中国科学院微生物研究所、中国微生物学会主办
文章信息
- 王杰, 逯晓云, 刁爱坡, 江会锋
- Wang Jie, Lu Xiaoyun, Diao Aipo, Jiang Huifeng
- 一种高效的多点组合突变克隆构建方法
- A high efficiency cloning approach of multi-points combinational mutagenesis
- 生物工程学报, 2020, 36(2): 309-319
- Chinese Journal of Biotechnology, 2020, 36(2): 309-319
- 10.13345/j.cjb.190147
-
文章历史
- Received: April 19, 2019
- Accepted: May 14, 2019
2. 中国科学院天津工业生物技术研究所,天津 300308
2. Tianjin Institute of Industrial Biotechnology, Chinese Academy of Sciences, Tianjin 300308, China
Due to the high specificity and selectivity, enzymes were extensively researched in the fields of metabolic engineering and synthetic biology[1]. Directed evolution is a powerful and widely used enzyme engineering method which has been found in industrial scale applications[2]. With the continuous improvement of computational programs and computer capabilities, computational design has been frequently applied to enzyme engineering[3]. The combination of directed evolution and computational design for generating a high-quality mutant library is increasingly important[4-5]. Computational design has advantage to identify the responsible positions or residues for protein function[6]. The structure and function of proteins depends on the cooperative interactions among amino acids[7], therefore, there is an increasing need to develop a directed evolution method to facilitate the joint mutation of different amino acid targets obtained by computational design.
Recent years, a variety of multi-point combination mutagenesis methods have been developed to improve the efficiency of directed evolution, including PCR based (such as Error-prone PCR[8], DNA shuffling[9], Rapidly Efficient Combinatorial Oligonucleotides for Directed Evolution (RECODE)[10], Combinatorial Codon Mutagenesis (CCM)[11], etc.) and non-PCR based methods (such as Multiplex Automated Genome Engineering (MAGE)[12], Multiplex Iterative Plasmid Engineering (MIPE)[13], etc). The advantage of RECODE and CCM is that multiple mutations can be introduced simultaneously, but a large number of colonies can not be guaranteed. MAGE and MPGE can get a large number of colonies, however, the mutation rate per cycle is very low and it is necessary to repeat many times for more mutations. Therefore, it is necessary to achieve a large number of colonies to cover all possible mutant populations to ensure the diversity of mutations.
In this research, a high-efficient MCM method was established to achieve combinational mutations and enough colonies simultaneously by combination of DNA assembly[14], fusion PCR[15] and hybridization[16-17]. In the DATEL method (DNA assembly with thermostable exonuclease and ligase, DATEL) the phosphorylated fragments were ligated using Taq DNA polymerase (5′–3′ exonuclease activity), Pfu DNA polymerase (3′–5′ exonuclease activity) and Taq DNA ligase without introducing any scar sequences[14]. DNA assembly guaranteed the access to get the targeted gene with multiple mutagenesis. Fusion PCR offering an improvement on combination of long PCR and overlap extension PCR was used to assemble up several fragments simultaneously[15]. Genes and vectors assembled into linear plasmids by fusion PCR. After simple denaturation and renaturation, linear plasmids hybridized into ideal circular plasmids. The hybridization didn’t require special equipment or enzyme and sequence-independent[16]. MCM method took the advantage of DNA assembly, fusion PCR and hybridization to construct the high-quality mutation library. Then, MCM method was optimized and tested with benzoylformate decarboxylase (BFD) from Pseudomonas putida which can catalyze the synthesis of glycolaldehyde from formaldehyde[18]. Indeed, MCM method was proved that can guarantee high mutation rate and number of clones.
1 Materials and methods 1.1 MaterialsEscherichia coli TreliefTM 5α competent cells were used in this research for cloning and characterization of the library of pET-28a-bfd. The E. coli BL21 (DE3) was used for the expression of BFD. Luria-Bertani medium (LB medium, 10 g/L tryptone, 5 g/L yeast extract, 10 g/L NaCl) was used for cloning and cell culture and 2×YT Growth Medium (2YT medium, 16 g/L tryptone, 10 g/L yeast extract, 5 g/L NaCl) was used for overexpression of proteins. If necessary, 1‰ 100 μg/mL kanamycin and 0.5 mmol/L IPTG were supplemented. Spectrophotometric detection of glycolaldehyde chromogenic reagent: 1.5 g diphenylamine was dissolved into 100 mL acetic acid, then added 1.5 mL concentrated sulfuric acid. Protein buffer A: 50 mmol/L K3PO4, 5 mmol/L MgSO4, pH 7.4; Protein buffer B: 50 mmol/L K3PO4, 5 mmol/L MgSO4, 1 mol/L imidazole, pH 7.4. All primers (Tsingke, China) were designed with Snap Gene and used in this study. Strains and plasmids, tool enzymes and reagents used in this work are listed in Table 1 and Table 2.
