中国科学院微生物研究所、中国微生物学会主办
文章信息
- 王凯婕, 安文静, 刘亚菲, 刘迪, 冯连杰, 王俊杰, 黄俊骏, 刘肖飞, 梁卫红
- Wang Kaijie, An Wenjing, Liu Yafei, Liu Di, Feng Lianjie, Wang Junjie, Huang Junjun, Liu Xiaofei, Liang Weihong
- CRISPR/Cas9技术编辑OsRhoGDI2基因导致水稻半矮化
- Disruption of OsRhoGDI2 by CRISPR/Cas9 technology leads to semi-dwarf in rice
- 生物工程学报, 2020, 36(4): 707-715
- Chinese Journal of Biotechnology, 2020, 36(4): 707-715
- 10.13345/j.cjb.190333
-
文章历史
- Received: July 24, 2019
- Accepted: September 11, 2019
- Published: October 10, 2019
CRISPR/Cas9技术是近些年发展起来的一种简捷高效的基因编辑技术[1]。该技术通过sgRNA与基因组靶序列结合,引导Cas9内切酶对靶序列进行切割产生DSB,在DSB修复过程中可能产生碱基的插入、缺失等[2],从而实现基因的编辑。近年来,CRISPR/Cas9技术因其操作简单、成本低、效率高,迅速发展并广泛应用于动物、植物、微生物等研究中[3-6]。例如高彩霞课题组率先利用该技术实现了水稻基因组编辑[7];邵高能等利用CRISPR/Cas9技术成功编辑了水稻Badh2香味基因,加速香稻品种的选育进程[8];朱健康课题组相关研究表明,CRISPR/Cas9系统可高效编辑水稻特异基因,并且基因突变能够稳定遗传[9]。
植物Rho/Rop蛋白,属于小G蛋白家族,在信号转导中起“分子开关”的作用。已明确,植物RhoGDIs是一类与Rho/Rop相互作用的互作蛋白。RhoGDIs已知的功能涉及调节根毛[10]、花粉管生长、维持花粉管细胞稳态[11]、幼苗发育以及叶片细胞形态发生[12]等过程。OsRhoGDI2是水稻Rho蛋白OsRacD互作蛋白的编码基因[13]。OsRhoGDI2启动子功能鉴定以及表达模式分析发现,OsRhoGDI2基因在叶鞘与节中高效表达[14];OsRhoGDI2蛋白亚细胞定位分析表明,该蛋白主要分布在细胞膜、细胞质与细胞核[15]。目前,OsRhoGDI2基因的功能尚不明确。为此,本研究利用CRISPR/Cas9技术对水稻OsRhoGDI2进行编辑,旨在筛选纯合敲除植株,通过表型分析,确定该基因可能涉及的生物学过程,为后续以敲除水稻为材料开展OsRhoGDI2的功能研究奠定基础。
1 材料与方法 1.1 植物材料本研究所用实验材料为粳稻栽培品种日本晴(Oryza sativa L. Japonica cv. Nipponbare)。
1.2 菌株与载体大肠杆菌Escherichia coli DH5α与根癌农杆菌Agrobacterium tumefaciens EHA105由本实验室保存,载体pw-A-cas9与pw-B由武汉伯远公司提供。
1.3 主要试剂及试剂盒限制性内切酶、T4 DNA连接酶、质粒提取试剂盒、胶回收试剂盒均购自大连TaKaRa公司。卡那霉素、潮霉素、酵母浸膏等试剂均购自北京鼎国生物技术有限公司。
1.4 引物本文所用引物均采用Primer Premier 5软件设计,委托英潍捷基(上海)贸易有限公司合成(表 1)。
| Primer name | Primer sequence (5′–3′) | Usage |
| P1 | CAGTGGTCTCAGGCACGATGACGAGGAAGATGAG | Synthesis of sgRNA1 |
| P2 | CAGTGGTCTCAAAACCTCATCTTCCTCGTCATCG | |
| P3 | CAGTGGTCTCAGGCACCCCTCCGTGAGCTTCTCG | Synthesis of sgRNA2 |
| P4 | CAGTGGTCTCAAAACCGAGAAGCTCACGGAGGGG | |
| P5 | ACGGTGTCGTCCATCACAGTTTGCC | Amplification of Hyg gene |
| P6 | TTCCGGAAGTGCTTGACATTGGGGA | |
