中国科学院微生物研究所、中国微生物学会主办
文章信息
- 沈应博, 史晓敏, 沈建忠, 汪洋, 王少林
- Shen Yingbo, Shi Xiaomin, Shen Jianzhong, Wang Yang, Wang Shaolin
- 全基因组测序与生物信息学分析在细菌耐药性研究中的应用
- Application of whole genome sequencing technology and bioinformatics analysis in antimicrobial resistance researches
- 生物工程学报, 2019, 35(4): 541-557
- Chinese Journal of Biotechnology, 2019, 35(4): 541-557
- 10.13345/j.cjb.180350
-
文章历史
- Received: August 30, 2018
- Accepted: December 17, 2018
二十世纪末期人们对细菌耐药性的忽略及抗生素的大量使用导致耐药细菌的广泛流行,对临床抗感染治疗造成了极大的威胁。而且耐药细菌能够在动物、环境、食品等环节间相互传播,使细菌耐药性逐渐成为全世界医学、政界、媒体等各界广泛关注的重大公共卫生安全问题[1-3]。细菌耐药性问题日趋严重,目前每年约有70万人死于耐药菌造成的感染性疾病,英国经济学家Jim O’Neill预测2050年该数据可能会上升到每年1 000万人,并累计给全世界GDP (Gross domestic product)造成高达100万亿美元的损失[4]。随着多重耐药菌甚至泛耐药菌的广泛流行,尤其是对目前临床治疗细菌感染的两类“最后一道防线”药物——碳青霉烯和多黏菌素耐药的菌株,严重威胁了动物及人类的健康。其中最重要的两个基因碳青霉烯耐药基因blaNDM-1[5]和多黏菌素耐药基因mcr-1[6]均是由质粒介导的可转移耐药基因,可在不同种属细菌或不同媒介间相互传播扩散,同时还可与其他耐药基因共存而成为“超级细菌”[7-12],使得治疗这些细菌造成的感染性疾更加困难。面对越来越严峻的细菌耐药性形势,加强耐药菌株的监控以及耐药菌株特征的分析显得尤为重要。精确、快速、便捷地获得耐药菌株的详尽信息(包括耐药基因、毒力因子、质粒分型等特征)以及菌株间亲缘关系等数据对于对抗细菌耐药性将会有很大的帮助。随着科学技术的发展,全基因组测序技术(Whole genome sequencing,WGS)或称为二代测序技术(Next-generation sequencing,NGS),同时结合生物信息学分析技术(Bioinformatics analysis)已逐渐成为科学界研究细菌耐药性的重要技术手段[13]。
全基因组测序技术不仅可获得单一菌落的基因组信息,还可获得混合基因组的信息(Metagenomics,宏基因组),即包括非常规培养和不可培养的细菌DNA信息。相较于Sanger测序(一代测序),WGS无需针对不同DNA片段或细菌种属设计特定引物,而是测序获得随机的序列后装配成相对完整的基因组。然而不同测序平台对每段序列的读取长度的差异,导致测序结果的不同[14-15]。通过WGS不仅可获得近乎完整的细菌DNA信息,包括种属(Species)、耐药基因(Antimicrobial resistance genes)、毒力因子(Virulence-associated genes)、转移元件(Mobile elements)等信息,还可对多个细菌间的基因组信息进行比较,对于耐药菌株的分子流行病学和传播机制研究至关重要。
WGS自问世以来因其价格昂贵,一直未被广泛使用,近年来,各大测序平台的发展致使价格持续下降,该技术被广泛运用于基础和临床细菌耐药性的研究中[16-17]。虽然WGS技术在通量方面有优势,但因其测序读长有一定的局限性(小于600 bp),而无法获得细菌完整的DNA信息。然而三代测序技术(Third-generation sequencing)则突破了WGS在读长方面的壁垒,实现了无需PCR扩增对每条DNA分子进行单独测序,平均读长可达10 kb甚至更长,可越过一些二代测序技术难以测通的重复序列[18]。对于二代及三代测序技术而言,从样品DNA提取到数据分析(图 1),每一个步骤使用的方法不同获得的数据质量也大相径庭,最终都将对结果造成不同程度的影响。
近年来,随着WGS技术在细菌耐药性领域的应用,科学家们借助该技术取得了重大的研究进展。例如在质粒介导的多黏菌素耐药基因mcr-1-mcr-8[6, 19-23]以及碳青霉烯耐药基因blaNDM-17[24]等新型耐药基因的发现过程中,WGS技术发挥着不可替代的作用。Hadziabdic等[25]、Kröger等[26]、Porse等[27]利用WGS技术与生物信息学分析方法阐明了耐药基因、耐药质粒的进化过程;Holt等[28-29]、Wong等[30]、Duchêne等[31]通过该技术对耐药菌或病原菌(志贺杆菌、多重耐药沙门菌)在某个地区或全球范围内的分子进化过程进行了详细的研究,分析了其起源及进化因素,为细菌耐药性的控制提出了新的科学依据。另一方面,WSG数据不仅可以在微观世界给予我们很多信息,而且可以结合宏观数据在耐药性的进化以及其相关风险因素的分析中发挥着重要作用。例如,Shen等[32]、Wedley等[33]将WGS数据与宏观数据进行联合分析,获得了耐药菌/基因的产生或传播的相关风险因素。综上所述,WGS技术助力了细菌耐药性领域的重大发展,已逐渐成为推动该研究领域发展的重要技术手段,但是由于该技术存在较高的技术壁垒,导致其受众面相对较窄。因此,我们将针对二代全基因组测序技术(由于三代测序技术价格颇高暂未广泛应用,所以本文仅作简单介绍)从文库构建、平台测序、数据分析(组装拼接、基本特征分析、核苷酸多样性分析等)三大方面系统地评估不同环节中不同方法的特点,为细菌耐药性中单一细菌的研究提供帮助。此外,宏基因组技术目前在耐药性研究方面的应用相对较少,且与单基因组测序在文库构建、平台测序两方面基本相同,仅在数据分析方面有所不同,因此本文在此不再单独详细介绍。
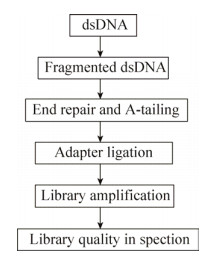
1 文库构建对于WGS技术而言,虽然对最终的结果影响最大的是样品DNA的提取质量[34],但DNA文库构建的影响也不可忽视。随着技术的发展,二代测序DNA文库的构建方法越来越多,虽各有千秋,但其原理和流程基本一致(图 2)。
|
| 图 2 DNA文库构建流程图 Fig. 2 Flowchart of DNA library preparation. |
| |
DNA文库构建的核心步骤是DNA片段化,常用的方法有物理破碎(Physical)、酶切(Enzymatic)和化学打断(Chemical)。其中应用最广的是物理破碎(如超声破碎法)或酶切法(如非特异性核酸内切酶和转座酶Tagmentation)[35]。超声破碎通常使用Covaris仪器(Covaris,Woburn,MA)和Bioruptor仪器(Diagenode,Belgium)。酶切法主要包括使用DNaseⅠ或者Fragmentase或者两者混合物(New England Biolabs,Ipswich,MA)将DNA片段化。虽然两种方法均可以将DNA片段化并获得有效的测序结果,但相较于物理破碎方法,酶切法会产生更多人为的DNA片段插入或缺失。新的酶切法——Illumina’s Nextera tagmentation (Illumina,San Diego,CA)的出现,不仅减少了人为引入的错误,而且大大缩短了DNA文库构建的时间[35]。然而文库质量与DNA的GC含量和片段化方法有着很大关系,对于GC含量高或低的样品,使用物理片段化方法效果更好。
目前市面上存在很多商业化的DNA文库构建试剂盒,随着试剂盒的快速更新换代,其价格下降的同时文库质量在提高,并且初始DNA量的要求也在降低。尽管如此,一个重要的原则是DNA初始量越大,后续需要扩增的循环越少,得到基因组信息越接近真实。KAPA Hyper Prep Kit Illumina® platforms (Kapa Biosystems,Wilmington,MA)试剂盒在DNA初始量足够的情况下可实现无PCR扩增完成DNA文库的构建。众多的商业化试剂盒中均有详细的流程,在具体过程中有些许差别,在此我们仅对几款运用广泛的试剂盒在关键的几个步骤上进行整体的比较(表 1)。末端修复、加A尾、接头连接以及PCR扩增是DNA文库构建中关键的步骤,其中主要的优化程序是将末端修复和加A尾这两步进行合并以缩短总体的时间。然而只有少部分如Truseq®DNA PCR-free和KAPA的两款试剂盒可以做到PCR-free,这不仅能减少文库构建时间,也能最大程度地保证DNA的真实性,减少PCR扩增带来的偏差。此外,磁珠纯化是各类试剂盒中不可缺少的步骤,该步骤费时费力,且磁珠的价格昂贵,若能在此步骤上进行优化将会对DNA文库的构建效率有很大的提高。
| DNA library preparation kit | Company | DNA (ng) | Index | End repair | Post-ligation cleanup | A-tailing | Post-ligation cleanup | Adapter ligation | Post-ligation cleanup | PCR amplication & post-ligation cleanup |
| NEBNext® | New England Biolabs®inc. | 500 | Sanger | √ | √ | √ | √ | √ | √ | √ |
| NEBNext® UltraTM | New England Biolabs®inc. | 500 | Sanger | a | √ | √ | √ | |||
| SureSelectXT | Agilent | 500 | Sanger | √ | √ | √ | √ | √ | √ | √ |
| Truseq®Nano | Illumina® | 500 & 100 | Sanger & Illumina | √ | √ | √ | √ | √ | √ | |
| Turseq®DNA PCR-free | Illumina® | 500 | Sanger & Illumina | √ | b | √ | √ | √ | ||
| KAPA Hyper | KAPA Biosystems | 500 | Sanger | a | √ | √ | c | |||
| KAPA HyperPlusd | KAPA Biosystems | 500 & 20 | Sanger | √ | √ | √ | c | |||
| a: end repair & A-tailing are performed in the same reaction system; b: size selection after end repair; c: the PCR amplification step is selective. | ||||||||||
