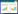中国科学院微生物研究所、中国微生物学会主办
文章信息
- 于丽华, 滕飞, 蒋明, 郭佳
- Yu Lihua, Teng Fei, Jiang Ming, Guo Jia
- 一种检测kras基因突变的新型TB-ARMS qPCR方法的建立
- Establishment of a novel TB-ARMS qPCR method for kras mutation detection
- 生物工程学报, 2019, 35(5): 880-891
- Chinese Journal of Biotechnology, 2019, 35(5): 880-891
- 10.13345/j.cjb.180434
-
文章历史
- Received: November 19, 2018
- Accepted: March 4, 2019
kras基因突变是结直肠癌、肺癌和胰腺癌等癌症中最常见的突变类型之一[1-2]。kras突变与肿瘤靶向治疗的耐药性密切相关,例如非小细胞肺癌(Non-small cell lung cancer,NSCLC)中的kras基因突变会导致对吉非替尼、厄洛替尼等EGFR靶向药物耐药,因此kras突变检测可用于指导靶向药物的个体化治疗和用于治疗过程中的耐药监测[3-4]。目前的突变检测技术大都以肿瘤组织为主要检测标本,由于肿瘤组织的异质性[5],突变检测往往需要在较高野生型基因背景下进行,因此需要一种高特异性的检测方法,以减少假阳性检测结果的产生。液态活检样本例如血液由于取样方便是更为理想的突变检测样本[6],已有研究[7-10]发现血浆游离DNA中存在肿瘤来源的DNA,其突变情况与肿瘤组织高度一致,循环血突变检测被认为是一种较有前途的无创检测方法,既适用于不适宜手术的肺癌和结直肠癌等癌症患者,也适用于对病人的突变情况进行跟踪监测,具有更广的应用范围。但癌症病人血浆游离DNA水平较正常人往往明显升高,kras突变基因所占比例通常较低,很多情况下达不到1%,为了增加突变检测的准确性,需要灵敏度和特异性高的检测方法。目前已有的kras突变检测方法包括Sanger测序法、等位基因特异性PCR (AS PCR,也称ARMS PCR)[11]、高分辨率溶解曲线分析法[12]、scorpion ARMS[13]、包含焦磷酸测序在内的第二代测序[14-15]、BEAMing技术[16]以及数字PCR技术[17]。除了最后3种检测技术,大部分突变检测技术的突变检测灵敏度都在1%−5%。BEAMing技术、第二代测序和数字PCR技术虽然突变检测灵敏度较高(甚至可达到0.000 5%),但在实际应用中存在一些限制,需要一般临床实验室所不具备的特殊仪器,价格昂贵,样品制备、操作比较费时。荧光定量qPCR方法作为快速简便的检测方法,仍然具有较大的临床应用价值,ARMS PCR是目前比较常用的qPCR检测方法,有研究[18-21]通过在ARMS PCR的基础上引入野生型等位基因特异性封闭引物的方法来抑制野生型基因的扩增,从而改善点突变检测的选择性可以达到0.1%。文中在现有技术的基础上综合采用多种技术方法对ARMS技术作了进一步改进,命名为Tm值相关的封闭ARMS (Tm-related blocking ARMS,TB-ARMS)技术,建立了针对kras基因8种常见点突变的检测方法,通过与广泛使用的商品化试剂盒进行比较,及对临床组织和配对血浆样本的检测来验证该方法的实际应用价值。
1 材料与方法 1.1 基因组DNA和质粒标准品的制备研究采用经测序证实为野生型kras基因的健康人全血基因组DNA作为野生型模板。基因组DNA的提取采用天根生化科技有限公司的血液、细胞、组织基因组DNA提取试剂盒(目录号:DP304-02),具体操作按照说明书进行。本研究所采用的突变标准品为采用基因重组技术构建的kras突变质粒,设计并采用pCR Blunt Ⅱ TOPO质粒载体(Invitrogen),构建8种常见突变类型的kras突变质粒,包括G12C、G12V、G12D、G12R、G12S、G12A、G13C和G13D,并经测序证实突变序列。kras 8种突变质粒分别采用BamHⅠ限制性内切酶单酶切使其线性化,并割胶回收,采用PCR胶回收试剂盒纯化。基因组DNA和线性化质粒分别采用分光光度计测定浓度,并对其进行质量控制OD260/280=1.6−1.8。
1.2 标准品的制备本研究中通过将线性化突变质粒系列稀释至10 000、1 000、100和10 copies/μL,然后分别取10 μL与90 μL野生型人基因组DNA (30 ng/μL)混合制备10%、1%、0.1%和0.01%不同突变率的样品。
1.3 肿瘤组织样品及血浆样品的收集和处理40例NSCLC病人组织样品由上海市肺科医院提供,均为术后新鲜肿瘤组织样品;收集20例NSCLC病人术前血浆样本和配对组织样本,样品收集后均−80 ℃保存备用。组织DNA的提取采用天根生化科技有限公司的血液、细胞、组织基因组DNA提取试剂盒,血浆DNA的提取采用天根生化科技有限公司的血清/血浆游离DNA提取试剂盒(目录号:DP339),具体操作按照说明书进行。
1.4 新的kras突变检测方法的建立为了准确有效地检测到大量野生型基因背景下稀少的kras突变基因,组合使用多种手段,以增强ARMS-PCR对较低突变率突变基因的检测灵敏度,增大突变型等位基因特异性引物对突变型等位基因模板与野生型等位基因模板的区分能力。使用的手段包括:1)应用野生型等位基因特异性封闭引物,其被设计为与野生型等位基因互补,3′端经磷酸化修饰;2)突变型等位基因特异性引物3′端倒数第4−6位引入错配碱基;3)突变富集扩增的反应条件,采用较高退火温度的预循环,该温度条件下仅有突变模板能被结合扩增,突变模板被富集,然后采用较低退火温度的循环进行高效扩增。