| P7 | CCACCTATTCTTCTCGCACATTC | Amplification of OsRhoGDI2 editing target region |
| P8 | AGGCTCTCATCCTCCTTCCAAGT | |
| off-1-F | CCTATGCCCTGGACCTGAAT | Amplification of off-target |
| off-1-R | GGGACCAAGAAAAGGAAAGAGT | |
| off-2-F | CATTTGAGCCTGCCTATCTG | |
| off-2-R | CCTATCCCGACCTTAAACCAA | |
| off-3-F | GCAAACAAACAACCTAACTGACCG | |
| off-3-R | CAAAGCCCAGCCGAATAAAC | |
| off-4-F | AATCTGAACGCCCAAATGC | |
| off-4-R | GGATGAACTGAACAGGTACAAGC | |
| off-5-F | GGCTTCCCCTGGACTGTAAA | |
| off-5-R | ATCTCGCTTCCCTGATGACC | |
| Note:BsaⅠ restriction site is underlined. | ||
将OsRhoGDI2 (AY364312)提交至NCBI (https://www.ncbi.nlm.nih.gov/)进行检索,找到全基因序列,获得外显子信息。根据gRNA设计原则,在1号外显子上设计2个gRNA靶位点,并将其与水稻基因组blast比对分析验证,确认靶序列的特异性。
1.6 中间载体的构建通过P1/P2引物退火,制备gRNA1片段,反应体系如下:P1/P2各5 µL,H2O 40 µL;反应条件如下:95 ℃变性10 min、55 ℃退火10 min。gRNA2片段通过P3/P4引物退火制备,体系同上。
将制备的2个gRNA片段分别与载体pw-A-cas9和pw-B连接,酶切连接体系如下:gRNA片段2 µL,载体1.5 µL,BsaⅠ 0.5 µL,T4 DNA连接酶0.5 µL,T4缓冲液1 µL,H2O 4.5 µL,混匀后于37 ℃反应2 h。
以热激法分别将上述2个连接产物转化DH5α感受态细胞,经阳性转化子的筛选、检测,将测序验证后的重组中间载体分别命名为GDI2-A与GDI2-B。
1.7 植物表达载体pW-T-OsRhoGDI2的构建取适量的GDI2-A与GDI2-B质粒,按照以下体系进行酶切连接反应。体系如下:GID2-A 1 µL,GID2-B 1.5 µL,LguⅠ 0.5µL,T4 DNA连接酶0.5 µL,T4缓冲液1 µL,加ddH2O至10 µL,混匀后于37 ℃反应2 h。以热激法进行连接产物的转化,进行阳性转化子的筛选、检测并测序,确认后的载体命名为pW-T-OsRhoGDI2。
1.8 水稻的遗传转化及筛选以农杆菌介导法将pW-T-OsRhoGDI2载体转化日本晴水稻愈伤组织,常规方法筛选。待再生水稻(T0代)长至四叶期时,提取叶片基因组DNA,以P5/P6引物组合进行基因组DNA的PCR检测,扩增体系为10 µL:2×Es Taq Master Mix 5 µL,P5/P6各0.4 µL,DNA 0.4 µL,ddH2O 3.8 µL;扩增条件为:94 ℃预变性2 min;94 ℃ 30 s,57 ℃ 30 s,72 ℃ 20 s,进行34个循环;72 ℃终延伸2 min,1%琼脂糖凝胶电泳检测扩增结果。采用相同体系,以P7/P8引物组合扩增包含编辑靶点序列,并对PCR产物胶回收测序,测序结果采用Primer Premier、DNAMAN与Chromas软件分析。
转基因水稻T1代筛选方法同上。
1.9 OsRhoGDI2编辑水稻的表型分析选择3株T1代纯合的OsRhoGDI2编辑水稻植株,观察其与野生型水稻日本晴的差异。待生长至成熟期时,测量其株高、穗与茎节的长度,获得的数据用Microsoft Excel 2018进行统计学分析,并用DPS软件进行数据差异性分析,用Origin85软件作图。
2 结果与分析 2.1 OsRhoGDI2编辑靶位点的选择及载体构建OsRhoGDI2序列检索显示,该基因由5个外显子和4个内含子构成。根据编辑靶点设计原则,在1号外显子选择两个靶位点(图 1)。其中,靶位点1 (Target 1)位于1号外显子第230−252 bp处;靶位点2 (Target 2)位于1号外显子第153−175 bp处的互补链上,且两个靶位点相隔54 bp。
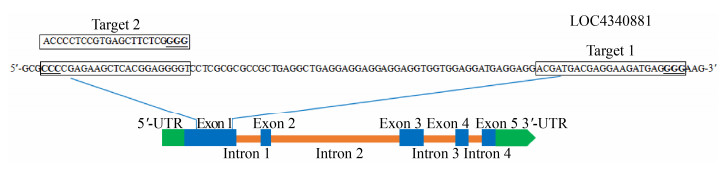
|