目前,三代全基因组测序技术还未得到广泛的应用,其相应的文库试剂盒较少,主要包括PacBio公司和Oxford Nanopore Technology公司推出的一系列试剂盒(SMRTbell Barcoded Adapter Prep Kit、SMRTbell Damage Repair Kit-SPv3、Rapid Barcoding Kit、Rapid Sequencing Kit、Ligation Sequencing Kit、1D^2 Sequencing Kit等)。PacBio公司推出试剂盒的建库流程主要包括DNA片段化、末端修复、接头连接、文库片段纯化、杂交引物和聚合酶绑定,而Oxford Nanopore Technology公司的试剂盒建库流程更加简化。对于两款快速试剂盒(Rapid Barcoding Kit/Rapid Sequencing Kit)而言其工作流程主要包括酶切和接头连接,对于Ligation Sequencing Kit和1D^2 Sequencing Kit这两款试剂盒而言,其工作流程主要包括可选片段化、末端制备、接头连接、磁珠吸附。相较于二代全基因组测序文库试剂盒,三代测序文库试剂盒最大的特点是无需PCR扩增,文库制备耗时较短。相较于PacBio公司,Oxford Nanopore Technology公司推出的试剂盒实现了片段化可选,且无需对文库进行纯化,耗时更少,例如使用Rapid Barcoding Kit/Rapid Sequencing Kit制备测序文库仅需要10 min。
2 测序平台目前市场上的全基因组测序平台主要由Illumina (San Diego)、ThermoFisher (Waltham)、PacBio (Pacific Biosciences)和Oxford Nanopore Technology开发的一系列平台如Miseq、Hiseq、X Ten、Ion、PacBio RSⅡ、PacBio Sequel以及MinION等,不同平台的测序技术大相径庭,Illumina测序平台主要通过读取不同色荧光标记的可逆终止核苷酸的图像得到最终的序列结果[37];ThermoFisher测序平台主要是使用半导体芯片计算每添加一个核苷酸pH值的改变而推算得到最后的序列结果[38];PacBio测序平台主要通过纳米技术和现代光学系统对单分子合成中的碱基磷酸基团上的荧光信号进行识别,并将荧光信号转化为序列结果[39];Oxford Nanopore Technology测序平台主要通过将单分子碱基穿过纳米孔蛋白的电流信号转换为序列结果[40]。在数据产出、运行时间、序列读长等指标上不同公司的测序平台甚至同一公司不同系列的平台均存在较大的差别,可根据具体的情况选择最适合的平台(表 2)。
| Company | Sequencing platform | Reads length (bp) |
The number of flow cell/Run | Run time (h)a |
Output |
| Illumina | HiSeq X Ten | 2×150a | 2 | < 72 | 110 Gb |
| Illumina | NovaSeq 6000 | 2×150a | 2 | 40 | 1.2-1.4 Tb |
| Illumina | NextSeq | 2×50 & 2×75a | 1 | 29 | 20-120 Gb |
| Illumina | MiSeq | 2×300a | 1 | 56 | 0.3-15 Gb |
| ThermoFisher | Ion PGMTM | 200a 400a |
1c | 2.3e 3.7e | 10M-1 Gb |
| ThermoFisher | Ion S5TM | 200a 400a |
1c | 4.5f 10.5f | 0.6-15 Gb |
| ThermoFisher | Ion S5TMXL | 200 & 400a | 1c | 2.4-4f | 0.6-15 Gb |
| PacBio | PacBio RSII (P6-C4) | > 2×104 a | 16d | 57.4 | 8-16 Gbg |
| PacBio | PacBio Sequel system | > 2×104 a | 16d | 574.2 | 80-160 Gbg |
| Oxford Nanopore Technology | Oxford Nanopore MinION Mk (1D) | > 882 000b | 1 | 71.8 | 10-20 Gb |
| Oxford Nanopore Technology | Oxford Nanopore MinION Mk (2D) | > 882 000b | 1 | 71.8 | 10-20 Gb |
| a Manufacture’s data. b Previously reported data. c The number of chip/run. d The number of SMRT cells. e Ion 314TMChip v2 or Ion 314TMChip v2 BC. f Ion 510Chip. g For 16 SMRT cells. | |||||
每一条读长(Reads)的DNA信息将存储于FASTQ格式的文件中,并通过组装拼接算法将原始序列(Raw reads)拼接成更长的片段(Contig或者Node)。尽管我们希望拼接得到的序列可以代表完整的基因组信息,但由于二代测序技术在测序读长上的劣势,而细菌基因组上又存在许多比单一读长还长的重复序列无法被测序拼接,因此导致基因组序列拼接后出现多个裂缝(Gap)而不完整。三代测序技术具有读长上的优势,可以不被重复序列所限制,所以能够得到完整的基因组信息。对于太小的质粒(小于10 kb),Pacbio平台在文库构建时反而容易将其忽略[39, 41],所以对于含有小质粒的菌株需要注意所构建的文库的片段大小。由于序列拼接需要处理庞大的数据占用较多的计算机资源,因此对计算机的硬件要求相对较高,且多数软件属于命令类型(Command line)需要通过终端(Terminal)代码运行,不易操作。此外,商业化的软件可提供可视化的多功能操作平台,但并非所有实验室能够承受其昂贵的价格。
对于二代测序短读长(Short reads)的结果,最常用的拼接方法是基于德布鲁因图(de Bruijn Graph,DBG)算法开发的软件,而该算法最常见的问题则是当不同短序列间有重复时很难分辨其中错误的碱基,这将导致重复部分的序列在拼接过程中被排除[42]。为解决这个问题,在该算法的基础上进一步将原始短序列分割成更小的序列,即为k-mers,随后被降低到k-1 mers (图 3),并通过欧拉算法(Eulerian walk)获得最短的k-1 mers的可能路径,从而将序列拼接起来,减少重复区域的错误拼接。

|
| 图 3 DBG算法与k-mers的简易示意图 Fig. 3 Simple schematic diagram of DBG algorithm and k-mers. |
| |
Velvet是一款基于DBG算法的软件,用于重测序(de novo)数据的组装拼接[43]。该软件包括velveth和velvetg两个组件,前者用于k-mer的构建,后者用于k-mer阵列的图形搭建。VelvetOptimizer是一个Perl脚本由Simon Gladman和Torsten Seemann开发,用于参数的自动优化(http://www.vicbioinformatics.com/software.velvetoptimiser.shtml)。通过k-mers和DBG算法的合用,Velvet可增加拼接富含重复序列菌株的可能性。
SPAdes也是一款基于DBG算法开发的适用于多种测序平台重测序数据的拼接软件[44]。该算法通过以下4个步骤:1)组装图通过错误校准算法后被构建为多片段大小的图;2)通过k-mer和DBG两种方法进行估计;3)构建配对的组装图被构建;4)得到拼接序列的合集后通过原始序列与该结果比对校正后得到最终结果。此外,该软件还推出了一种新的针对质粒拼接的算法[49]。
IDBA-UD同样是基于DBG算法针对短读长序列拼接而开发的软件[45]。该软件使用相邻序列的测序深度(Depth)来改进相关序列测序深度的阈值,然后通过本地配对序列的组装来减少重复序列造成的间隔(Gap),通过这样的方法可以减少在短读长序列中遗失的k-mers中的序列信息。但由于该程序只提供命令类型的版本,用户界面不够友好,操作较困难而不推荐使用。
RAY依旧是一款基于DBG算法的程序,但其特殊之处在于依赖欧拉距离算法,这种算法定义某一特殊序列子集为种子(Seeds)并通过添加这些种子对拼接的序列进行延伸[42]。这样的延伸过程被启发法(Heuristics)或命令法(Commands)控制,将在种子与序列无交叉重叠时停止。这样拼接出来的序列长度相对较短,但是错误较少。表 3列出了几种常用的序列拼接软件及其各自的特点。
| Software | Hardware requirements | Calculation speed | Assembly quality | Source of input data | Types of software |
| Velvet[43] | Low | Medium | Low | Illumina | Command-line |
| SPAdes[44] | Low | Low | Medium | Illumina, Ion Torrent, PacBio, Oxford Nanopore | Command-line |
| IDBA-UD[45] | Low | Low | Medium | Illumina | Command-line |
| RAY[42] | Low | Fast | Low | 454, Illumina, Ion Torrent | Command-line |
| HGAP[46] | High | Medium | High | PacBio | Command-line/Webpage |
| Canu[47] | Low | Low | High | PacBio, Oxford Nanopore | Command-line |
| Unicycler[48] | Low | Low | High | Illumina, PacBio, Oxford Nanopore | Command-line |
对于三代测序结果而言,拼接方法多是基于Overlap Layout Consensus (OLC)算法或DBG算法[50]。三代测序技术的超长reads,导致其单碱基随机错误率较高,所以三代测序的原始reads不推荐直接用于组装。为解决该问题,三代测序结果在组装前需对reads进行校正。部分三代测序结果的组装程序需要用二代测序结果对三代测序结果进行清洗,例如PacBioToCA[51]、LSC[52]等,而另一部分程序则可通过三代测序结果中的短序列对长序列进行校正,如HGAP (Hierarchical genome-assembly process)在小型基因组的校正和组装中表现良好[46]。HGAP首先对三代测序序列按照一定规则进行有序排序,选出其中较长的序列作为“种子”序列,然后用三代数据中的较短序列校正较长序列的错误,将校正后的“种子”序列进行组装[46]。
由于MinION测序的发展晚于Pacbio,所以关于MinION测序数据的组装软件多数是在PacBio组装软件的基础上发展而来或是同时适用于两者,例如Canu[47]和unicycler[48]等。
Canu是专为单分子序列拼接而设计,由Celera Assembler优化而来,通过三代测序数据进行自身校正拼接[47]。Canu的优化源于新的重叠和拼接算法,包括一种基于tf-idf加权MinHash的适宜重叠策略以及一种避免折叠分叉重复和单倍型的装配算法[47]。Canu运行包括3个阶段:reads校正、reads修剪和单序列构建(Unitig construction)。Unitig指许多短片段交叉重叠后装配的正确有效的长片段。在所有阶段中,第一步均是建立一个短序列重叠数据库(Read and overlap database)。1)校正阶段主要为估计正确的reads,生成正确的reads和选出用于校正的最佳短序列重叠群;2)修剪阶段主要是标识出输入序列重叠群中重叠区域,对其进行修剪,获取基于重叠群延伸开的最长序列;3)装配阶段首先进行错误序列的识别,然后构建重叠序列的最佳overlap直方图(Best overlap graph,BOG),最终输出组装好的序列[47]。