| Strains and plasmids | Description | Sources |
| Strains | ||
| Escherichia coli TreliefTM 5α | F– Φ80dlacZ△M15, △(lacZYA-argF) U169, deoR, recA1, endA1, hsdR17 (rK–, mK–), phoA, supE44, λ-, thi-1, gyrA96, relA1 | Tsingke, China |
| E. coli BL21 (DE3) | F– ompT hsdSB (rB-mB-) gal dcm (DE3) | Transgen, China |
| Plasmids | ||
| pET-28a | Kan+ His-Tag T7-Tag T7/lac promoter | Our laboratory |
| Tool enzymes and reagents | Sources |
| Tool enzymes | |
| I-5TM 2× High-Fidelity Master Mix | Molecular Cloning Labs, Inc |
| T4 DNA ligase buffer with 10 mmol/L ATP | New England Biolabs, USA |
| Taq DNA ligase | New England Biolabs, USA |
| Taq DNA ligase buffer | New England Biolabs, USA |
| Cut SmartTM Buffer | New England Biolabs, USA |
| T4 Polynucleotide kinase | New England Biolabs, USA |
| Trans Fast Taq DNA polymerase | Transgen, China |
| Trans Start Fast Pfu DNA polymerase | Transgen, China |
| DMT enzyme | Transgen, China |
| Reagents | |
| 5×DNA loading buffer | Solarbio, China |
| DNA marker | Solarbio, China |
| Kanamycin | Solarbio, China |
| Iopropyl-β-D-thiogalactoside (IPTG) | Solarbio, China |
| Hydroxyacetaldehyde | Solarbio, China |
| Tryptone | Oxoid, British |
| Yeast extract | Oxoid, British |
| NaCl | Solarbio, China |
| MgSO4 | Solarbio, China |
| K2HPO4 | Solarbio, China. |
| KH2PO4 | Solarbio, China |
| Diphenylamine | Sinopharm, China |
| NADH | Solarbio, China |
| Acetic acid | Sinopharm, China |
| TPP | Solarbio, China |
| Parafilm | BEMIS, USA |
According to the previous research results, the five sites of BFD protein were L109, L110, H281, Q282 and A460, and the mutation of amino acid residues at these sites had a significant effect on BFD protease activity[18-19]. This study performed a combined mutation for the selected 5 sites. The MCM method introduced mutations by using the degenerate codon NDT. There are 12 possible amino acid mutations based on E. coli codon preference (F, R, H, S, N, Y, V, G, I, C, D, L).
Mutant primers were designed according to the DATEL method[14]. The mutated primers comprised a 5′-end overlap region and a 3′-end extension region and the overlaps of the adjacent primers 5′-end overlap region is 30 bp (Fig. 1). The relatively uniform optimal annealing temperature for all overlapping sequences of primers is approximately 55 ℃. Primers used in this work are listed in Table 3.
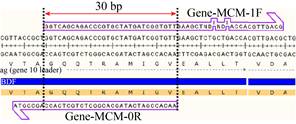
|
| Fig. 1 Primers design with introduced mutation. The size of overlap between gene bfd and vector pET-28a as an influencing factor. |
| |
| Primer name | Primer sequence (5′–3′) |
| Various length of overlaps between genes and vectors | |
| Gene1-F | TGATGCCGGCCACGATGCGTCCGGCGTAGAG |
| Gene1-R | CGGGGAAAGCCGGCGAACGTGG |
| pET-28a-50F | CCTTTCGCTTTCTTCCCTTCCTTTCTCGCC |
| pET-28a-50R | CCGGCGCCACAGGTGCGGTTGCTG |
| pET-28a-100F | TGGTTACGCGCAGCGTGACCGC |
| pET-28a-100R | CCGGCGCCACAGGTGCGGTTGCTG |
| pET-28a-200F | TAAACGGGTCTTGAGGGGTTTTTTGCTGAAAGGAGG |
| pET-28a-200R | CCGGCGCCACAGGTGCGGTTGCTG |
| pET-28a-300F | ACCACCACCACTGAGATCCGGCTGCTAAC |
| pET-28a-300R | CCGGCGCCACAGGTGCGGTTGCTG |
| Various length of overlaps between linear plasmids | |
| Linear Plasmid-A-F | TGATGCCGGCCACGATGCGTCCGGCGTAGAG |
| Linear Plasmid-A-R | CGGGGAAAGCCGGCGAACGTGG |
| Linear Plasmid-B-400F | GGCTCCGCAGGGTCCGGTTTACCTGTCTGTTC |
| Linear Plasmid-B-400R | ATAGAAGCCATGTGGATAGCACGAGACATAGCGTGC |
| Linear Plasmid-B-500F | TTCTGTTCGTCTGAACGACCAGGACCTGGACATCCTG |