| 图 1 OsRhoGDI2基因结构及编辑靶点的位置 Fig. 1 Schematic diagram of OsRhoGDI2 gene structure and the sites of target sequences. PAM sequences are underlined. |
| |
通过引物退火制备gRNA1与gRNA2,它们分别与载体pw-A-cas9和pw-B连接,制备中间载体GDI2-A与GDI2-B。通过2个中间载体酶切连接,最终构建植物表达载体pW-T-OsRhoGDI2(图 2),该载体中的Hyg基因和Cas9基因均由CaMV35S启动子驱动,gRNA1与gRNA2均由OsU3启动子驱动,以NOS终止子终止。
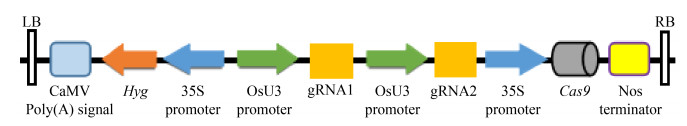
|
| 图 2 pW-T-OsRhoGDI2表达载体的构建 Fig. 2 Construction of pW-T-OsRhoGDI2 vector. LB: vector left border; RB: vector right border; Hyg: the resistance gene of hygromycin; OsU3 promoter: rice U3 promoter; 35S promoter: Cauliflower mosaic virus (CaMV) 35S promoter; Nos terminator: Agrobacterium tumefaciens nopaline synthase gene (Nos) terminator; gRNA1 and gRNA2: guide RNA; Cas9: the gene of Cas9. |
| |
经农杆菌介导的水稻愈伤转化及筛选,在T0代共获得11株独立的再生植株。潮霉素基因引物扩增结果显示,11株均为转基因水稻。编辑靶点序列的扩增结果显示,均扩增出与预期598 bp大小相符的目的片段(图 3)。

|
| 图 3 编辑靶点序列扩增产物的琼脂糖凝胶电泳 Fig. 3 Agarose gel electrophoresis of the editing target sequences amplification products. M: DL2000 marker; 1–11: T0 transgenic lines. |
| |
将上述PCR产物测序,并进行序列分析。结果显示(表 2),T0代获得杂合株系6个(占54.5%)、双等位突变株系2个(占18.2%)、纯合株系2个(占18.2%)、未被编辑株系1个(占9.1%)。T0代11个株系与野生型进行编辑靶点序列比对,发现6个杂合株系中有2个(#1、#5)在靶位点2处发生突变,4个(#2、#3、#8、#11)在靶位点1处发生突变;2个双等位突变株系(#7、#9)均在靶位点1处发生突变;2个纯合株系(#6、#10)均在靶位点1处发生突变(图 4)。
| Mutation type | T0 generation (n=11) | T1 generation (n=153) (from T0 Heterozygous and Bi-allielic lines) |
T1 generation (n=24) (from T0 Homozygous lines) |
| Heterozygous mutation | 6 (54.5%) | 65 (42.5%) | 0 |
| Bi-allielic mutation | 2 (18.2%) | 6 (3.9%) | 0 |
| Homozygous mutation | 2 (18.2%) | 45 (29.4%) | 24 (100%) |
| No mutation | 1 (9.1%) | 37 (24.2%) | 0 |

|
| 图 4 T0代与T1代转基因植株编辑靶点序列比对 Fig. 4 Comparison of the editing target sequences between T0 and T1 transgenic plants. (A) Comparison of the editing target sequences between T0 transgenic lines and WT. (B) Comparison of the editing target sequences between T1 homozygous transgenic lines and WT. The targeted sequences are highlighted with underline and PAM sequences in shadow. The deleted sequences are shown by black hyphens, and red fonts are mutated bases. m: mutation; d: deletion; WT: wild type; #1 to #11: T0 transgenic lines; #1-1 to #11-1: T1 homozygous transgenic lines. |