Unicycler是将二代测序技术与三代测序技术的数据结合起来,生成更加精确、完整的基因序列[48]。首先,将短reads进行拼接获得精确、连续的拼接序列。然后,通过长reads确定短reads的最佳拼接方式[48]。主要分为以下7个步骤:1) SPAdes (v3.6.2 or later)构建de Bruijn图形程序集,然后通过Unicycler的相关算法,平衡contigs数与死端(Dead ends)数,从而获得最优的装配图;2)将测序深度和contigs依据贪心(Greedy)算法确定contigs的多重性;3)通过SPAdes获得短reads间的链接序列,从而链接序列将单拷贝contigs连起来;4)将长reads与多个单拷贝contigs进行比对,可以获得单拷贝contigs之间的链接序列,并用单拷贝contigs对长reads进行矫正;5)确定单拷贝contig之间的链接序列的可信度,根据其可信度高低获得短contigs的最佳排列方式;6)基于高质量的链接序列,将contigs进行拼接获得长contigs;7)通过短contigs对长序列进行比对,以减少碱基错配、小序列插入或缺失[48]。
4 数据分析软件通过拼接好的序列可获得细菌的许多信息,对于细菌耐药性研究而言,以下几方面的信息是我们所需要的:1)细菌种属(Species);2)细菌携带耐药基因(Antimicrobial resistance genes,ARGs)、毒力因子(Virulence-associated genes,VAGs)以及插入序列(Insert sequence,IS)的情况等;3)细菌携带质粒(Incomplete types,Inc types)类型;4)细菌的多位点序列分型(Multi-locus sequence type,MLST)等。为得到菌株的这些信息,通常是将拼接好的序列或原始数据用不同的软件与特定的数据库进行比对,寻找对应的基因信息,相应的软件可以按照操作方式划分为网页类型(Web-based tools)和命令类型(Command-line)。
网页类型工具可提供更为直观的用户界面和简便的操作环境。对于细菌种属鉴定来说,KmerFinder是一个很好的工具,如果使用其命令版本处理拼接好的序列(Contigs)只需约9 s,而对于原始数据(Raw reads)则需要大约190 s53]。且该工具已在Center for Genomic Epidemiology (CGE,https://cge.cbs.dtu.dk/services/KmerFinder/)网站上开放使用。另一个使用较多的网页工具是NCBI (National Center for Biotechnology Information)提供的比对工具BLAST (Basic Local Alignment Search Tool,https://blast.ncbi.nlm.nih.gov/Blast.cgi)。该工具由NCBI提供,可使用NCBI上所有的数据库,具有较全面的信息,但解释结果时需要通过对各种参数和结果严格地筛选并结合相应的背景知识去判断,相对繁琐。另一款程序Rapid Annotation using Subsystem Technology (RAST,http://rast.nmpdr.org)[54]能够快速注释基因组信息,但由于参数的设置不同且算法的相对固定导致结果的准确性有所下降。此外,在耐药基因、毒力因子、质粒分型以及菌株分子分型等特征的鉴定方面可分别使用CGE网站中的ResFinder[55]、VirulenceFinder[56]、PlasmidFinder[57]和MLST[58]等工具完成。该网站拥有方便快捷的独立数据分析工具而且还有一个完整的批量数据分析流程(Bacterial Analysis Pipeline)[59],但是有些工具的数据库并不全,例如VirulenceFinder中的数据库仅有李斯特菌Listeria、金黄色葡萄球菌Staphylococcus aureus、大肠杆菌Escherichia coli和肠球菌Enterococcus四个数据库。
在命令类型的工具方面,PathoScope可以直接通过原始数据鉴别细菌的种属而不需要对序列进行拼接组装[60],该分析软件多用于宏基因组数据的菌群结构分析,如肠道微生物[61-62]、皮肤菌群结构[63]的研究。Clinical PathoScope作为该工具为适应临床样本分析而延伸出的附加程序,可在25 min内从多种属的样品中检测到致病菌,且准确率可达94.7%[64]。PROKKA是一款快速注释原核生物基因组信息的软件,该软件可以预测基因的位置和其相应的功能[65]。该软件在4核的电脑上完成大肠杆菌K-12的基因组注释仅需6 min,且准确率可达99.63%,此外一项试验表明PROKKA在预测基因的数量上优于RAST[65]。在Stoesser等[66]、Yang等[67]的研究中,利用该软件对blaNDM基因定位,并对其所在的质粒进行了注释。此外,在耐药基因、毒力因子、质粒类型及分子分型方面,一款快速高效的软件SRST2 (Short read sequencing typing)可通过原始短序列对任何序列的数据库进行比对计算并得到准确的结果[68]。SRST2推荐使用耐药基因数据库ARDB[69]、毒力因子数据库VFDB[70]以及分子分型数据库PubMLST (https://pubmlst.org/databases.shtml)。由于该软件可利用不同数据库完成耐药菌的多种分子特征分析,从而使其成为细菌耐药性研究中应用最广泛的软件之一。例如Shen等[32]、Wang等[71]在其各自的研究中(细菌耐药性的分子流行病学、耐药基因的传播途径等)中均使用了该软件进行分子特征的定义。各类软件信息见表 4。
| Application | Software | Operator interface | Type of input data | Advantage | Disadvantage |
| Assembly | Velvet[43] | Commands | Raw sequences | Suitable for high coverage, short read data sets; automatic parameterization; detailed guidelines | Small N50 contig size; ignore potential correct low coverage sequences; hard to use; large memory usage |
| SPAdes[44, 49] | Commands | Raw sequences | Suitable for multiple platform data; small memory requirement; large N50 contig size; quality control; option to merge contigs from other assemblers; plasmids can be assembled; detailed guidelines | Long-time operation | |
| IDBA-UD[45] | Commands | Raw sequences | Suitable for single-cell sequencing or metagenomic sequencing technologies with uneven sequencing depths; small memory requirement; longer contigs with high accuracy; | Hard to use; no guidelines | |
| RAY[42] | Commands | Raw sequences | Suitable for multiple platform data; high accuracy; automatic parameterization; detailed guidelines | Small N50 contig size; poor performance with lower-quality reads | |
| Species identification | K-merFinder | Webpage | Raw sequences, contigs | No bioinformatics skills required; easy to use; easy to read; possible to detect contamination | Method must be set properly |
| PathoScope[60] | Commands | Raw sequences | Able to detect contamination; quality control; complete workflow; detailed guidelines | Some bacteria may be missing from metagenomic sample; coverage requires more than 20% to distinguish similar strain; long-time operation | |
| NCBI BLAST | Webpage | Contigs | Have largest database; many available tools; | Hard to read; blast knowledge is necessary | |
| Gene annotation | Prokka[65] | Commands | Contigs | Short-time operation; five tools can be run in the same workflow; detailed guidelines | Annotations will be reduced for incomplete sequence; suitable only for single-cell sequencing |
| RAST[54] | Webpage | Contigs | KEGG connection; easy to read | Long-time operation; data must be uploaded to a data server; | |
| Character analysis | Center for Genomic Epidemiology | Webpage | Raw sequences, contigs | Easy to use; easy to read | Long-time operation; some databases are incomplete |
| SRST2[68] | Commands | Raw sequences | Able to Combine with other databases; High accuracy; detailed guidelines | Long-time operation; the database needs to be localized | |
