Kashiwagi T, Yokoyama K, Ishikawa K, et al. Crystal structure of microbial transglutaminase from Streptoverticillium mobaraense. J Biol Chem, 2002, 277(46): 44252-44260. DOI:10.1074/jbc.M203933200 |
| [22] |
Kanaji T, Ozaki H, Takao T, et al. Primary structure of microbial transglutaminase from Streptoverticillium sp. strain s-8112. J Biol Chem, 1993, 268(16): 11565-11572. |
| [23] |
Yokoyama K, Nio N, Kikuchi Y. Properties and applications of microbial transglutaminase. Appl Microbiol Biotechnol, 2004, 64(4): 447-454. DOI:10.1007/s00253-003-1539-5 |
| [24] |
Zhu Y, Rinzema A, Tramper J, et al. Microbial transglutaminase-a review of its production and application in food processing. Appl Microbiol Biotechnol, 1995, 44(3/4): 277-282. |
| [25] |
Deweid L, Avrutina O, Kolmar H. Microbial transglutaminase for biotechnological and biomedical engineering. Biol Chem, 2019, 400(3): 257-274. DOI:10.1515/hsz-2018-0335 |
| [26] |
Zhou JQ, He T, Wang JW. Pegylation of cytochrome c at the level of lysine residues mediated by a microbial transglutaminase. Biotechnol Lett, 2016, 38(7): 1121-1129. DOI:10.1007/s10529-016-2083-6 |
| [27] |
Khameneh B, Jaafari MR, Hassanzadeh-Khayyat M, et al. Preparation, characterization and molecular modeling of pegylated human growth hormone with agonist activity. Int J Biol Macromol, 2015, 80: 400-409. DOI:10.1016/j.ijbiomac.2015.06.037 |
| [28] |
da Silva Freitas D, Mero A, Pasut G. Chemical and enzymatic site specific pegylation of hGH. Bioconjug Chem, 2013, 24(3): 456-463. |
| [29] |
Scaramuzza S, Tonon G, Olianas A, et al. A new site-specific monopegylated filgrastim derivative prepared by enzymatic conjugation: production and physicochemical characterization. J Controlled Release, 2012, 164(3): 355-363. DOI:10.1016/j.jconrel.2012.06.026 |
| [30] |
Zhao X, Shaw AC, Wang JH, et al. A novel high-throughput screening method for microbial transglutaminases with high specificity toward Gln141 of human growth hormone. J Biomol Scr, 2010, 15(2): 206-212. DOI:10.1177/1087057109356206 |
| [31] |
Dennler P, Chiotellis A, Fischer E, et al. Transglutaminase-based chemo-enzymatic conjugation approach yields homogeneous antibody-drug conjugates. Bioconjug Chem, 2014, 25(3): 569-578. DOI:10.1021/bc400574z |
| [32] |
Anami Y, Xiong W, Gui X, et al. Enzymatic conjugation using branched linkers for constructing homogeneous antibody-drug conjugates with high potency. Organic Biomol Chem, 2017, 15(26): 5635-5642. DOI:10.1039/C7OB01027C |
| [33] |
Ohtsuka T, Ota M, Nio N, et al. Comparison of substrate specificities of transglutaminases using synthetic peptides as acyl donors. Biosci Biotechnol Biochem, 2000, 64(12): 2608-2613. DOI:10.1271/bbb.64.2608 |
| [34] |
Shimba N, Yokoyama KI, Suzuki E. NMR-based screening method for transglutaminases: rapid analysis of their substrate specificities and reaction rates. J Agric Food Chem, 2002, 50(6): 1330-1334. DOI:10.1021/jf010995k |
| [35] |
Sugimura Y, Hosono M, Wada F, et al. Screening for the preferred substrate sequence of transglutaminase using a phage-displayed peptide library: identification of peptide substrates for tgase 2 and factor xⅢa. J Biol Chem, 2006, 281(26): 17699-17706. DOI:10.1074/jbc.M513538200 |
| [36] |