| |
对153株(来自T0代的杂合与双等位突变种子#1、#2、#3、#5、#7、#8、#9、#11) T1代水稻筛选,结果显示(表 2)有65个杂合株系(占42.5%)、6个双等位突变株系(占3.9%)、45个纯合株系(占29.4%)、37个未被编辑株系(占24.2%)。T1代纯合株系与野生型进行编辑靶点序列比对,发现靶位点2处发生突变的有1种(#5-1);靶位点1处发生突变的有6种(#5-2与#9-1、#7-1、#7-2、#8-1、#8-2、#11-1);靶位点1与靶位点2均发生突变的有1种(#1-1) (图 4)。
对24株(来自T0代的纯合突变种子#10) T1代水稻筛选,结果显示,24株(#10-1)均为靶位点1处缺失4个碱基“AGAT”,突变位点与T0代#10一致。序列分析证明,T1代产生移码突变的株系共有6种(#1-1、#5-1、#5-2与#9-1与#10-1、#8-1、#8-2、#11-1),另外,在对编辑靶点扩增和测序的分析中发现,后代没有出现大片段删除事件。
选取3个产生移码突变的T1代纯合株系与野生型进行预期OsRhoGDI2蛋白的氨基酸序列比对(图 5)。由于3个纯合敲除株系靶位点发生碱基缺失,导致该基因产生移码突变与无义突变,翻译提前终止,生成的截短蛋白中保守的RhoGDI结构域丢失,因此转基因水稻中将无法产生正常的OsRhoGDI2蛋白。

|
| 图 5 野生型与T1代纯合敲除株系内源OsRhoGDI2氨基酸序列的对比 Fig. 5 Comparison of amino acid sequences of endogenous OsRhoGDI2 between wild type and T1 homozygous knockout lines. The conserved domain of RhoGDI is underlined. WT: wild type; #1-1, #5-1, #5-2: T1 homozygous transgenic lines. |
| |
为评估2个靶位点的脱靶效应,本研究选取T1代的3株纯合突变体(#1-1、#5-1、#5-2)进行评估。对5个潜在的脱靶位点进行PCR扩增,测序比对发现,所有被检测植株的潜在脱靶位点均未出现突变(表 3)。
| Putative off-target | Putative off-target locus | Sequence of putative off-target site | Mismatching bases No. | Detected plant No. | Mutation plant No. |
| Target1-off-1 | Chr1:15308-15330 | GCGAGCACGAGGAAGATGAGGGG | 3 | 3 | 0 |
| Target1-off-2 | Chr11:114855-114877 | AACGGATCGAGGAAGATGAGGGG | 6 | 3 | 0 |
| Target1-off-3 | Chr12:45131-45109 | GAAGGGGCGAGGAAGATGAGGGG | 6 | 3 | 0 |
| Target2-off-4 | Chr2:87871-87893 | CCATCTCCTTGAGCTTCTCGGGG | 4 | 3 | 0 |
| Target2-off-5 | Chr8:47065-47087 | CCCTCTCCGCGAGCTCCTCGGGG | 4 | 3 | 0 |
| Note:Lmismatching bases are in red fonts; PAM sequences are underlined. | |||||
对大田种植的OsRhoGDI2敲除水稻和野生型水稻进行观察。在成熟期,3个敲除水稻株系株高相比野生型明显降低,统计学分析显示,3个敲除株系株高平均下降9.3%,且差异达到极显著水平(图 6A, C)。对成熟期水稻茎节与穗长度进行测量,结果显示,3个敲除株系第Ⅱ茎节与第Ⅲ茎节长度相比野生型平均降低19.0%与27.0%,在统计学上,差异均达到极显著水平(图 6B, 6D)。但是3个敲除株系的穗长与野生型相比无统计学上的差异。