| PubMLST | Webpage | Raw sequences, contigs | All databases can be downloaded; new types of ST can be generated by users | Hard to find correct data; need to share data |
获得菌株内在信息(基本特征)后,进一步研究不同菌株之间的外部联系(亲缘关系),对于是否存在克隆传播或是判断流行株的暴发来源有着重要的意义。上文提及的MLST可作为菌株间关联性的一种指标,但因为这种方法只针对菌株的7个基因进行种类的划分,在精确性上有着一定的局限性,所以我们将针对其他精确性更高的基于细菌间单核苷酸多态性(Single nucleotide polymorphism,SNP)的方法进行讨论。不同的方法在精确性、操作性等方面有着较大的差异,但基本都需要通过代码进行操作且对计算机硬件要求高。
CSI Phylogeny是一款在CGE网站上可用的基于参考序列检测SNP的工具,具有高保守性和准确性高的特点[72]。这款软件使用Burrows-Wheeler Aligner (BWA)将目标序列与参考序列进行比对后通过用户设置的参数提取出SNPs,并可检查SNP是否在所有序列中均存在,最终通过FastTree软件[73]构建最大释然法树。
NDtree是另一款在CGE网站上可用的工具,它可将原始序列生成k-mers后与参考序列进行比对,然后通过公式计算菌株间SNP的数量。这些数据将生成一个矩阵(Matrix)文件并使用Phylip (http://evolution.genetics.washington.edu/phylip.html)计算各菌株间的进化关系。值得一提的是该方法具有保守程度的参数设定,可能会导致不正确的结果。
kSNP3是一款可不使用参考序列并不需提供多序列校准文件的工具[74]。该软件使用k-mer分析去推断SNPs并适用于任何种属细菌。kSNP3可选择对核心SNPs进行分析,生成多个文件包括核心和非核心SNPs信息的文件、Newick格式的简约法(Parsimony)、邻接法(Neighbor-joining)和最大释然法树的文件。Wilson等利用该软件绘制了来源于澳大利亚食品生产链条中耐药李斯特菌的系统发育树,确定了其耐药性的关键遗传标记[75]。
Roary是一款命令类型工具,可快速检测各菌株泛基因组(Pan-genome)中SNPs的情况。由于该软件每个样本需使用注释的拼接序列,所以所有的分析菌株必须来自同一个种[76]。序列中的编码区将会被CD-HIT工具转换为蛋白序列,然后使用BLASTP工具对所有菌株中的蛋白序列进行搜索比对,最终通过Markov cluster algorithm (MCL)算法[77]分为不同组别后与CD-HIT先前转换的序列合并为最后的结果。来自1 000株鼠伤寒沙门菌Salmonella typhimurium序列的分析在单核CPU的计算机上只运行了4.3 h,并且泛基因组的正确率达到了100%[76]。因其快速高效,该软件多用于大量数据的分析,例如Moradigaravand等通过该软件对10年内分离的205株粘质沙雷氏菌进化分析后发现,该菌在进化过程中多次获得不同耐药基因,揭示了多重耐药粘质沙雷氏菌的阳性率逐渐上升的原因[78]。
Pan-Seq是另一款比较序列间泛基因组差异的软件,包括3个组件:Novel region finder (NRF)、Core and accessory genome finder (CAGF)和Locus selector (LS)[79]。NRF工具使用MUMmer[80]鉴别序列间的差异位点后CAGF通过MUMmer alignment将这些差异位点序列添加到初始的泛基因组中,再把泛基因组分成碎片与原始序列进行校对,通过BLASTn算法将不同的碎片分为核心和附加基因组,最终通过LS工具可识别输入序列间不同基因的SNPs。López-Camacho等人利用该软件分析了肺炎克雷伯菌在烧伤重症监护病房内暴发期间的耐药性演变,确定了抗生素选择压力在此次暴发的出现和进化过程中发挥了重要作用[81]。
Lyve-SET是最近新报道的基于参考序列SNP的高质量SNP分析工具。该工具通过序列最小最大覆盖度(Coverage)分辨碱基一致性,并舍弃那些只在单向序列里出现的SNPs,同时可选择性排除特殊的噬菌体(Phage-specific regions)和重复区域(Repeat regions)以提高SNPs的准确性和可靠性[82]。
Harvest是一款基于核心基因组SNP序列分析的软件,可快速提取菌株间核心基因组的SNPs信息并构建进化树,同时包含插件Gingr可动态的可视化数据。该软件既可指定参考序列,也可随机选择参考序列,但是两个菌株间差异过大的话可能会被排除[83]。Shen等基于该软件的系统发育分析,结合种群结构贝叶斯分析,发现来源于中国30个省市287株mcr-1-positive Escherichia coli测序分离株具有4个明显的谱系,且4个谱系与省份无明显相关性[32]。
获取SNPs信息后可通过不同的算法对菌株间的进化关系构建进化树,其中贝叶斯(Bayesian)和最大释然法是最常用的方法,它们相较于邻接法和简约法具有更高的精确性,但是对于大量的样本分析需要耗费过多的计算机资源,存在一定的缺陷[84]。Holt等在2013年利用该软件对越南地区索氏志贺菌的系统发育进行了分析,为病原体在新宿主群体中的微进化提供了一种独特的,高分辨率的研究思路[29]。
Randomized Axelerated Maximum Likelihood (RAxML)是一款以最大释然法为基础的进化树构建软件[85]。该算法首先生成一个假定的进化树,然后经过几步的优化调整后得到新的进化树,当经过优化后的进化树不再增加进化关系的合理性时便停止重复该过程。当用户使用该软件处理核苷酸和SNP数据时,必须使用GTRCAT (GTR,General time-reversible)模型通过添加命令“-m GTRCAT”来校准进化树,但当样本数小于50时,不推荐使用该模型。该软件主要用于大样本数据的系统发育分析,例如Casali等利用该软件完成了1 035株耐药结核杆菌的系统发育分析,阐明了耐药结核杆菌在俄罗斯人群中的演变过程[86]。
FastTree是另一款基于最大释然法开发的软件[73],通过4个阶段完成进化树的构建。1)创建一个起始的进化树和存储内部节点的文件;2)初始进化树的长度通过调换相邻节点和重排分枝后逐渐减少;3)通过数学模型得到最大释然树(CAT);4)进化树的可信度需通过Shimodaira-Hasegawa (SH)测试[87]。同样类似于RAxML软件,FastTree在处理核苷酸和SNP数据时也需要使用GTR+CAT模型通过添加命令“-gtr”来完成进化树的校准。该软件常用于宏基因组数据的“系统发育重建”,对揭示微生态菌群结构的进化过程有一定的作用。
相较于最大释然法,MrBayes则是基于贝叶斯算法设计的一款软件,该软件使用Markov chain Monte Carlo (MCMC)算法,自定义选项和参数设定较多,因此难以操作[88]。
进化树的文件最常用的是NEXUS和Newick格式,许多图形用户界面(GUI,Graphical User Interface)的软件比如FigTree (http://tree.bio.ed.ac.uk/software/figtree/)、MEGA[89]、Archaeopteryx[90]以及网页类型的工具iTOL[91]等均可对进化树进行可视化的分析。相关基因组比对软件以及进化树构建软件见表 5。
| Software | The type of software | Software features | Analysis method | Type of input data | Format of output file |
| CSI Phylogeny 1.4[72] | Webpage | Extract high quality SNPs after comparison with reference sequences | Reference-based | Original sequence, assembled sequence | − |
| NDtree 1.2[72] | Webpage | Create k-mers of reads and map them to a reference sequence; detection of the number of SNPs through a single model | Statistical method | Original sequence | Newick |
| kSNPs [74] | Commands | Obtain SNPs between strains by k-mers analyses | Non-reference-based | Original sequence, assembled sequence | Newick, MSA (Multiple-sequence alignment) |
| Roary [76] | Commands | SNPs analyses for Pan genome | Pan genome | Assembled sequence | FASTA, TXT, CSV, Rtab |
| Pan-Seq [79] | Commands | Pan genome analysis tools; available for core or additional genomes respectively. | Pan genome | Assembled sequence | TXT, FASTA |
| Lyve-SET [82] | Commands | Extract high quality SNPs after comparison with reference sequences | Reference-based | Original sequence, assembled sequence | Matrix, FASTA, Newick, VCF |
| Harvest [83] | Commands | Rapidly extract core genome SNPs from large number of strains and construct a phylogenetic tree | Core genome | Assembled sequence | FASTQ, VCF, XMFA, tree |
| RAxML [85] | Commands | Maximum likelihood phylogenetic tree; long-time operation; high accuracy | Maximum Likelihood | PHYLIP or FASTA | Newick |
| FastTree [73] | Commands | Approximately maximum likelihood phylogenetic tree; fast but slightly less accurate | Maximum Likelihood | PHYLIP or FASTA | Newick |
| MrBayes [88] | Commands | Bayesian-base phylogenetic tree; model definition is complex and difficult to use | Bayesian-based | NEXUS | NEXUS |