| Linear Plasmid-B-500R | GAAGAAACGTGACGGTCGAACAGGTGGTGAGACTG |
| Linear Plasmid-B-600F | AACGCTAACGCTGACTGCGTTATGCTGGCTGAACG |
| Linear Plasmid-B-600R | AGCAGCGTCAACGTCCGGACCCAGAACGATAG |
| Linear Plasmid-B-800F | CCAGTACGACCCGGGTCAGTACCTGAAACC |
| Linear Plasmid-B-800R | TGGTAACGGAAAACCGGAGCACCGATAACCAGAAC |
| Linear Plasmid-B-1200F | CTGGCTGAACCGGAACGTCAGGTTATCGCTG |
| Linear Plasmid-B-1200R | CTGAACACCGATAGCAGCCGGCAGAGCGAAAC |
| Linear Plasmid-B-1500F | GGTCCGGTTCTGATCGAAGTTTCTACCGTTTCTCCGG |
| Linear Plasmid-B-1500R | TTTAGCAGACAGAGCTTCCTGCAGAGAACCTTTCAGCTG |
| Linear Plasmid-B-1800F | CGCCCGCTCCTTTCGCTTTCTTCCCTTCC |
| Linear Plasmid-B-1800R | CTAGGGCGCTGGCAAGTGTAGCGGTCAC |
| Linear Plasmid-B-2000F | ATCGCCCTGATAGACGGTTTTTCGCC |
| Linear Plasmid-B-2000R | GGCCCACTACGTGAACCATC |
| Mutant introduction primers design | |
| Gene-MCM-0F | TGATGCCGGCCACGATGCGTCCGGCGTAGAG |
| Gene-MCM-0R | AACACCGATCATAGCACGGGTCTGCTGACCAGCGGTA |
| Gene-MCM-1F | GGTCAGCAGACCCGTGCTATGATCGGTGTTGAAGCTNDTNDTACCAACGTTGACG |
| Gene-MCM-1R | GAAAACCGGAGCACCGATAACCAGAACAACGTCGTGACCTTCCAG |
| Gene-MCM-2F | GTTGTTCTGGTTATCGGTGCTCCGGTTTTCCGTTACNDTNDTTACGACCCGGGTCAGT |
| Gene-MCM-2R | GGTACCGTTGTTCATGATAACGAAGATGGTCGGGATGTTGTACTGAGCAG |
| Gene-MCM-3F | ACCATCTTCGTTATCATGAACAACGGTACCTACGGTNDTCTGCGTTGGTTCGCTG |
| Gene-MCM-3R | CGGGGAAAGCCGGCGAACGTGG |
| Vector-pET-28a-1F | TCAAGCTCTAAATCGGGGGCTCCC |
| Vector-pET-28a-1R | GGTTACCGAAAACGGTGTCGATACCCTGAC |
| Vector-pET-28a-2F | CTGCAGGAAGCTCTGTCTGCTAAAGGTCCG |
| Vector-pET-28a-2R | CCGGCGCCACAGGTGCGGTTGCTG |
The size of the overlaps between the gene bfd and the vector pET-28a was used as an influencing factor, and the size of overlaps on corresponding primers were designed to be 50, 100, 200, 300 bp. Primers used in this work are listed in Table 3. The size of the overlaps between the two types of linear plasmids pET-28a-bfd were designed to 400, 500, 600, 800, 1 200, 1 500, 1 800, 2 000 bp. Generally, the size was close to the intact genes. There was a nick on both sides of the gene bfd of two types of linear plasmid pET-28a-bfd, but no base gaps (Fig. 2). Primers used in this work are listed in Table 3.
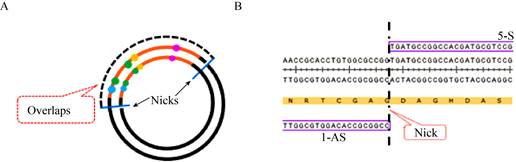
|
| Fig. 2 Detailed diagram of nicks. A: The location of nicks. The dotted line refers to the overlaps, including the overlaps at both ends of the gene. The blue line refers to the location where nicks exist. B: Detail of nick. The dotted line indicates the position of nick, where has no base gaps. |
| |
Primers should be phosphorylated before PCR amplification for the mutated gene fragments. The phosphorylation reaction mixture (50 µL) contained 100 pmol of each primer (primers with different mutations), 10 U T4 polynucleotide kinase and 1× T4 DNA ligase buffer with 10 mmol/L ATP. The reaction system was incubated at 37 ℃ for 30 min and subsequently terminated by heating 10 min at 75 ℃.
All the DNA fragments were amplified with I-5TM 2× High-Fidelity Master Mix, according to the manufacturer’s protocol. And then the PCR products were purified using a Zymoclean Gel DNA Recovery Kit. The DATEL method used for gene fragments assembly was performed. In the reaction system, phosphorylated fragments were ligated using Taq DNA polymerase (5′–3′ exonuclease activity), Pfu DNA polymerase (3′–5′ exonuclease activity) and Taq DNA ligase without introducing any scar sequences[14]. According to the protocol of DATEL method, the intact genes were obtained with mutations.