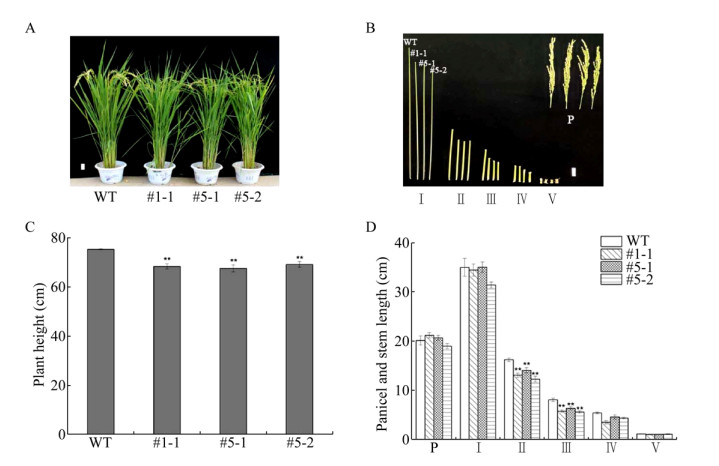
|
| 图 6 野生型与T1代转基因株系表型比较 Fig. 6 Phenotypes of wild type and T1 homozygous transgenic lines. (A) Morphologies in the mature stage. (B) Morphologies of panicles and internodes. There are WT, #1-1, #5-1 and #5-2 lines from left to right. (C) Height of plants in the mature stage. (D) Length of panicles and the first to fourth internodes. Bar=5 cm; * P < 0.05, ** P < 0.01, n=10; Ⅰ: first internode; Ⅱ: second internode; Ⅲ: third internode; Ⅳ: forth internode; Ⅴ: fifth internode; P: panicle; #1-1, #5-1, #5-2: T1 homozygous transgenic lines; WT: wild type. |
| |
有报道显示,利用CRISPR/Cas9技术编辑水稻,当碱基插入或缺失引起移码突变或蛋白结构改变时,靶基因的功能就会被破坏[16-17]。本研究利用双靶点CRISPR/Cas9表达载体对水稻OsRhoGDI2基因进行定点编辑,实现了OsRhoGDI2基因的敲除。
有研究显示,利用CRISPR/Cas9技术编辑水稻产生的突变可以稳定遗传到后代植株[18-19]。本研究在T0代获得2种纯合敲除水稻,分别为缺失3个碱基株系(#6)与4个碱基株系(#10)。由于缺失3或3倍数的碱基突变不会造成移码突变,株系#6是无效的,因此需要在T1代继续进行纯合株系的筛选,获得足够的突变材料。对T1代水稻(24株来自T0代纯合种子#10,153株来自T0代杂合突变与双等位突变种子#1、#2、#3、#5、#7、#8、#9、#11)继续筛选,共获得8种纯合敲除水稻。对T1代纯合敲除水稻编辑靶点的分析发现,在T0代纯合的#10株系其后代均保持与T0代相同的突变类型,说明T0代获得的纯合株系在T1代可以稳定遗传,也说明采用CRISPR/Cas9技术在T0代便有可能筛选到纯合敲除水稻,这与王美娜等[20]的研究结果一致,证明CRISPR/Cas9技术具有高效性与稳定性。
利用T1代纯合敲除株系进行初步的表型分析,发现最显著的差异体现在株高这一性状上。株高是直接影响水稻品种抗倒性能与产量的重要农艺学性状[21],研究株高对于更好地了解植物的生长发育规律以及植物分子育种具有重要意义[22]。但是,株高的调控是一个复杂的过程。Liang等将达到正常植株高度50%与50%−100%的植株分别定义为矮化和半矮化植株[23],本研究所分析的3种OsRhoGDI2基因T1代纯合敲除水稻株系相比野生型,株高均处于野生型株高50%−100%之间,属于半矮化类型。半矮化植株在第一次“绿色革命”时被用来培育抗倒伏作物新品种,从而大幅度提高了粮食产量[24]。已知水稻节间数目与节间长度影响株高[25]。Takahashi与Takeda等根据水稻各茎节的长短,将水稻矮化突变体分为dn、dm、sh、d6与nl 5种类型,本文获得的突变体均出现第Ⅱ和第Ⅲ茎节显著缩短的现象,属于dm类。目前,植物RhoGDI与株高相关研究尚未见报道,而OsRhoGDI2敲除水稻的株高相比野生型均有极显著降低,提示OsRhoGDI2基因与株高控制关系密切,值得深入研究其作用机制。
| [1] |
Arazoe T, Miyoshi K, Yamato T, et al. Tailor-made CRISPR/Cas system for highly efficient targeted gene replacement in the rice blast fungus. Biotechnol Bioeng, 2015, 112(12): 2543-2549. |
| [2] |
Shen B, Zhang J, Wu HY, et al. Generation of gene-modified mice via Cas9/RNA-mediated gene targeting. Cell Res, 2013, 23(5): 720-723. |