随着细菌耐药性问题的日益严重,针对致病性耐药菌暴发甚至共生菌的耐药性监控等相关研究显得越来越重要,尤其是这些研究可作为提出暴发控制、耐药性逆转等理论的基础。值得关注的是,随着全基因组测序技术的迅速发展,测序质量不断提高的同时,测序费用呈显著下降趋势。该技术相较于传统的技术手段,可获得更全面、更准确的结果,将会在未来研究中得到广泛的应用[92]。
除了DNA提取质量上的差别外,文库质量对于测序结果的影响也十分显著,不同DNA文库构建试剂盒有着不同的特点[93]。很显然物理破碎DNA的方法相较于化学法能带来更少的误差对样品GC含量的嗜好也相对较少。令人振奋的是,目前已有许多公司支持机器自动化的操作来构建DNA文库,大量减少了人力和物力,同时减少了人工操作处理样本时带来的实验误差。此外,将文库构建步骤简化、趋向无PCR步骤以及减少磁珠纯化等均可减少文库构建过程中所耗费的时间,也可获得更加趋近于真实的基因组信息。
目前主流的测序平台还是由Illumina公司推出的几款平台,很显然,在数据的产出量上这几款平台有着明显的优势,并且测序结果中的碱基错误率也相对较低,对于大量单菌或是宏基因组的测序推荐选择这类平台。相反的ThermoFisher公司开发的几款测序平台在测序读长上有着很大的优势,但是相对的产出量上则低了很多,这对于紧急情况单菌的测序有着潜在的应用前景。目前,二代测序技术的测序读长仅在300-600 bp之间,对于富含重复序列的细菌来说,难以得到一个完整的结果。针对这样的情况,三代测序技术——PacBio平台[39]以及MinION平台[94]在其测序读长(平均 > 10 kb)上有着很大的优势,能够解决上述二代测序技术所遇到的问题,可以得到细菌的完整基因组信息即完成图。然而测序读长长的同时单碱基的错误率便会升高,且测序深度和产出量均不高,所以若能结合二代测序的数据进行校对,那么得出的结果将会十分可靠。目前Pilon软件即可用二代数据对三代数据进行拼接结果的校准[94]。与此同时,在PacBio平台测序价格高居不下的情况下,测序成本也将增加,虽然MinION平台的价格相对低一些,但Nanopore测序技术还处于初级阶段,该技术及后续的生物信息学分析还需进一步的开发。目前,已有不少研究者选用MinION平台开展耐药性的研究,Li等利用该测序技术结合二代测序技术不仅获得了质粒完整序列[96],而且对插入序列ISCR1在沙门菌中的质粒异质性进行了研究[97]。Ludden等利用同样的技术证明了污水中携带碳青霉烯耐药基因质粒可能在不同细菌之间交换,为环境中耐药基因传播机制的研究提供了一种新的研究方法[87]。
虽然目前有许多商业化的软件可对WGS数据进行分析,例如BioNumerics (Applied Maths,Biomérieux)、CLC Genomic Workbench (Qiagen)和SeqSphere (Ridom)等,这些软件虽可完成多种生物信息学分析,如序列组装拼接、SNPs提取以及进化树构建等,但每个模块需要单独购买使用权。虽然商业化软件价格高昂,但它们具有友好的用户界面,加上有非常详尽的用户手册可以帮助用户去了解和使用它们的功能,降低了使用者的门槛。此外,本文也介绍了少数几款基于网页类型的工具,如包含了众多分析模块的CGE网站,其开源免费的特点造成了巨大的访问量,导致其效率偏低,且由于其属于公共网络,数据的保密措施还有待商榷。与上述工具不同的是,我们主要介绍了多款命令类型的软件,这些软件均为开源免费的,大多数都需要在Linux、Ubuntu、Mac OS X等系统环境下运行,少数可在Window系统下运行,因此限制了这些软件在非生物信息学背景的研究人员中使用。另外在批量处理数据时对系统硬件要求相对较高,所有操作均需使用代码完成,且无可视化的图形操作界面,给无生物信息学背景的分析人员又增添了许多困难。
7 展望全基因组测序技术尤其是长片段测序技术的发展,极大地推动了细菌耐药性的研究进程,尤其是在新耐药基因的发现、耐药基因的传播机制、耐药菌/耐药基因的进化分析以及细菌耐药性的风险因素分析等方面都起到了至关重要的作用。基于“One Health”的理念,耐药性在人、动物与环境之间相互传播的研究还需要更进一步更深入的证据,而WGS技术已逐渐展露出其在细菌耐药性研究中的潜力,可提供全面精确的数据,并将会成为未来多年内最为重要的耐药性研究手段之一。希望通过本文对文库构建、测序平台、数据分析完整流程的详细介绍,能够让更多的研究者对该技术有更全面的了解,并推进此项技术在耐药性领域的应用。
| [1] | World Health Organization. Worldwide country situation analysis: response to antimicrobial resistance. Switzerland: World Health Organization, 2015. |
| [2] | Laxminarayan R, Sridhar D, Blaser M, et al. Achieving global targets for antimicrobial resistance. Science, 2016, 353(6302): 874–875. DOI: 10.1126/science.aaf9286 |
| [3] | Woolhouse M, Ward M, van Bunnik B, et al. Antimicrobial resistance in humans, livestock and the wider environment. Philos Trans Roy Soc B Biol Sci, 2015, 370(1670): 20140083. DOI: 10.1098/rstb.2014.0083 |
| [4] | O'Neil J. Antimicrobial resistance: tackling a crisis for the health and wealth of nations. London: The Review on Antimicrobial Resistance, 2014. |
| [5] | Kumarasamy KK, Toleman MA, Walsh TR, et al. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis, 2010, 10(9): 597–602. DOI: 10.1016/S1473-3099(10)70143-2 |
| [6] | Liu YY, Wang Y, Walsh TR, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis, 2016, 16(2): 161–168. DOI: 10.1016/S1473-3099(15)00424-7 |
| [7] | Dalmolin TV, Castro L, Mayer FQ, et al. Co-occurrence of mcr-1 and blaKPC-2 in a clinical isolate of Escherichia coli in Brazil. J Antimicrob Chemother, 2017, 72(8): 2404–2406. DOI: 10.1093/jac/dkx142 |
| [8] | Sun P, Bi ZW, Nilsson M, et al. Occurrence of blaKPC-2, blaCTX-M, and mcr-1 in Enterobacteriaceae from well water in rural China. Antimicrob Agents Chemother, 2017, 61(4): e02569–16. |
| [9] | Tacão M, dos Santos Tavares R, Teixeira P, et al. mcr-1 and blaKPC-3in Escherichia coli sequence type 744 after meropenem and colistin therapy, Portugal. Emerg Infect Dis, 2017, 23(8): 1419–1421. DOI: 10.3201/eid2308.170162 |
| [10] | Wang Y, Tian GB, Zhang R, et al. Prevalence, risk factors, outcomes, and molecular epidemiology of mcr-1-positive Enterobacteriaceae in patients and healthy adults from China: an epidemiological and clinical study. Lancet Infect Dis, 2017, 17(4): 390–399. DOI: 10.1016/S1473-3099(16)30527-8 |
| [11] | Wang Y, Zhang RM, Li JY, et al. Comprehensive resistome analysis reveals the prevalence of NDM and MCR-1 in Chinese poultry production. Nat Microbiol, 2017, 2: 16260. DOI: 10.1038/nmicrobiol.2016.260 |
| [12] | Zhong LL, Zhang YF, Doi Y, et al. Coproduction of MCR-1 and NDM-1 by colistin-resistant Escherichia coli isolated from a healthy individual. Antimicrob Agents Chemother, 2017, 61(1): e01962–16. |
| [13] | Bertelli C, Greub G. Rapid bacterial genome sequencing: methods and applications in clinical microbiology. Clin Microbiol Infect, 2013, 19(9): 803–813. DOI: 10.1111/1469-0691.12217 |
| [14] | Jünemann S, Sedlazeck FJ, Prior K, et al. Updating benchtop sequencing performance comparison. Nat Biotechnol, 2013, 31(4): 294–296. |
| [15] | Loman NJ, Misra RV, Dallman TJ, et al. Performance comparison of benchtop high-throughput sequencing platforms. Nat Biotechnol, 2012, 30(5): 434–439. DOI: 10.1038/nbt.2198 |
| [16] | Hasnain SE, O'Toole RF, Grover S, et al. Whole genome sequencing: a new paradigm in the surveillance and control of human tuberculosis. Tuberculosis (Edinb), 2015, 95(2): 91–94. DOI: 10.1016/j.tube.2014.12.007 |