The intact genes with overlaps at both end and linear vectors, respectively. By using fusion PCR[15], two types of linear plasmids were obtained. The influencing factors of the fusion PCR reaction focused on the size of the overlaps between the genes and the vectors (50, 100, 200, 300 bp), the annealing temperature, the number of thermal cycles, and whether the non-nested primers were added. Performed the thermal cycling under the following program: (a) 98 ℃, 2 min; (b) 98 ℃, 10 s; (c) 55 ℃, 15 s; (d) 72 ℃, 1 min 45 s; (e) 50 ℃, 5 min; (f) Go to step b and repeat 10 times; (h) 4 ℃, hold. If necessary, primers at both ends were used to amplify linear plasmids.
The overlaps between the two linear plasmids pET-28a-bfd were focused on, which the size was 400, 500, 600, 800, 1 200, 1 500, 1 800, 2 000 bp, respectively. Equimolar linear plasmids were mixed in Cut SmartTM buffer for hybridization and immersed in boiling water (about 95–100 ℃) until the temperature dropped to near room temperature (25–35 ℃)[16]. It usually took about 2.5 hours. Subsequently, the hybrid recombinant products were transformed into E. coli by electroporation transformation.
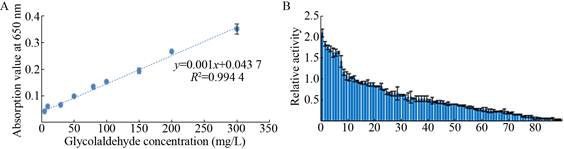
1.2.3 Establishment of high-throughput detection method for glycolaldehydeAn efficient method to detect glycolaldehyde for high-throughput screening was established. 30 μL variousconcentrations of glycolaldehyde were prepared, and then 150 μL spectrophotometric chromogenic reagent was added, keeping at 90 ℃ for 15 min. At last, glycolaldehyde concentration was measured by spectrophotometrically monitoring at 650 nm. A standard curve of glycolaldehyde was made.
The BFD-variants were picked into 96-well plates and cultured in 2YT medium containing 100 μg/mL kanamycin at 37 ℃. When the OD600 reached 0.6, the final concentration was 0.5 mmol/L IPTG at 16 ℃ for 16–18 hours. The cell pellets were rinsed using protein buffer A, subsequently, the whole cell catalytic system was carried out in protein buffer A containing 5 g/L formaldehyde at 30 ℃ for 2 h. The amount of glycolaldehyde produced was detected by the above.
1.2.4 Activity assay and kinetic properties of BFD and mutantsEnzyme kinetics with formaldehyde as substrate were determined in assays with formaldehyde concentrations of 0.1–1 000 mmol/L. An initial continuous assay included 0.5 mmol/L TPP, 5 mmol/L MgSO4, 0.8 mmol/L NADH, 50 μg/mL glycerol dehydrogenase and 50 mmol/L potassium phosphate buffer (pH 7.4). The reaction was initiated by the addition of purified BFD or mutants (0.05 mg/mL) at 37 ℃, and then an initial linear decrease in absorbance at 340 nm was observed. One unit of enzyme activity was defined as the amount of enzyme catalyzing the conversion of 1 μmol NADH per minute. Kinetic parameters kcat and Km were estimated by measuring the initial velocities of enzymic reaction and curve-fitting according to the Michaelis-Menten equation, using GraphPad Prism 5 software. All experiments were conducted in triplicate.
2 Results and discussion 2.1 Design of the MCM methodIn recent years, many DNA assembly methods in vitro have been designed and developed, including overlap extension PCR methods[20], introduction of mutagenesis DNA oligonucleotides[21-22], hybridization of DNA fragments[16-17], assembly method dependent on exonucleases and ligase[14], Golden Gate[23] and Gibson assembly[23-24], etc. We proposed a multi-point combinatorial mutagenesis method based on DNA assembly. In order to achieve both high mutation rates and enough colonies, we proposed to combine DNA assembly[14], fusion PCR[15, 25] and hybridization[16-17, 26] together (Fig. 3). Firstly, multiple mutations were introduced into gene fragments by PCR amplification. Then an intact gene with multi-point mutations can be assembled by the DATEL method[14]. Subsequently, since both ends of the intact gene contain overlaps with the vector backbone (V1/V2), we could generate two types of linear plasmids by using fusion PCR. Finally, the two types of linear plasmids (V-1/V-2) were hybridized to form circular double-stranded DNA molecules with two nicks. The circular plasmids were transformed into E. coli to construct a library with abundant mutation diversity for directed evolution of enzyme.