| [3] |
Wang T, Wei JJ, Sabatini DM, et al. Genetic screens in human cells using the CRISPR-Cas9 system. Science, 2014, 343(6166): 80-84. |
| [4] |
Wang Y, Geng LZ, Yuan ML, et al. Deletion of a target gene in Indica rice via CRISPR/Cas9. Plant Cell Rep, 2017, 36(8): 1333-1343. |
| [5] |
Zong Y, Wang YP, Li C, et al. Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion. Nat Biotechnol, 2017, 35(5): 438-440. |
| [6] |
Jakočiūnas T, Bonde I, Herrgård M, et al. Multiplex metabolic pathway engineering using CRISPR/Cas9 in Saccharomyces cerevisiae. Metab Eng, 2015, 28: 213-222. |
| [7] |
Shan QW, Wang YP, Li J, et al. Genome editing in rice and wheat using the CRISPR/Cas system. Nat Protoc, 2014, 9(10): 2395-2410. |
| [8] |
Shao GN, Xie LH, Jiao GA, et al. CRISPR/CAS9-mediated editing of the fragrant gene Badh2 in rice. Chin J Rice Sci, 2017, 31(2): 216-222 (in Chinese). 邵高能, 谢黎虹, 焦桂爱, 等. 利用CRISPR/CAS9技术编辑水稻香味基因Badh2. 中国水稻科学, 2017, 31(2): 216-222. |
| [9] |
Zhang H, Zhang JS, Wei PL, et al. The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol J, 2014, 12(6): 797-807. |
| [10] |
Carol RJ, Takeda S, Linstead P, et al. A RhoGDP dissociation inhibitor spatially regulates growth in root hair cells. Nature, 2005, 438(7070): 1013-1016. |
| [11] |
Feng QN, Kang H, Song SJ, et al. Arabidopsis RhoGDIs are critical for cellular homeostasis of pollen tubes. Plant Physiol, 2016, 170(2): 841-856. |
| [12] |
Wu YX, Zhao SJ, Tian H, et al. CPK3-phosphorylated RhoGDI1 is essential in the development of Arabidopsis seedlings and leaf epidermal cells. J Exp Bot, 2013, 64(11): 3327-3338. |
| [13] |
Liang WH, Tang CR, Wu NH. Isolation and characterization of two GDP dissociation inhibitor genes from Oryza sativa L. Chin J Biochem Mol Biol, 2004, 20(6): 785-791 (in Chinese). 梁卫红, 唐朝荣, 吴乃虎. 两种水稻GDP解离抑制蛋白基因的分离及特征分析. 中国生物化学与分子生物学报, 2004, 20(6): 785-791. |
| [14] |