| [17] | Lecuit M, Eloit M. The diagnosis of infectious diseases by whole genome next generation sequencing: a new era is opening. Front Cell Infect Microbiol, 2014, 4: 25. |
| [18] | Choi SC. On the study of microbial transcriptomes using second- and third-generation sequencing technologies. J Microbiol, 2016, 54(8): 527–536. DOI: 10.1007/s12275-016-6233-2 |
| [19] | Xavier BB, Lammens C, Ruhal R, et al. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Euro Surveill, 2016, 21(27): pii=30280. DOI: 10.2807/1560-7917.ES.2016.21.27.30280 |
| [20] | Yin WJ, Li H, Shen YB, et al. Novel plasmid-mediated colistin resistance gene mcr-3 in Escherichia coli. mBio, 2017, 8(3): e00543–17. |
| [21] | Carattoli A, Villa L, Feudi C, et al. Novel plasmid-mediated colistin resistance mcr-4 gene in Salmonella and Escherichia coli, Italy 2013, Spain and Belgium, 2015 to 2016. Eurosurveillance, 2017, 22(31): 30589. DOI: 10.2807/1560-7917.ES.2017.22.31.30589 |
| [22] | Borowiak M, Fischer J, Hammerl JA, et al. Identification of a novel transposon-associated phosphoethanolamine transferase gene, mcr-5, conferring colistin resistance in d-tartrate fermenting Salmonella enterica subsp. enterica serovar Paratyphi B. J Antimicrob Chemother, 2017, 72(12): 3317–3324. DOI: 10.1093/jac/dkx327 |
| [23] | Wang XM, Wang Y, Zhou Y, et al. Emergence of a novel mobile colistin resistance gene, mcr-8, in NDM-producing Klebsiella pneumoniae. Emerg Microbes Infect, 2018, 7: 122. |
| [24] | Liu ZH, Wang Y, Walsh TR, et al. Plasmid-mediated novel blaNDM-17 gene encoding a carbapenemase with enhanced activity in a sequence type 48 Escherichia coli strain. Antimicrob Agents Chemother, 2017, 61(5): e02233–16. |
| [25] | Hadziabdic S, Fischer J, Malorny B, et al. In vivo transfer and microevolution of avian native IncA/C2blaNDM-1-Carrying Plasmid pRH-1238 during a broiler chicken infection study. Antimicrob Agents Chemother, 2018, 62(4): e02128–17. |
| [26] | Kröger C, Kary SC, Schauer K, et al. Genetic regulation of virulence and antibiotic resistance in Acinetobacter baumannii. Genes, 2017, 8(1): 12. |
| [27] | Porse A, Schønning K, Munck C, et al. Survival and evolution of a large multidrug resistance plasmid in new clinical bacterial hosts. Mol Biol Evol, 2016, 33(11): 2860–2873. DOI: 10.1093/molbev/msw163 |
| [28] | Holt KE, Baker S, Weill FX, et al. Shigella sonnei genome sequencing and phylogenetic analysis indicate recent global dissemination from Europe. Nat Genet, 2012, 44(9): 1056–1059. DOI: 10.1038/ng.2369 |
| [29] | Holt KE, Thieu Nga TV, Thanh DP, et al. Tracking the establishment of local endemic populations of an emergent enteric pathogen. Proc Natl Acad Sci USA, 2013, 110(43): 17522–17527. DOI: 10.1073/pnas.1308632110 |
| [30] | Wong VK, Baker S, Pickard DJ, et al. Phylogeographical analysis of the dominant multidrug-resistant H58 clade of Salmonella typhi identifies inter- and intracontinental transmission events. Nat Genet, 2015, 47(6): 632–639. DOI: 10.1038/ng.3281 |
| [31] | Duchêne S, Holt KE, Weill FX, et al. Genome-scale rates of evolutionary change in bacteria. Microb Genom, 2016, 2(11): e000094. |
| [32] | Shen YB, Zhou HW, Xu J, et al. Anthropogenic and environmental factors associated with high incidence of mcr-1 carriage in humans across China. Nat Microbiol, 2018, 3(9): 1054–1062. DOI: 10.1038/s41564-018-0205-8 |
| [33] | Wedley AL, Dawson S, Maddox TW, et al. Carriage of antimicrobial resistant Escherichia coli in dogs: prevalence, associated risk factors and molecular characteristics. Vet Microbiol, 2017, 199: 23–30. DOI: 10.1016/j.vetmic.2016.11.017 |
| [34] | Costea PI, Zeller G, Sunagawa S, et al. Towards standards for human fecal sample processing in metagenomic studies. Nat Biotechnol, 2017, 35(11): 1069–1076. |
| [35] | Marine R, Polson SW, Ravel J, et al. Evaluation of a transposase protocol for rapid generation of shotgun high-throughput sequencing libraries from nanogram quantities of DNA. Appl Environ Microb, 2011, 77(22): 8071–8079. DOI: 10.1128/AEM.05610-11 |
| [36] | https://image.baidu.com |
| [37] | Bentley DR, Balasubramanian S, Swerdlow HP, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature, 2008, 456(7218): 53–59. DOI: 10.1038/nature07517 |
| [38] | Yergeau E, Lawrence JR, Sanschagrin S, et al. Next-generation sequencing of microbial communities in the Athabasca River and its tributaries in relation to oil sands mining activities. Appl Environ Microb, 2012, 78(21): 7626–7637. DOI: 10.1128/AEM.02036-12 |
| [39] | Rhoads A, Au KF. PacBio sequencing and its applications. Genomics, Proteomics & Bioinformatics, 2015, 13(5): 278–289. |
| [40] | Jain M, Olsen HE, Paten B, et al. The oxford nanopore minION: delivery of nanopore sequencing to the genomics community. Genome Biol, 2016, 17: 239. DOI: 10.1186/s13059-016-1103-0 |
| [41] | Ku CS, Roukos DH. From next-generation sequencing to nanopore sequencing technology: paving the way to personalized genomic medicine. Expert Rev Med Devices, 2013, 10(1): 1–6. DOI: 10.1586/erd.12.63 |
| [42] | Boisvert S, Laviolette F, Corbeil J. Ray: simultaneous assembly of reads from a mix of high-throughput sequencing technologies. J Comput Biol, 2010, 17(11): 1519–1533. DOI: 10.1089/cmb.2009.0238 |