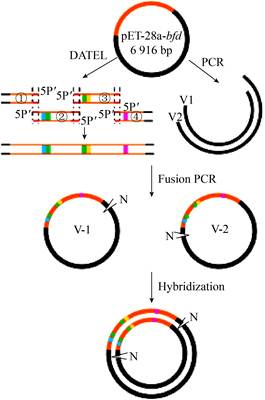
|
| Fig. 3 Schematic diagram of the MCM method. ①, ②, ③ and ④ represent gene fragments with different mutation obtained by PCR. V1 and V2 represent linearized vectors. V-1 and V-2 refer to linear plasmid containing mutated genes. Orange-red lines refer to genes, black lines refer to vectors, and blocks or dots of different colors refer to different mutants. N stands for Nick, indicating that the part is not connected but has no base gap. |
| |
To confirm and optimize the MCM method, we performed a series of optimization experiments with BFD which can catalyze formaldehyde to form glycolaldehyde. The previous results confirmed that five positions (L109, L110, H281, Q282 and A460) displayed important roles in enzyme activities[18-19]. Using the degenerate NDT codons to introduce mutations, we obtained four gene fragments with saturated mutations by PCR. Phosphorylated fragments (①②③④) were ligated using Taq (5′–3′ exonuclease activity) and Pfu (3′–5′ exonuclease activity) DNA polymerase and Taq DNA ligase[14]. Then intact genes with multiple mutations were generated in two steps by using DATEL method (Fig. 4).
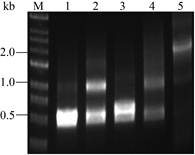
|
| Fig. 4 Assembly efficiency of intact genes containing mutations. M: marker; lane 1 or 2: DNA mixture of fragments ①② before or after assembly, respectively; lane 3 or 4: DNA mixture of fragments ③④ before or after assembly, respectively; lane 5: DNA mixture of fragments ①②③④ after assembly. |
| |
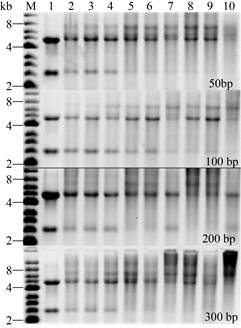
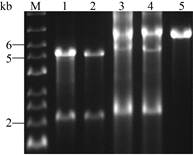
In order to improve the fusion efficiency between the intact genes with multiple mutations and the vector backbones (V1/V2), we optimized the size of the overlaps between the genes and the vectors (50, 100, 200, 300 bp), annealing temperature (45 ℃, 55 ℃, 65 ℃), number of thermal cycles (10, 20, 30 cycles), and whether the non-nested primers were added. Without adding non-nested primers, we chose 100 bp overlaps with an annealing temperature of 55 ℃ after comparison (Fig. 5). The agarose gel electrophoresis analysis results show thata large and 10 thermal cycles for subsequent fusion PCR number of non-target fragments were introduced in other conditions. Subsequently, when non-nested primers Gene-MCM-0F/Vector-pET-28a-2R or Vector-pET- 28a-2F/Gene-MCM-0R were added, the annealing temperature was set to 60 ℃ in the latter 20 cycles, which were close to the Tm value of primer design. As expected, we obtained linear plasmids (V-1/V-2) (Fig. 6).
|
| Fig. 5 Effect of different size of overlaps, annealing temperature and number of thermal cycles on fusion PCR efficiency. M: marker; lane 1: the DNA mixtures before fusion PCR; lane 2, 3 and 4: the DNA mixtures after 10, 20, 30 fusion PCR thermal cycles, respectively, when the annealing temperature was 45 ℃; lane 5, 6 and 7: the DNA mixtures after 10, 20, 30 fusion PCR thermal cycles, respectively, when the annealing temperature was 55 ℃; lane 8, 9 and 10: the DNA mixtures after 10, 20, 30 fusion PCR thermal cycles, respectively, when the annealing temperature was 65 ℃. |
| |
|
| Fig. 6 Ligation efficiency of fusion PCR. M: marker; lane 1 or 3: DNA mixture for V-1 before or after fusion PCR, respectively; lane 2 or 4: DNA mixture for V-2 before or after fusion PCR, respectively; lane 5: the linear plasmids of pET-28a-bfd (6 916 bp). |
| |
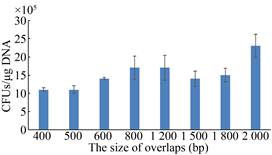
Finally, we verified the effect of different size overlaps between two types of linear plasmidson the colony forming units in hybridization. We found that the CFUs exceeded 106 CFUs/μg DNA, when the size of overlaps ranges from 400 bp to 2 000 bp (Fig. 7). Here, we successfully constructed the MCM method, which can be used for the directed evolution of enzymes. In addition, we have obtained enough colonies to cover the abundant mutants.