Huang JJ, Zhang J, Hao YF, et al. Distinct expression patterns of the GDP dissociation inhibitor protein gene (OsRhoGDI2) from Oryza sativa during development and abiotic stresses. Biologia, 2016, 71(11): 1230-1239. |
| [15] |
Peng WF, Liang WH. Bioinformatics analysis and subcellular localization study on rice OsRhoGDI2 gene. Biotechnol Bull, 2010(5): 82-86 (in Chinese). 彭威风, 梁卫红. 水稻OsRhoGDI2蛋白生物信息学分析及亚细胞定位研究. 生物技术通报, 2010(5): 82-86. |
| [16] |
Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems. Science, 2013, 339(6121): 819-823. |
| [17] |
Zhou HB, Liu B, Weeks DP, et al. Large chromosomal deletions and heritable small genetic changes induced by CRISPR/Cas9 in rice. Nucleic Acids Res, 2014, 42(17): 10903-10914. |
| [18] |
Feng ZY, Mao YF, Xu NF, et al. Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc Natl Acad Sci USA, 2014, 111(12): 4632-4637. |
| [19] |
Minkenberg B, Xie KB, Yang YN. Discovery of rice essential genes by characterizing a CRISPR-edited mutation of closely related rice MAP kinase genes. Plant J, 2017, 89(3): 636-648. |
| [20] |
Wang MN, Peng JJ, Wang KJ, et al. Editing ROP gene OsRac5 of rice by CRISPR/Cas9 technique. Chin J Biochem Mol Biol, 2018, 34(12): 1350-1357 (in Chinese). 王美娜, 彭静静, 王凯婕, 等. 利用CRISPR/Cas9技术编辑水稻ROP基因OsRac5. 中国生物化学与分子生物学报, 2018, 34(12): 1350-1357. |
| [21] |
Wang YH, Li JY. Molecular basis of plant architecture. Annu Rev Plant Biol, 2008, 59: 253-279. |
| [22] |
Wang YJ, Zhao J, Lu WJ, et al. Gibberellin in plant height control: old player, new story. Plant Cell Rep, 2017, 36(3): 391-398. |
| [23] |
Liang F, Xin XY, Hu ZJ, et al. Genetic analysis and fine mapping of a novel semidominant dwarfing gene LB4D in rice. J Integr Plant Biol, 2011, 53(4): 312-323. |
| [24] |
Hedden P. The genes of the green revolution. Trends Genet, 2003, 19(1): 5-9. |
| [25] |
Yang XC, Hwa CM. Genetic modification of plant architecture and variety improvement in rice. Heredity (Edinb), 2008, 101(5): 396-404. |
 2020, Vol. 36
2020, Vol. 36