| [43] | Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res, 2008, 18(5): 821–829. DOI: 10.1101/gr.074492.107 |
| [44] | Bankevich A, Nurk S, Antipov D, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol, 2012, 19(5): 455–477. DOI: 10.1089/cmb.2012.0021 |
| [45] | Peng Y, Leung HCM, Yiu SM, et al. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics, 2012, 28(11): 1420–1428. DOI: 10.1093/bioinformatics/bts174 |
| [46] | Chin CS, Alexander DH, Marks P, et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods, 2013, 10(6): 563–569. DOI: 10.1038/nmeth.2474 |
| [47] | Koren S, Walenz BP, Berlin K, et al. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res, 2017, 27(5): 722–736. DOI: 10.1101/gr.215087.116 |
| [48] | Wick RR, Judd LM, Gorrie CL, et al. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol, 2017, 13(6): e1005595. DOI: 10.1371/journal.pcbi.1005595 |
| [49] | Antipov D, Hartwick N, Shen M, et al. plasmidSPAdes: assembling plasmids from whole genome sequencing data. Bioinformatics, 2016, 32(22): 3380–3387. |
| [50] | De Lannoy C, De Ridder D, Risse J. The long reads ahead: de novo genome assembly using the MinION. F1000Res, 2017, 6: 1083. |
| [51] | Karalius J. Resources for advanced bioinformaticians working in plant and animal genomes with SMRT sequencing. Menlo Park, CA: Pacific Biosciences, 2015. |
| [52] | Au KF, Underwood JG, Lee L, et al. Improving PacBio long read accuracy by short read alignment. PLoS ONE, 2012, 7(10): e46679. DOI: 10.1371/journal.pone.0046679 |
| [53] | Larsen MV, Cosentino S, Lukjancenko O, et al. Benchmarking of methods for genomic taxonomy. J Clin Microbiol, 2014, 52(5): 1529–1539. DOI: 10.1128/JCM.02981-13 |
| [54] | Aziz RK, Bartels D, Best AA, et al. The RAST server: rapid annotations using subsystems technology. BMC Genomics, 2008, 9: 75. DOI: 10.1186/1471-2164-9-75 |
| [55] | Zankari E, Hasman H, Cosentino S, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother, 2012, 67(11): 2640–2644. DOI: 10.1093/jac/dks261 |
| [56] | Joensen KG, Scheutz F, Lund O, et al. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J Clin Microbiol, 2014, 52(5): 1501–1510. DOI: 10.1128/JCM.03617-13 |
| [57] | Carattoli A, Zankari E, García-Fernández A, et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother, 2014, 58(7): 3895–3903. DOI: 10.1128/AAC.02412-14 |
| [58] | Larsen MV, Cosentino S, Rasmussen S, et al. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol, 2012, 50(4): 1355–1361. DOI: 10.1128/JCM.06094-11 |
| [59] | Thomsen MCF, Ahrenfeldt J, Cisneros JLB, et al. A bacterial analysis platform: an integrated system for analysing bacterial whole genome sequencing data for clinical diagnostics and surveillance. PLoS ONE, 2016, 11(6): e0157718. DOI: 10.1371/journal.pone.0157718 |
| [60] | Hong CJ, Manimaran S, Shen Y, et al. PathoScope 2.0: a complete computational framework for strain identification in environmental or clinical sequencing samples. Microbiome, 2014, 2: 33. DOI: 10.1186/2049-2618-2-33 |
| [61] | Faith JJ, Guruge JL, Charbonneau M, et al. The long-term stability of the human gut microbiota. Science, 2013, 341(6141): 1237439. DOI: 10.1126/science.1237439 |
| [62] | Cox LM, Yamanishi S, Sohn J, et al. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell, 2014, 158(4): 705–721. DOI: 10.1016/j.cell.2014.05.052 |
| [63] | Oh J, Byrd AL, Deming C, et al. Biogeography and individuality shape function in the human skin metagenome. Nature, 2014, 514(7520): 59–64. DOI: 10.1038/nature13786 |
| [64] | Byrd AL, Perez-Rogers JF, Manimaran S, et al. Clinical PathoScope: rapid alignment and filtration for accurate pathogen identification in clinical samples using unassembled sequencing data. BMC Bioinformatics, 2014, 15: 262. DOI: 10.1186/1471-2105-15-262 |
| [65] | Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics, 2014, 30(14): 2068–2069. DOI: 10.1093/bioinformatics/btu153 |
| [66] | Stoesser N, Giess A, Batty EM, et al. Genome sequencing of an extended series of NDM-producing Klebsiella pneumoniae isolates from neonatal infections in a Nepali hospital characterizes the extent of community- versus hospital-associated transmission in an endemic setting. Antimicrob Agents Chemother, 2014, 58(12): 7347–7357. DOI: 10.1128/AAC.03900-14 |
| [67] | Yang P, Xie Y, Feng P, et al. blaNDM-5 carried by an IncX3 plasmid in Escherichia coli sequence type 167. Antimicrob Agents Chemother, 2014, 58(12): 7548–7552. DOI: 10.1128/AAC.03911-14 |
| [68] | Inouye M, Dashnow H, Raven LA, et al. SRST2: rapid genomic surveillance for public health and hospital microbiology labs. Genome Med, 2014, 6: 90. DOI: 10.1186/s13073-014-0090-6 |
| [69] | Liu B, Pop M. ARDB—antibiotic resistance genes database. Nucleic Acids Res, 2009, 37(S1): D443–D447. |
| [70] | Chen LH, Zheng DD, Liu B, et al. VFDB 2016: hierarchical and refined dataset for big data analysis—10 years on. Nucleic Acids Res, 2016, 44(D1): D694–D697. DOI: 10.1093/nar/gkv1239 |