Coussons PJ, Price NC, Kelly SM, et al. Factors that govern the specificity of transglutaminase-catalysed modification of proteins and peptides. Biochem J, 1992, 282: 929-930. DOI:10.1042/bj2820929 |
| [37] |
Maullu C, Raimondo D, Caboi F, et al. Site-directed enzymatic pegylation of the human granulocyte colony-stimulating factor. FEBS J, 2009, 276(22): 6741-6750. DOI:10.1111/j.1742-4658.2009.07387.x |
| [38] |
Spolaore B, Raboni S, Satwekar AA, et al. Site-specific transglutaminase-mediated conjugation of interferon α-2b at glutamine or lysine residues. Bioconjug Chem, 2016, 27(11): 2695-2706. DOI:10.1021/acs.bioconjchem.6b00468 |
| [39] |
Klaus W, Gsel B, Labhardt AM, et al. The three-dimensional high resolution structure of human interferon α-2a determined by heteronuclear NMR spectroscopy in solution. J Mol Biol, 1997, 274(4): 661-675. DOI:10.1006/jmbi.1997.1396 |
| [40] |
Gautier R, Camproux AC, Tufféry P. Scit: web tools for protein side chain conformation analysis. Nucleic Acids Res, 2004, 32(S2): W508-W511. |
| [41] |
Folk JE, Cole PW. Transglutaminase: Mechanistic features of the active site as determined by kinetic and inhibitor studies. Biochim Biophys Acta (BBA)-Enzymol Biol Oxidat, 1966, 122(2): 244-264. |
| [42] |
Petersen B, Petersen TN, Andersen P, et al. A generic method for assignment of reliability scores applied to solvent accessibility predictions. BMC Struct Biol, 2009, 9: 51. DOI:10.1186/1472-6807-9-51 |
| [43] |
Taguchi S, Nishihama K, Igi K, et al. Substrate specificity analysis of microbial transglutaminase using proteinaceous protease inhibitors as natural model substrates. J Biochem, 2000, 128(3): 415-425. DOI:10.1093/oxfordjournals.jbchem.a022769 |
| [44] |
Ohtsuka T, Sawa A, Kawabata R, et al. Substrate specificities of microbial transglutaminase for primary amines. J Agric Food Chem, 2000, 48(12): 6230-6233. DOI:10.1021/jf000302k |
| [45] |
Caliceti P, Veronese FM. Pharmacokinetic and biodistribution properties of poly(ethylene glycol)- protein conjugates. Adv Drug Deliv Rev, 2003, 55(10): 1261-1277. DOI:10.1016/S0169-409X(03)00108-X |
| [46] |
Veronese FM. Peptide and protein pegylation: a review of problems and solutions. Biomaterials, 2001, 22(5): 405-417. DOI:10.1016/S0142-9612(00)00193-9 |
| [47] |
Houmard J, Drapeau GR. Staphylococcal protease: a proteolytic enzyme specific for glutamoyl bonds. Proc Natl Acad Sci USA, 1972, 69(12): 3506-3509. DOI:10.1073/pnas.69.12.3506 |
| [48] |
Williams BR. Signal transduction and transcriptional regulation of interferon-alpha-stimulated genes. J Interferon Res, 1991, 11(4): 207-213. DOI:10.1089/jir.1991.11.207 |
| [49] |
Larocque L, Bliu A, Xu RR, et al. Bioactivity determination of native and variant forms of therapeutic interferons. J Biomed Biotechnol, 2015, 2011: 174615. |
| [50] |
Foser S, Schacher A, Weyer KA, et al. Isolation, structural characterization, and antiviral activity of positional isomers of monopegylated interferon α-2a (PEGASYS). Protein Expr Purif, 2003, 30(1): 78-87. DOI:10.1016/S1046-5928(03)00055-X |
| [51] |
Grace M, Youngster S, Gitlin G, et al. Structural and biologic characterization of pegylated recombinant IFN-α2b. J Interf Cytok Res, 2001, 21(12): 1103-1115. DOI:10.1089/107999001317205240 |
 2020, Vol. 36
2020, Vol. 36