1.4.1 引物设计及扩增本研究针对kras基因12和13号外显子上8种常见突变类型,包括c. 34G > A (p. G12S)、c. 34G > T (p. G12C)、c. 34G > C (p. G12R)、c. 35G > T (p. G12V)、c. 35G > A (p. G12D)、c. 35G > C (p. G12A)、c. 37G > T (p. G13C)和c. 38G > A (p. G13D)的点突变,采用Oligo 7软件辅助设计ARMS-PCR等位基因特异性引物,并控制其Tm值,以便筛选出与富集扩增反应条件相适应的最佳引物。采用的富集扩增反应条件较高退火温度为64 ℃,设计突变特异性引物时控制引物的Tm值范围为50−60 ℃。封闭引物序列与野生型等位基因位点完全匹配,包括引入修饰的碱基,封闭引物Tm值要大于等于退火温度,确保其在退火时与野生型模板结合起封闭作用,并且封闭引物3′末端经修饰,使其在DNA聚合酶作用下不能被延伸。同时设计锁核酸修饰封闭引物,可使引物Tm值增加5−10 ℃,修饰碱基为kras突变等位基因对应的野生型基因,以增强封闭引物对野生型等位基因的区分能力。封闭引物3′末端碱基采用磷酸化修饰。由于8种点突变位点在外显子上的位置靠近,因此可采用相同的封闭引物。同时采用beta-actin基因作为内参照基因,对其设计常规引物探针,用以对反应的质量进行监控。所有引物、探针均由生工生物工程(上海)股份有限公司合成。本研究所采用的引物探针序列见表 1。
| Primer name | Primer sequence (5′–3′) | Tm (℃) |
| G34A-R1 | TTGCCTACGCCACT | 50.0 |
| G34A-R2 | CTCTTGCCTACGCCACT | 55.6 |
| G34A-R3 | CACTCTTGCCTACGaCACT | 55.4 |
| G34T-R1 | CcgCTTGCCTACGCCACA | 52.7 |
| G34T-R2 | CTTGCCTACGCCACA | 54.9 |
| G34T-R3 | GCACTCTTGCCTACGaCACA | 58.9 |
| G34T-R4 | CACTCTTGCCTACGaCACA | 52.7 |
| G34C-R1 | CTTGCCTACGCCACG | 56.7 |
| G35T-R1 | CACTCTTGCCTACGCCAA | 54.8 |
| G35A-R1 | gTCTTGCCTACGCCAT | 52.7 |
| G35C-R1 | CTCTTGCCTACGCCAG | 54.1 |
| G37T-R1 | CACTCTTGCCTACGCA | 53.4 |
| G38A-R1 | CACTCTTGCCTACGT | 50.6 |
| G38A-R2 | GCACTCTTGCCTACGT | 54.5 |
| G38A-R3 | AGGCACTCTTGCaTACGT | 56.1 |
| G38A-R4 | GGCACTCTTGCaTACGT | 52.4 |
| G38A-R5 | AGGCACTCTTGCCcACGT | 56.1 |
| G38A-R6 | AAGGCACTCTTGCCcACGT | 56.0 |
| kras-F | TGACATGTTCTAATATAGTCACATT | 54.1 |
| kras-P | FAM-ATTCAGTCATTTTCAGCAGGCCTT-BHQ1 | 61.0 |
| kras-B1 | TGCCTACGCCACCAGCTC-PO4 | 62.2 |
| kras-B2 | CCTACG+CCA+C+CAGC-PO4 | 73.0 |
| IPC-F | ATCGCCGCGCTCGTC | 60.2 |
| IPC-R | CCGGGGGGCATCGTCG | 62.8 |
| IPC-P | JOE-CAACGGCTCCGGCATGTGCA-BHQ1 | 67.6 |
| Note: kras-F is shared forward primer of 8 kras mutations, and it is combined with different allele specific primers of c. G34A, c. G34T, c. G34C, c. G35T, c. G35A, c. G35C, c. G37T, c. G38A mutations for genotype assay. kras-P is detection probe modified with fluorescence dye and quencher dye at 5′-end and 3′-end. kras-B1 and kras-B2 are blockers. “+” denotes LNA modified nucleotide and kras-B2 was only used in G38A mutation detection. IPC-F, IPC-R and IPC-P are primers and probe for internal positive control gene. Bases in lower case are mismatched bases adopted in this study. | ||
检测仪器采用ABI 7500荧光定量PCR仪。荧光定量PCR反应采用25 μL反应体系,包括1×Taq HS酶(TaKaRa) PCR反应液、200 nmol/L上游引物和对应的下游引物、400 nmol/L封闭引物、200 nmol/L的Taqman探针、100 nmol/L内参照引物、200 nmol/L内参照探针,突变富集扩增反应条件设定为:95 ℃变性5 min,然后10个预循环(95 ℃ 10 s,64 ℃ 1 min),再运行35个循环(95 ℃ 10 s,60 ℃ 1 min)。第三步退火时检测荧光信号。内参照引物探针包含在每个反应体系中。
1.4.2 封闭引物增强kras点突变检测特异性的验证以kras基因G34A、G35A突变检测为例,分别采用含有和不含有封闭引物的反应体系对突变率0、0.01%、0.1%、1%和10%的样品进行检测,除封闭引物其余组分完全相同,通过△Ct值(即Ctwild−Ctmut)的比较确定封闭引物对kras突变检测特异性是否有改善,△Ct值越大说明检测特异性越好。