|
| Fig. 7 Effect of different size of overlaps on hybridization efficiency. The different size overlaps between two types of linear plasmids range from 400 bp to 2 000 bp. CFUs, the colony forming units. |
| |
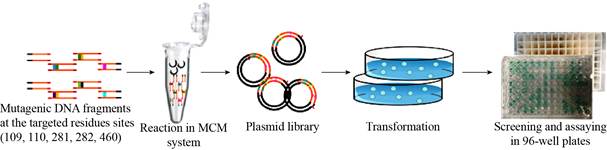
BFD was chosen as a candidate for rapid evolution by MCM method. After DNA assembly and hybridization, all the mutants were transformed into BL21 (DE3). Subsequently, 1 000 colonies were picked in 96 deep-well plates for cultivation and assay of BFD activity. And then, variants with significant difference compared to controls require further investigated (Fig. 8). Remarkably, many variants with different activities toward BFD were successfully created by the MCM method. We found a mutant with a 2-fold increase in the production of glycolaldehyde (Fig. 9B).
|
| Fig. 8 Illustration of the construction and screening for the BFD mutant library. |
| |
|
| Fig. 9 Enzyme activity assay of variants. (A) Standard curve of glycolaldehyde. (B) The relative activity of random BFD variants generated by MCM method (Parts). The y-axis label represents the relative catalytic activity of different mutants. The relative activity was defined as the ratio of the concentration of glycolaldehyde in the mutants to wild BFD using whole-cell system. |
| |
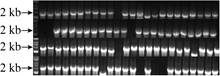
After obtaining enough colonies, whether these colonies contain abundant mutants has become the focus of our concern. To validate the efficiency of the MCM method, 100 colonies were randomly selected for PCR amplification and DNA sequencing analysis. 90 out of 100 colonies are precisely assembled according to PCR verification (Fig. 10).
|
| Fig. 10 PCR amplification for verifying assembly efficiency. Target gene is about 2 kb. |
| |
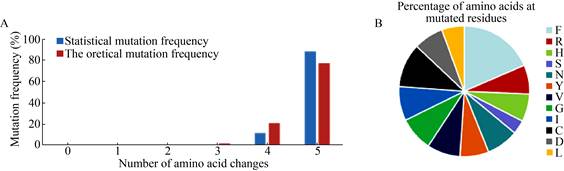
After DNA sequencing analysis, we found that the simultaneous mutation frequency at 5 positions was up to 88%, which was a little higher than the theoretical estimation. The reason may be that the number of samples for DNA sequencing analysis is not large enough; Biases exist for the mutation of continuous base[11]. In addition, the frequency of simultaneous mutation at 4 positions is 12% (Fig. 11A). Through further sequence analysis, we found that these mutants contain 12 possible amino acid derived by the NDT degenerate codon (Fig. 11B). These results indicate that the MCM method was high mutation diversity, which is beneficial for directed evolution.
|
| Fig. 11 The results of DNA sequencing analysis. (A) Comparison of theoretical and statistical mutation diversity. (Theoretical mutation frequency: Primers were designed using the NDT degenerate codon at 5 mutated sites (L109, L110, H281, Q282 and A460). Considering the codon preference of E. coli, the 12 possible amino acid residues at mutated sites were included (F, R, H, S, N, Y, V, G, I, C, D and L), and the simultaneous mutation frequency at 5 positions: 11*11*11*12*12/125≈77%; the simultaneous mutation frequency at 4 positions: 3*1*11*11*12*12/125≈21%; the simultaneous mutation frequency at 3 positions: 3*1*1*11*12*12/125≈1.9%; the simultaneous mutation frequency at 2 positions: 1*1*1*12*12/125≈0.1%. (B) Percentage of 12 possible amino acidresidues at mutated sites. Different colors indicate the percentage of each amino acid encoded by NDT in all mutants. |
| |
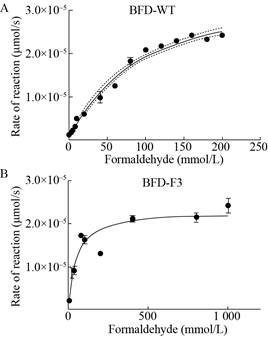
We found a mutant with a 2-fold increase in the production of glycolaldehyde, subsequently, we extracted the purified protein and characterized the function of wild type and mutant. The kcat of the best mutant BFD-F3 (L109Y, L110D, H281G, Q282V and A460M) is improved about 6-fold and the final catalytic efficiency is 1.35 L/(mol·s), which is roughly 10-fold than the starting enzyme (Table 4, Fig. 12). These results confirm that the MCM method can be used to rapidly improve enzyme function.