| [71] | Wang Y, Zhang RM, Li JY, et al. Comprehensive resistome analysis reveals the prevalence of NDM and MCR-1 in Chinese poultry production. Nat Microbiol, 2017, 2: 16260. |
| [72] | Kaas RS, Leekitcharoenphon P, Aarestrup FM, et al. Solving the problem of comparing whole bacterial genomes across different sequencing platforms. PLoS ONE, 2014, 9(8): e104984. DOI: 10.1371/journal.pone.0104984 |
| [73] | Price MN, Dehal PS, Arkin AP. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol, 2009, 26(7): 1641–1650. DOI: 10.1093/molbev/msp077 |
| [74] | Gardner SN, Slezak T, Hall BG. kSNP3.0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference genome. Bioinformatics, 2015, 31(17): 2877–2878. DOI: 10.1093/bioinformatics/btv271 |
| [75] | Wilson A, Gray J, Chandry PS, et al. Phenotypic and genotypic analysis of antimicrobial resistance among Listeria monocytogenes isolated from Australian food production chains. Genes, 2018, 9(2): 80. DOI: 10.3390/genes9020080 |
| [76] | Page AJ, Cummins CA, Hunt M, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics, 2015, 31(22): 3691–3693. DOI: 10.1093/bioinformatics/btv421 |
| [77] | Enright AJ, van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res, 2002, 30(7): 1575–1584. DOI: 10.1093/nar/30.7.1575 |
| [78] | Moradigaravand D, Boinett CJ, Martin V, et al. Recent independent emergence of multiple multidrug-resistant Serratia marcescens clones within the United Kingdom and Ireland. Genome Res, 2016, 26(8): 1101–1109. DOI: 10.1101/gr.205245.116 |
| [79] | Laing C, Buchanan C, Taboada EN, et al. Pan-genome sequence analysis using Panseq: an online tool for the rapid analysis of core and accessory genomic regions. BMC Bioinformatics, 2010, 11: 461. DOI: 10.1186/1471-2105-11-461 |
| [80] | Kurtz S, Phillippy A, Delcher AL, et al. Versatile and open software for comparing large genomes. Genome Biol, 2004, 5(2): R12. DOI: 10.1186/gb-2004-5-2-r12 |
| [81] | López-Camacho E, Gómez-Gil R, Tobes R, et al. Genomic analysis of the emergence and evolution of multidrug resistance during a Klebsiella pneumoniae outbreak including carbapenem and colistin resistance. J Antimicrob Chemother, 2014, 69(3): 632–636. DOI: 10.1093/jac/dkt419 |
| [82] | Katz LS, Griswold T, Williams-Newkirk AJ, et al. A comparative analysis of the Lyve-SET phylogenomics pipeline for genomic epidemiology of foodborne pathogens. Front Microbiol, 2017, 8: 375. |
| [83] | Treangen TJ, Ondov BD, Koren S, et al. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol, 2014, 15(11): 524. DOI: 10.1186/s13059-014-0524-x |
| [84] | Strimmer K, von Haeseler A. Likelihood-mapping: a simple method to visualize phylogenetic content of a sequence alignment. Proc Natl Acad Sci USA, 1997, 94(13): 6815–6819. DOI: 10.1073/pnas.94.13.6815 |
| [85] | Stamatakis A, Ludwig T, Meier H. RAxML-Ⅲ: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics, 2005, 21(4): 456–463. DOI: 10.1093/bioinformatics/bti191 |
| [86] | Casali N, Nikolayevskyy V, Balabanova Y, et al. Evolution and transmission of drug-resistant tuberculosis in a Russian population. Nat Genet, 2014, 46(3): 279–286. DOI: 10.1038/ng.2878 |
| [87] | Price MN, Dehal PS, Arkin AP. FastTree 2-approximately maximum-likelihood trees for large alignments. PLoS ONE, 2010, 5(3): e9490. DOI: 10.1371/journal.pone.0009490 |
| [88] | Ronquist F, Teslenko M, van der Mark P, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol, 2012, 61(3): 539–542. |
| [89] | Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol, 2016, 33(7): 1870–1874. DOI: 10.1093/molbev/msw054 |
| [90] | Han MV, Zmasek CM. phyloXML: XML for evolutionary biology and comparative genomics. BMC Bioinformatics, 2009, 10: 356. DOI: 10.1186/1471-2105-10-356 |
| [91] | Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res, 2016, 44(W1): W242–W245. DOI: 10.1093/nar/gkw290 |
| [92] | Land M, Hauser L, Jun SR, et al. Insights from 20 years of bacterial genome sequencing. Funct Integr Genomics, 2015, 15(2): 141–161. DOI: 10.1007/s10142-015-0433-4 |
| [93] | Aigrain L, Gu Y, Quail MA. Quantitation of next generation sequencing library preparation protocol efficiencies using droplet digital PCR assays—a systematic comparison of DNA library preparation kits for Illumina sequencing. BMC Genomics, 2016, 17: 458. DOI: 10.1186/s12864-016-2757-4 |
| [94] | van der Helm E, Imamovic L, Hashim Ellabaan MM, et al. Rapid resistome mapping using nanopore sequencing. Nucleic Acids Res, 2017, 45(8): e61. |
| [95] | Walker BJ, Abeel T, Shea T, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE, 2014, 9(11): e112963. DOI: 10.1371/journal.pone.0112963 |
| [96] | Li R, Xie M, Dong N, et al. Efficient generation of complete sequences of MDR-encoding plasmids by rapid assembly of MinION barcoding sequencing data. GigaScience, 2018, 7(3): 1–9. |
| [97] | Li R, Chen K, Chan E W-C, et al. Resolution of dynamic MDR structures among the plasmidome of Salmonella using MinION single-molecule, long-read sequencing. J Antimicrob Chemother, 2018, 73(10): 2691–2695. DOI: 10.1093/jac/dky243 |
 2019, Vol. 35
2019, Vol. 35