1.4.3 引入适当突变碱基增强等位基因特异性引物检测特异性的验证通过在kras G34A、G34T和G38A等位基因特异性引物3′端倒数第4–6位引入适当突变碱基并调整Tm值对其进行优化,并与未引入突变碱基的等位基因特异性引物对同样的野生型模板、0.01%和0.1%突变率的样品进行检测,通过△Ct值(即Ctwild−Ctmut)的比较确定每种突变类型的最佳突变等位基因特异性引物,△Ct值越大说明引物的检测特异性越好。
1.4.4 突变富集扩增反应条件与常规反应条件的比较对kras-G34A、kras-G35A的引物探针组合分别采用富集扩增的反应条件和常规反应条件对相同的135 ng野生型样品、0.01%和1%突变率的样品进行检测比较。富集扩增反应条件为:95 ℃预变性5 min,然后10个预循环(95 ℃ 10 s,64 ℃ 1 min),再运行35个循环(95 ℃ 10 s,60 ℃ 1 min),第三步退火时检测荧光信号。对比反应条件为:95 ℃预变性5 min,然后10个预循环(95 ℃ 10 s,60 ℃ 1 min),再运行35个循环(95 ℃ 10 s,60 ℃ 1 min),第三步退火时检测荧光信号。通过△Ct值(即Ctwild−Ctmut)的比较确定最佳反应条件,△Ct值越大说明检测特异性越好。
1.5 方法学评估 1.5.1 突变检测特异性分析采用经测序证实为野生型的人全血基因组DNA样本,分别对3个不同量(1 ng、20 ng、135 ng)的野生型模板进行kras突变分型检测,每样品重复3次。PCR扩增反应实施方法和条件如前所述。反应结束,获得每个样品的突变检测Ct值和内控Ct值。
1.5.2 突变检测灵敏度的评价本研究通过在135 ng野生型基因组DNA背景下,对kras 8种突变的每种突变类型采用TB-ARMS qPCR方法检测其0.1%和0.01%突变率的样品,以野生型样品作对照,来分析kras突变检测的灵敏度。
1.5.3 两种检测方法对组织样品的检测比较对40例NSCLC患者组织样品提取DNA,每样品分别取10 ng DNA,采用本研究的TB-ARSM技术和商品化ADx-ARMS kras突变检测试剂盒进行分型检测,并对检测结果进行比较。对两种方法检测结果不一致的样品进一步进行测序验证。
1.5.4 血浆和配对组织样品的对照分析为验证本研究方法对血浆DNA kras突变检测的准确性,对20例NSCLC患者血浆及其配对组织样品提取的DNA进行对照分析。采用本研究的TB-ARMS技术方法对血浆DNA进行8种kras突变分型检测。同时采用测序方法对组织DNA样品进行kras突变检测。对血浆和组织样品的检测结果进行对比分析,判断血浆突变检测的准确性。
2 结果与分析 2.1 封闭引物增强kras基因点突变检测的特异性通过对kras G34A和G35A点突变的检测,结果显示含有或不含有封闭引物内控检测Ct值相似,而采用封闭引物对野生型模板突变检测的Ct值增大,对突变样品突变检测的Ct值与不采用封闭引物相似或更低(图 1),因而采用封闭引物的△Ct值即Ctwild−Ctmut会变大,说明封闭引物可改善kras点突变的检测特异性。
|
| 图 1 封闭引物增强突变检测的特异性 Fig. 1 Enhanced specificity with wild type blocker in mutation detection. (A) Comparison of the results with and without blocker in detection of kras G34A mutation. (B) Comparison of the results with and without blocker in detection of kras G35A mutation. Mutation analysis was performed on wild and mutant samples with reagents (with and without blocker), and each reagent included the primer set for internal positive control. Reactions were run in triplicate and data are shown as average Ct values. Legend "M" represents mutation detection without blocker; "M+B" represents mutation detection with blocker; "IPC" represents internal positive control detection without blocker; and "IPC+B" represents internal positive control detection with blocker. |
| |
对kras突变的引物优化结果(表 2)表明,kras-G34A等位基因特异性引物3′端倒数第5位引入突变碱基C > A的等位基因特异性引物G34A-R3较未引入突变碱基的等位基因特异性引物G34A-R1、G34A-R2对0.1%和0.01%样品突变检测的△Ct值明显增大;同样,3′端倒数第5位引入突变碱基C > A的等位基因特异性引物G34T-R4较3′端未引入突变碱基的等位基因特异性引物的G34T-R1、G34T-R2的△Ct值增大,较Tm值较高的G34T-R3突变检测的△Ct值亦增大;而对于kras-G38A倒数第6位引入突变碱基C > A的等位基因特异性引物G38A-R4较3′端未引入突变碱基的等位基因特异性引物G38A-R1、G38A-R2的△Ct值增大,较3′端倒数第5位引入突变碱基T > C的等位基因特异性引物G38A-R5、G38A-R6的△Ct值也增大,较Tm值较高的G38A-R3突变检测的△Ct值亦增大,G38A-R4检测特异性最好。
| Mutant type | Primer name | Ctwild | Ctmut0.01% (ΔCt) | Ctmut0.1% (ΔCt) |
| kras-G34A | G34A-R1 | 28.27 | 26.99 (1.28) | 23.66 (4.61) |
| G34A-R2 | 23.10 | 22.95 (0.15) | 22.29 (0.81) | |