| Variants | kcat(s–1) | Km(mol/L) | kcat/Km[L/(mol·s)] |
| BFD-WT | 0.01 | 0.09 | 0.14 |
| BFD-F3 | 0.06 | 0.05 | 1.35 |
|
| Fig. 12 Catalytic rate of BFD-WT and BFD-F3 under different formaldehyde concentrations. |
| |
In this study, the MCM method, which combined DNA assembly[14], fusion PCR[15, 25] and hybridization[16-17, 26], were successfully constructed for rapid and efficient multipoint combinatorial mutagenesis of enzymes. This method introduced different mutations by PCR, and the mutation diversity was ensured as much as possible by the DNA assembly method. The hybridization technology increases the number of final colonies, enabling high mutation richness and high colony counts to be achieved simultaneously. In the research, in order to confirm and optimize the MCM method, we performed a series of experiments with BFD. Previous experiments demonstrated that the 5 sites selected for this study had a significant effect on the activity of BFD enzymes[18-19]. The 5 sites selected in this study use the NDT degenerate codon to mutate the key amino acid sites, 12 possible amino acid residues were founded due to the codon preference of E. coli, the high mutation rate of MCM (4/5 points combination mutation) was confirmed by the actual statistical mutation rate (Fig. 11). If the key amino acid sites use the NNK degenerate codon to mutate, 20 possible amino acids at mutated sites will be found. The result would not cause a deviation in the mutation rate for the multi-point combination (1/2/3/4/5 point3 combination mutation), which could better explore the effects of different mutant amino acids at key mutated sites. Directed evolution of multi-site combinatorial mutations of enzymes using the MCM method does not mean that the more sites of the combined mutations, the better it is. Since multiple site-mutant amino acid interactions affect the structure and function of the enzyme, the effective combination of key sites is the most reasonable.
Directed evolution is a valuable tool for synthetic biology, enabling the identification or separation of desired functions from large libraries of variants[27-30]. It plays an important role in improvement of the activity and stability of biocatalyst, development of desired complex phenotypes in biological systems, engineering of biosynthetic pathways and tuning of functional regulatory systems and logic circuits[30-32]. In this study, we obtained mutants with significantly increased activity by screening 1 000 colonies. These results indicate that the MCM method can significantly shorten the period of enzyme directed evolution. If the MCM method combined with computer-aided design, beneficial mutations will be efficiently screened. There will be a huge impact on the development of directed evolution of enzymes.
| [1] |
Denard CA, Ren HQ, Zhao HM. Improving and repurposing biocatalysts via directed evolution. Curr Opin Chem Biol, 2015, 25: 55-64. DOI:10.1016/j.cbpa.2014.12.036 |
| [2] |
Neylon C. Chemical and biochemical strategies for the randomization of protein encoding DNA sequences: library construction methods for directed evolution. Nucleic Acids Res, 2004, 32(4): 1448-1459. DOI:10.1093/nar/gkh315 |
| [3] |
Packer MS, Liu DR. Methods for the directed evolution of proteins. Nat Rev Genet, 2015, 16(7): 379-394. DOI:10.1038/nrg3927 |
| [4] |
Amrein BASteffen-Munsberg F, Szeler I, et al. CADEE: computer-aided directed evolution of enzymes. IUCrJ, 2017, 4: 50-64. DOI:10.1107/S2052252516018017 |
| [5] |
Ward TR. Artificial enzymes made to order: combination of computational design and directed evolution. Angew Chem Int Ed, 2008, 47(41): 7802-7803. DOI:10.1002/anie.200802865 |
| [6] |
Wijma HJ, Floor RJ, Jekel PA, et al. Computationally designed libraries for rapid enzyme stabilization. Protein Eng Des Sel, 2014, 27(2): 49-58. |
| [7] |
Wang D, Wang J, Wang B, et al. A new and efficient colorimetric high-throughput screening method for triacylglycerol lipase directed evolution. J Mol Catal B-Enzym, 2012, 82: 18-23. DOI:10.1016/j.molcatb.2012.05.021 |
| [8] |
Gram H, Marconi LA, Barbas CF, et al. In vitro selection and affinity maturation of antibodies from a naive combinatorial immunoglobulin library. Proc Natl Acad Sci USA, 1992, 89(8): 3576-3580. DOI:10.1073/pnas.89.8.3576 |
| [9] |
Lehmann M, Wyss M. Engineering proteins for thermostability: the use of sequence alignments versus rational design and directed evolution. Curr Opin Biol, 2001, 12(4): 371-375. DOI:10.1016/S0958-1669(00)00229-9 |