| G34A-R3 | 31.49 | 24.77 (6.72) | 22.74 (8.75) | |
| kras-G34T | G34T-R1 | 32.44 | 29.25 (3.19) | 25.86 (6.58) |
| G34T-R2 | 33.20 | 27.65 (5.55) | 24.78 (8.42) | |
| G34T-R3 | 31.98 | 29.80 (2.18) | 25.24 (6.74) | |
| G34T-R4 | 34.24 | 28.08 (6.16) | 25.52 (8.72) | |
| kras-G38A | G38A-R1 | 31.39 | 29.78 (1.61) | 25.85 (5.54) |
| G38A-R2 | 29.15 | 27.46 (1.70) | 24.48 (4.67) | |
| G38A-R3 | 31.04 | 30.19 (0.85) | 24.74 (6.30) | |
| G38A-R4 | 31.97 | 29.68 (2.29) | 25.67 (6.30) | |
| G38A-R5 | 32.84 | 33.12 (–0.28) | 28.36 (4.48) | |
| G38A-R6 | 29.54 | 29.32 (0.22) | 24.60 (4.94) | |
| Note: qPCR reactions were performed in duplicate. Ct values were presented as mean Ct. ΔCt was calculated as Ctmut−Ctwild. Valnes representing highest specifity of each mutant type are highlighted in bold. | ||||
通过采用突变富集扩增反应条件(表 3)和常规扩增条件(表 4)对kras-G34A和kras-G35A的不同突变样品进行检测,检测结果表明采用突变富集扩增反应条件对野生型样品检测的Ct值比常规扩增条件的Ct值均增大;采用突变富集扩增反应条件对于10%高突变率样品检测,G34A突变检测△Ct值与常规检测条件相比增加,而G35A突变检测△Ct值与常规检测条件相比无明显改变;但采用突变富集扩增反应条件对0.1%和0.01%低突变率样品检测,G34A和G35A突变检测的△Ct值均高于常规扩增条件的△Ct值,数值至少可增加1以上。
| Mutant type | Mutant-enriched condition | ||||
| Ctwild | Ctmut0.01% (ΔCt) | Ctmut0.1% (ΔCt) | Ctmut1% (ΔCt) | Ctmut10% (ΔCt) | |
| kras-G34A | 33.83 | 29.06 (4.77) | 25.13 (8.70) | 22.67 (11.16) | 18.87 (14.95) |
| kras-G35A | 33.33 | 29.98 (3.35) | 25.93 (7.40) | 23.42 (9.91) | 20.76 (12.57) |
| Mutant type | Traditional condition | ||||
| Ctwild | Ctmut0.01% (ΔCt) | Ctmut0.1% (ΔCt) | Ctmut1% (ΔCt) | Ctmut10% (ΔCt) | |
| kras-G34A | 28.20 | 25.38 (2.82) | 22.19 (6.01) | 19.02 (9.18) | 16.01 (12.19) |
| kras-G35A | 29.53 | 28.86 (0.67) | 23.23 (6.30) | 19.46 (10.08) | 16.87 (12.66) |
| Note: qPCR reactions were performed in triplicate. Ct values were presented as mean Ct. ΔCt was calculated as Ctmut−Ctwild. | |||||
通过对1 ng、20 ng和135 ng野生型基因组DNA的检测,观察每个样本的内控Ct值均 < 30 (数据未列出),不同分型试剂对野生型样品突变检测的Ct值均大于30 (表 5),提示TB-ARMS技术kras突变分型检测在1−135 ng模板范围内,突变检测Ct值均小于30。
| Amount | G34A | G34T | G34C | G35T | G35A | G35C | G37T | G38A |
| 1 ng | 35.00±0.00 | 33.97±1.79 | 35.00±0.00 | 35.00±0.00 | 35.00±0.00 | 35.00±0.00 | 35.00±0.00 | 35.00±0.00 |
| 20 ng | 33.41±1.53 | 35.00±0.00 | 35.00±0.00 | 35.00±0.00 | 32.92±2.14 | 35.00±0.00 | 34.80±0.34 | 34.51±0.86 |
| 135 ng | 33.54±2.54 | 35.00±0.00 | 35.00±0.00 | 34.70±0.52 | 32.98±1.27 | 35.00±0.00 | 34.59±0.60 | 33.72±2.22 |
| Note: 1 ng, 20 ng and 135 ng wild type gDNA were tested with 8 typing reagents. qPCR reactions were run in triplicate. Ct value was presented as mean Ct±standard deviation for each mutation detection system. | ||||||||
kras 8种分型试剂对0.01%、0.1%突变率样品的检测结果(表 6)表明,不同分型试剂对相应的0.1%和0.01%突变率样品的突变检测Ct值均小于30,小于相应野生型模板检测Ct值,且与野生型模板检测的Ct值差值即△Ct值均大于3,初步判定本发明kras分型试剂突变检测的灵敏度均可达到0.01%,分型检测0.01%突变检测Ct临界值可取Ctwild和Ct 0.01%中间的数值。
| Typing reagent | Ct wild | Ct mut0.01% (ΔCt) | Ct mut0.1% (ΔCt) |
| kras-G34A | 31.83 | 28.16 (3.68) | 24.77 (5.32) |
| kras-G34T | 33.20 | 27.79 (7.21) | 25.21 (9.79) |
| kras-G34C | 35.00 | 26.93 (8.07) | 24.15 (10.85) |
| kras-G35T | 34.15 | 28.68 (6.32) | 24.68 (10.32) |
| kras-G35A | 33.33 | 28.98 (4.35) | 25.93 (7.40) |
| kras-G35C | 35.00 | 27.32 (7.68) | 24.78 (10.22) |
| kras-G37T | 35.00 | 27.62 (7.38) | 23.22 (11.78) |
| kras-G38A | 35.00 | 27.39 (7.61) | 24.89 (10.11) |
| Note: Ct values were calculated as average Ct from samples of triplex reactions. If there were no available Ct values for certain samples, the number 35 was used to calculate the Ct values in order to get informative data. | |||
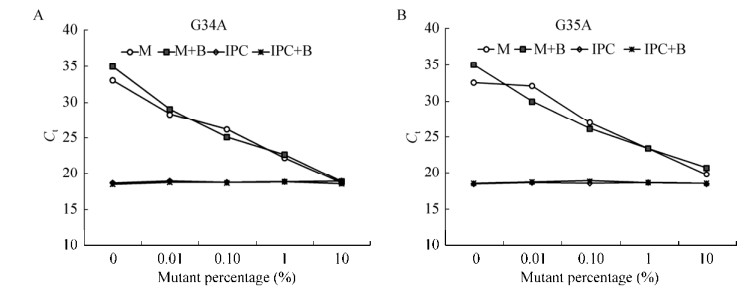
对40例NSCLC组织样本提取的DNA,分别采用TB-ARMS技术和ADx-ARMS技术进行检测,其中35例样品检测结果一致,5例样品两种方法检测结果不一致。5例检测不一致样品中2例样品分型结果不一致,3例样品ADx-ARMS商品化试剂盒未检测到突变。对该5例检测不一致样品进一步进行测序验证,结果(见表 7和图 2)表明TB-AMRS技术的检测结果与测序结果一致。
| Sample | ADx-ARMS | TB-ARMS | Sequencing |
| U6 | WT | G37T | G37T |
| U8 | WT | G35T | G35T |
| U16 | WT | G37T | G37T |
| #13 | G34A | G34T | G34T |
| #14 | G34A | G34T | G34T |
|
| 图 2 五例检测不一致样品的TB-ARMS qPCR检测结果和测序结果 Fig. 2 TB-ARMS qPCR results and sequencing results of five inconsistent samples. |
| |
血浆DNA检测结果与配对组织样品测序结果的比较见表 8和图 3,表明血浆DNA样品检测结果与配对组织样品测序结果完全一致。
| Sample ID | Ct | Results | |||||||
| G34A | G34T | G34C | G35T | G35A | G35C | G37T | G38A | ||
| S1 | 34.47 | 31.92 | 30.53 | – | 8.56 | 22.11 | 24.39 | 27.01 | G35A |
| S2 | 32.59 | – | – | 31.48 | 34.32 | – | – | – | N |
| S3 | 19.30 | 19.35 | – | – | 6.18 | 15.94 | 19.07 | 19.38 | G35A |
| S4 | – | – | – | 13.88 | – | – | – | – | G35T |
| S5 | – | – | – | – | – | – | – | – | N |
| S6 | – | – | – | – | – | – | – | – | N |
| S7 | – | – | – | 33.87 | 31.27 | – | – | – | N |
| S8 | – | – | – | – | – | 34.47 | 34.39 | – | N |
| S9 | – | – | – | – | – | – | – | – | N |
| S10 | – | – | – | – | – | – | – | – | N |
| S11 | 34.73 | – | – | – | 6.24 | 24.45 | 23.22 | 26.85 | G35A |
| S12 | – | – | – | – | 10.43 | – | 39.32 | – | G35A |
| S13 | – | – | – | – | 14.58 | – | – | – | G35A |
| S14 | – | – | – | – | – | – | – | – | N |
| S15 | – | – | 31.44 | 34.90 | 34.64 | – | – | – | N |
| S16 | – | – | – | – | – | – | – | – | N |
| S17 | – | – | – | 7.04 | – | – | 25.96 | – | G35T |
| S18 | – | – | – | – | – | – | – | – | N |
| S19 | – | – | – | – | – | – | – | – | N |
| S20 | – | – | – | – | – | – | – | – | N |