| [10] |
Jin P, Kang Z, Zhang JL, et al. Combinatorial evolution of enzymes and synthetic pathways using one-step PCR. ACS Synth Biol, 2016, 5(3): 259-268. |
| [11] |
Belsare KD, Andorfer MC, Cardenas FS, et al. A simple combinatorial codon mutagenesis method for targeted protein engineering. ACS Synth Biol, 2017, 6(3): 416-420. DOI:10.1021/acssynbio.6b00297 |
| [12] |
Wang HH, Isaacs FJ, Carr PA, et al. Programming cells by multiplex genome engineering and accelerated evolution. Nature, 2009, 460(7257): 894-898. DOI:10.1038/nature08187 |
| [13] |
Li YF, Gu Q, Lin ZQ, et al. Multiplex iterative plasmid engineering for combinatorial optimization of metabolic pathways and diversification of protein coding sequences. ACS Synth Biol, 2013, 2(11): 651-661. DOI:10.1021/sb400051t |
| [14] |
Jin P, Ding WW, Du GC, et al. DATEL: a scarless and sequence-independent DNA assembly method using thermostable exonucleases and ligase. ACS Synth Biol, 2016, 5(9): 1028-2032. DOI:10.1021/acssynbio.6b00078 |
| [15] |
Shevchuk NA, Bryksin AV, Nusinovich YA, et al. Construction of long DNA molecules using long PCR-based fusion of several fragments simultaneously. Nucleic Acids Res, 2004, 32(2): e19. DOI:10.1093/nar/gnh014 |
| [16] |
Jiang XL, Yang JM, Zhang HB, et al. In vitro assembly of multiple DNA fragments using successive hybridization. PLoS ONE, 2012, 7(1): e30267. DOI:10.1371/journal.pone.0030267 |
| [17] |
Liang J, Liu ZH, Low XZ, et al. Twin-primer non-enzymatic DNA assembly: an efficient and accurate multi-part DNA assembly method. Nucleic Acids Res, 2017, 45(11): e94. DOI:10.1093/nar/gkx132 |
| [18] |
Cui B, Zhuo BZ, Lu XY, et al. Enzymatic synthesis of xylulose from formaldehyde. Chin J Biotech, 2018, 34(7): 1128-1136. 崔博, 卓炳照, 逯晓云, 等. 酶法催化甲醛合成木酮糖. 生物工程学报, 2018, 34(7): 1128-1136. |
| [19] |
Lu XY, Liu YW, Yang YQ, et al. Constructing a synthetic pathway for acetyl-coenzyme A from one-carbon through enzyme design. Nat Commun, 2019, 10: 1378. DOI:10.1038/s41467-019-09095-z |
| [20] |
Zhao HM, Zha WJ. In vitro 'sexual' evolution through the PCR-based staggered extension process (StEP). Nat Protoc, 2006, 1(4): 1865-1871. DOI:10.1038/nprot.2006.309 |
| [21] |
Herman A, Tawfik DS. Incorporating synthetic oligonucleotides via gene reassembly (ISOR): a versatile tool for generating targeted libraries. Protein Eng Des Sel, 2007, 20(5): 219-226. DOI:10.1093/protein/gzm014 |
| [22] |
de Kok S, Stanton LH, Slaby T, et al. Rapid and reliable DNA assembly via ligase cycling reaction. ACS Synth Biol, 2014, 3(2): 97-106. DOI:10.1021/sb4001992 |
| [23] |
Halleran AD, Swaminathan A, Murray RM. Single day construction of multigene circuits with 3G assembly. ACS Synth Biol, 2018, 7(5): 1477-1480. DOI:10.1021/acssynbio.8b00060 |
| [24] |
Gibson DG, Young L, Chuang RY, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods, 2009, 6(5): 343-345. DOI:10.1038/nmeth.1318 |
| [25] |
Urban A, Neukirchen S, Jaeger KE. A rapid and efficient method for site-directed mutagenesis using one-step overlap extension PCR. Nucleic Acids Res, 1997, 25(11): 2227-2228. DOI:10.1093/nar/25.11.2227 |
| [26] |
Jiang XL, Yang JM, Zhang HB, et al. In vitro assembly of multiple DNA fragments using successive hybridization. PLoS ONE, 2012, 7(1): e30267. DOI:10.1371/journal.pone.0030267 |
| [27] |
Bassalo MC, Liu RM, Gill RT. Directed evolution and synthetic biology applications to microbial systems. Curr Opin Biotechnol, 2016, 39: 126-133. DOI:10.1016/j.copbio.2016.03.016 |
| [28] |
Haseltine EL, Arnold FH. Synthetic gene circuits: design with directed evolution. Annu Rev Biophys Biomol Struct, 2007, 36: 1-19. DOI:10.1146/annurev.biophys.36.040306.132600 |
| [29] |
Porcar M. Beyond directed evolution: Darwinian selection as a tool for synthetic biology. Syst Synth Biol, 2010, 4(1): 1-6. |
| [30] |
Kang Z, Zhang JL, Jin P, et al. Directed evolution combined with synthetic biology strategies expedite semi-rational engineering of genes and genomes. Bioengineered, 2015, 6(3): 136-140. DOI:10.1080/21655979.2015.1011029 |
| [31] |
Cobb RE, Sun N, Zhao HM. Directed evolution as a powerful synthetic biology tool. Methods, 2013, 60(1): 81-90. DOI:10.1016/j.ymeth.2012.03.009 |
| [32] |
Cobb RE, Si T, Zhao HM. Directed evolution: an evolving and enabling synthetic biology tool. Curr Opin Chem Biol, 2012, 16(3/4): 285-291. |
 2020, Vol. 36
2020, Vol. 36