| Note: “–” in Ct value shows Ct value is not available due to the limited cycle number. Ct < 30 was used as cutoff value and the genotype with the minimum Ct value is determined as mutation type if there are more than one mutation types. Result “N” means mutation negative. The minimum Ct value for each sample is highlighted in bold, which corresponds to the relevant mutation type. | |||||||||
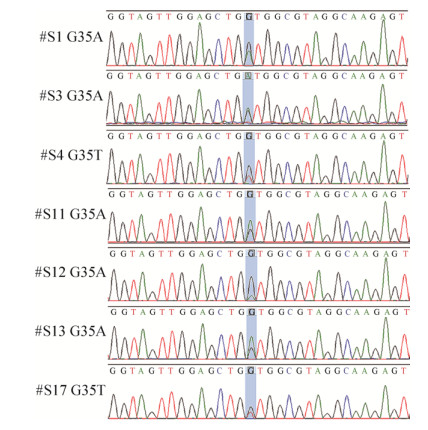
|
| 图 3 突变阳性组织样品测序结果 Fig. 3 Sequencing results of mutant tissue samples. |
| |
当使用基于PCR方法的kras基因突变检测时,样本中的突变型等位基因的扩增会被野生型基因干扰,尤其在高野生型基因背景下检测稀少的突变时,突变型等位基因特异性引物会错配至野生型等位基因并发生延伸,从而产生假阳性结果。目前常用的kras点突变检测PCR方法大都为ARMS方法及其改进的技术方法,原理都是通过等位基因特异性引物3′末端的特异性碱基区分突变和野生型kras基因。仅通过ARMS方法对点突变的检测灵敏度只能达到1%,即最低只能检测到1%突变率的样品。而对于大量野生型基因背景下的稀少突变,例如0.1%突变率的样品,ARMS技术方法的应用将受到限制。高野生型背景下稀少的突变检测依赖于抑制样品中野生型等位基因的扩增,不少研究[18-21]均已证明野生型等位基因特异性封闭引物有助于增加突变检测的特异性,我们的研究也作了进一步验证,采用封闭引物kras G34A和G35A突变检测的灵敏度和特异性均可获得改善,甚至提高到10倍。有研究对封闭引物进一步采用MGB修饰或锁核酸修饰来增加其Tm值从而增强其封闭效果,虽然这样可增强其结合能力但引物合成的成本较单纯磷酸化修饰大大增加并对突变检测亦产生抑制。例如Huang等[22]采用6个锁核酸修饰碱基的WTB封闭引物,明显提高了WTB的Tm值,对野生型模板的封闭能力显著增强,但亦需要采用较长的36碱基的等位基因特异性引物以增加其Tm值可与WTB竞争,减少WTB对突变检测的抑制,均使其应用成本增加。本研究的封闭引物仅3′末端进行磷酸化修饰或采用3个锁核酸修饰碱基,其亦可以达到较好的检测效果,具有更高的性价比。
同时对含有野生型等位基因样品中的突变型等位基因的特异性检测依赖于对突变型等位基因的选择性扩增,等位基因特异性引物3′端仅有一个区分碱基有时不足以抑制野生型模板的结合,在靠近3′端的适当位置再人为引入一个突变碱基,会进一步降低野生型模板的结合能力。Xue等[23]的研究即采用引入错配碱基的方法改善了基因分型检测中等位基因特异性引物的检测特异性,我们的研究也表明通过引入适当突变碱基可改善某些kras突变等位基因特异性引物的检测特异性,如果扩增效率按100%计算,最高可改善达10倍。
此外,通过富集扩增的反应条件我们亦改善了kras突变检测的特异性,与常规检测条件相比,我们的富集扩增反应条件对0.01%和0.1%低突变率样品检测的△Ct值可增加至少1,如果扩增效率按100%计算,则我们的富集扩增条件可使突变检测特异性改善2倍以上。
通过采用多种组合手段,文中的TB-ARMS技术kras点突变检测的灵敏度可达到0.01%,较单纯ARMS方法检测灵敏度可提高100倍以上。与现有商品化试剂盒相比具有更高的检测灵敏度和准确性,证明了TB-ARMS技术检测灵敏度优于单纯的ARMS技术方法。此外我们的研究方法对模板具有更广的检测范围,模板量10–135 ng不影响突变检测的特异性,并且由于检测特异性的改善可在同一反应体系中进行kras多种点突变的同时检测,因此在实际应用中将会更加方便。
液态活检作为伴随诊断的无创检测方法受到越来越多的关注,成为研究的热点之一,血液是最常用的液态活检样品。已有研究[24]表明肺癌组织样品中常常存在低突变率的kras突变,血液中的kras突变率可能会更低,甚至不到0.1%的突变率。现有的ARMS检测技术受灵敏度限制不适宜血液等样品的检测,TB-ARMS技术突变检测的灵敏度大大提高,通过对20例术前血浆样本和配对组织样品的对照检测,两者检测结果的一致亦验证了TB-ARMS技术方法的检测准确性,适用于液体活检样本的检测。
除了应用于kras基因点突变的检测,TB-ARMS突变检测方法只要遵循相应的引物设计原则和应用方法,理论上适用于所有野生型背景下的基因点突变检测,例如可应用于与癌症个体化治疗和预后预测相关的braf、pi3k、tp53等其他基因的点突变检测[25-26],因此具有广泛的应用前景。随着基因突变检测技术的发展,虽然出现了一系列高灵敏度的突变检测方法,包括第二代测序技术、数字化PCR等,相较于这些高成本的检测方法,改进的TB-ARMS荧光PCR点突变检测方法具有低成本、简单方便的应用特点,检测时间只需大约1.5 h,在荧光定量PCR仪临床应用已经十分普及的今天,更适宜在临床检测中广泛应用。
| [1] | Li SQ, Balmain A, Counter CM. A model for RAS mutation patterns in cancers: finding the sweet spot. Nat Rev Cancer, 2018, 18(12): 767–777. DOI: 10.1038/s41568-018-0076-6 |
| [2] | Inamura K. Clinicopathological characteristics and mutations driving development of early lung adenocarcinoma: tumor initiation and progression. Int J Mol Sci, 2018, 19(4): 1259. DOI: 10.3390/ijms19041259 |
| [3] | Lindsay CR, Jamal-Hanjani M, Forster M, et al. KRAS: reasons for optimism in lung cancer. Eur J Cancer, 2018, 99: 20–27. DOI: 10.1016/j.ejca.2018.05.001 |
| [4] | Chuang HC, Huang PH, Kulp SK, et al. Pharmacological strategies to target oncogenic KRAS signaling in pancreatic cancer. Pharmacol Res, 2017, 117: 370–376. DOI: 10.1016/j.phrs.2017.01.006 |
| [5] | Molinari C, Marisi G, Passardi A, et al. Heterogeneity in colorectal cancer: a challenge for personalized medicine?. Int J Mol Sci, 2018, 19(12): 3733. DOI: 10.3390/ijms19123733 |
| [6] | Marrugo-Ramírez J, Mir M, Samitier J. Blood-based cancer biomarkers in liquid biopsy: a promising non-invasive alternative to tissue biopsy. Int J Mol Sci, 2018, 19(10): 2877. DOI: 10.3390/ijms19102877 |
| [7] | Sorber L, Zwaenepoel K, Deschoolmeester V, et al. Circulating cell-free nucleic acids and platelets as a liquid biopsy in the provision of personalized therapy for lung cancer patients. Lung Cancer, 2017, 107: 100–107. DOI: 10.1016/j.lungcan.2016.04.026 |
| [8] | Furuki H, Yamada T, Takahashi G, et al. Evaluation of liquid biopsies for detection of emerging mutated genes in metastatic colorectal cancer. Eur J Surg Oncol, 2018, 44(7): 975–982. DOI: 10.1016/j.ejso.2018.01.224 |
| [9] | Tu M, Chia D, Wei F, et al. Liquid biopsy for detection of actionable oncogenic mutations in human cancers and electric field induced release and measurement liquid biopsy (eLB). Analyst, 2016, 141(2): 393–402. DOI: 10.1039/C5AN01863C |
| [10] | Guo NN, Lou F, Ma YF, et al. Circulating tumor DNA detection in lung cancer patients before and after surgery. Sci Rep, 2016, 6: 33519. DOI: 10.1038/srep33519 |
| [11] | Wang C, Chen XM, Wu YY, et al. Lateral flow strip for visual detection of K-ras mutations based on allele-specific PCR. Biotechnol Lett, 2016, 38(10): 1709–1714. DOI: 10.1007/s10529-016-2161-9 |
| [12] | Sun HY, Yang Y, Yang LX, et al. Snapback primer mediated clamping PCR for detection of EGFR and KRAS mutations in NSCLC patients by high resolution melting analysis. Biomed Res Int, 2014, 2014: 407537. |
| [13] | Matsunaga M, Kaneta T, Miwa K, et al. A comparison of four methods for detecting KRAS mutations in formalin-fixed specimens from metastatic colorectal cancer patients. Oncol Lett, 2016, 12(1): 150–156. DOI: 10.3892/ol.2016.4576 |
| [14] | Cortes U, Guilloteau K, Rouvreau M, et al. Development of pyrosequencing methods for the rapid detection of RAS mutations in clinical samples. Exp Mol Pathol, 2015, 99(2): 207–211. DOI: 10.1016/j.yexmp.2015.07.003 |
| [15] | Jing CW, Mao XH, Wang Z, et al. Next-generation sequencing-based detection of EGFR, KRAS, BRAF, NRAS, PIK3CA, Her2 and TP53 mutations in patients with non-small cell lung cancer. Mol Med Rep, 2018, 18(2): 2191–2197. |
| [16] | Garcia J, Forestier J, Dusserre E, et al. Cross-platform comparison for the detection of RAS mutations in cfDNA (ddPCR Biorad detection assay, BEAMing assay, and NGS strategy). Oncotarget, 2018, 9(30): 21122–21131. |
| [17] |
Luo YW, Li Y. Detection of kras mutation in colorectal cancer patients' cfDNA with droplet digital PCR.
Chin J Biotech, 2018, 34(3): 407–420.
(in Chinese). 罗宇文, 李瑶. 基于微滴式数字PCR检测肠癌病人游离环状DNA KRAS突变的新方法. 生物工程学报, 2018, 34(3): 407-420. |
| [18] | Huang MMC, Leong SM, Chua HW, et al. Highly sensitive KRAS mutation detection from formalin-fixed paraffin-embedded biopsies and circulating tumour cells using wild-type blocking polymerase chain reaction and Sanger sequencing. Mol Diagn Ther, 2014, 18(4): 459–468. DOI: 10.1007/s40291-014-0098-z |
| [19] | Jia Y, Sanchez JA, Wangh LJ. Kinetic hairpin oligonucleotide blockers for selective amplification of rare mutations. Sci Rep, 2014, 4: 5921. |
| [20] | Qu SF, Liu LC, Gan SZ, et al. Detection of low-level DNA mutation by ARMS-blocker-Tm PCR. Clin Biochem, 2016, 49(3): 287–291. DOI: 10.1016/j.clinbiochem.2015.07.012 |
| [21] | Kim H, Ruby AE, Shandilya HG, et al. T-blocker: a simple and robust probe-free quantitative PCR assay to detect somatic mutations down to 0.1% frequency. Biotechniques, 2018, 65(4): 205–210. DOI: 10.2144/btn-2018-0111 |
| [22] | Huang JF, Zeng DZ, Duan GJ, et al. Single-tubed wild-type blocking quantitative PCR detection assay for the sensitive detection of codon 12 and 13 KRAS mutations. PLoS ONE, 2015, 10(12): e0145698. DOI: 10.1371/journal.pone.0145698 |
| [23] |
Xue L, Ye YX, Yi GH, et al. Analysis of CYP2C19 genotypes in Chinese population using allele-specific PCR.
J Hainan Med Univ, 2010, 16(9): 1101–1105, 1110.
(in Chinese). 薛丽, 叶玉兴, 易国辉, 等. AS-PCR方法检测中国人群CYP2C19基因多态性. 海南医学院学报, 2010, 16(9): 1101-1105, 1110. |
| [24] | Myers MB, McKim KL, Meng FX, et al. Low-frequency KRAS mutations are prevalent in lung adenocarcinomas. Personal Med, 2015, 12(2): 83–98. DOI: 10.2217/pme.14.69 |
| [25] | Tsilimigras DI, Ntanasis-Stathopoulos I, Bagante F, et al. Clinical significance and prognostic relevance of KRAS, BRAF, PI3K and TP53 genetic mutation analysis for resectable and unresectable colorectal liver metastases: A systematic review of the current evidence. Surg Oncol, 2018, 27(2): 280–288. DOI: 10.1016/j.suronc.2018.05.012 |
| [26] | Salemi R, Falzone L, Madonna G, et al. MMP-9 as a candidate marker of response to BRAF inhibitors in melanoma patients with BRAFV600E mutation detected in circulating-free DNA. Front Pharmacol, 2018, 9: 856. DOI: 10.3389/fphar.2018.00856 |
 2019, Vol. 35
2019, Vol. 35